Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (12): 2756-2766.doi: 10.12122/j.issn.1673-4254.2025.12.23
Zhenni YU1( ), Jingzhe GAO1, Hui SUN1, Qin Feng2, Xiaoqi NA1, Ning ZHANG1(
), Jingzhe GAO1, Hui SUN1, Qin Feng2, Xiaoqi NA1, Ning ZHANG1( ), Kungshuang SHEN1, Yuanyuan WANG1, Xijun WANG1(
), Kungshuang SHEN1, Yuanyuan WANG1, Xijun WANG1( )
)
Received:2025-04-14
Online:2025-12-20
Published:2025-12-22
Contact:
Ning ZHANG, Xijun WANG
E-mail:yzhennni@163.com;zhangning0454@163.com;xijunw@sina.com
Zhenni YU, Jingzhe GAO, Hui SUN, Qin Feng, Xiaoqi NA, Ning ZHANG, Kungshuang SHEN, Yuanyuan WANG, Xijun WANG. Causal relationship between gut microbiota and T cell subsets in the development of colorectal cancer: a Mendelian randomization analysis[J]. Journal of Southern Medical University, 2025, 45(12): 2756-2766.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.12.23
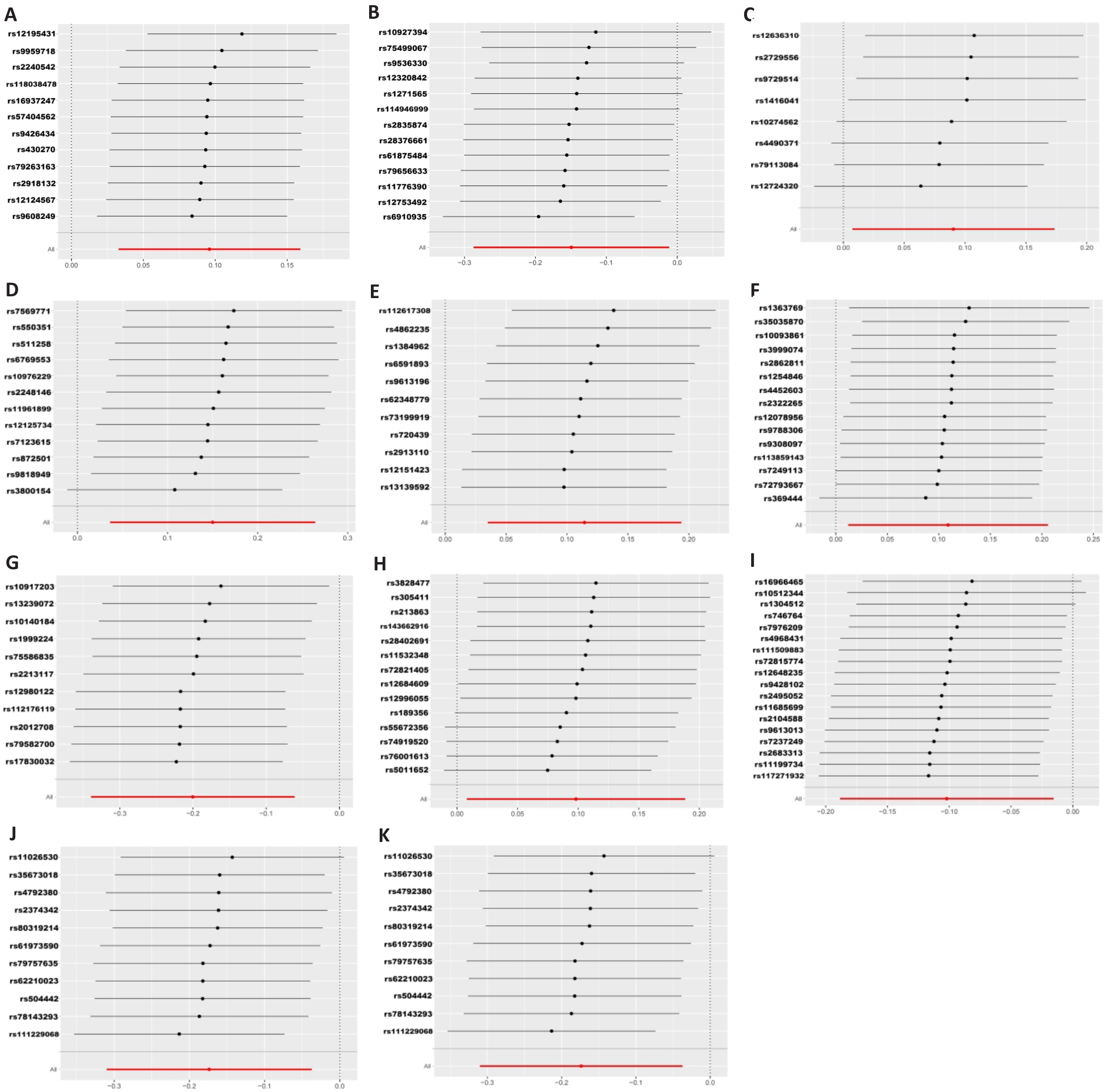
Fig.2 Leave-one-out plot of the association between gut microbiota and colorectal cancer under Mendelian randomization analysis. A: Prevotella7. B: Faecalibacterium. C: Ruminococcaceae UCG011. D: Ruminococcaceae UCG004. E: Eubacterium brachy group. F: Lachnospiraceae FCS020 group. G: Eubacterium xylanophilum group. H: Coprobacter. I: Prevotella9. J: Enterobacteriaceae. K: Enterobacteriales.
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Prevotella7 | 22 | 0.076 | 0.944 | 1.005 | 0.866-1.1167 |
| genus.Faecalibacterium | 23 | 0.040 | 0.216 | 1.051 | 0.972-1.136 |
| genus.Ruminococcaceae UCG011 | 22 | 0.057 | 0.347 | 0.948 | 0.848-1.06 |
| genus.Ruminococcaceae UCG004 | 23 | 0.036 | 0.406 | 0.970 | 0.904-1.042 |
| genus.Eubacterium brachy group | 22 | 0.054 | 0.734 | 1.019 | 0.916-1.132 |
| genus.Lachnospiraceae FCS020 group | 23 | 0.033 | 0.427 | 0.974 | 0.913-1.039 |
| genus.Eubacterium xylanophilum group | 23 | 0.035 | 0.774 | 0.990 | 0.924-1.061 |
| genus.Coprobacter | 23 | 0.052 | 0.468 | 1.038 | 0.938-1.149 |
| genus.Prevotella9 | 23 | 0.039 | 0.835 | 1.008 | 0.934-1.088 |
| family.Enterobacteriaceae | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| order.Enterobacteriales | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
Tab.1 Results of inverse variance weighted method analysis in Mendelian randomization study on gut microbiota and colorectal cancer
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Prevotella7 | 22 | 0.076 | 0.944 | 1.005 | 0.866-1.1167 |
| genus.Faecalibacterium | 23 | 0.040 | 0.216 | 1.051 | 0.972-1.136 |
| genus.Ruminococcaceae UCG011 | 22 | 0.057 | 0.347 | 0.948 | 0.848-1.06 |
| genus.Ruminococcaceae UCG004 | 23 | 0.036 | 0.406 | 0.970 | 0.904-1.042 |
| genus.Eubacterium brachy group | 22 | 0.054 | 0.734 | 1.019 | 0.916-1.132 |
| genus.Lachnospiraceae FCS020 group | 23 | 0.033 | 0.427 | 0.974 | 0.913-1.039 |
| genus.Eubacterium xylanophilum group | 23 | 0.035 | 0.774 | 0.990 | 0.924-1.061 |
| genus.Coprobacter | 23 | 0.052 | 0.468 | 1.038 | 0.938-1.149 |
| genus.Prevotella9 | 23 | 0.039 | 0.835 | 1.008 | 0.934-1.088 |
| family.Enterobacteriaceae | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| order.Enterobacteriales | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 3 | 0.032 | 0.035 | 0.935 | 0.878-0.995 |
| Weighted median | 3 | 0.031 | 0.006 | 0.919 | 0.866-0.976 |
| MR-Egger | 3 | 0.107 | 0.566 | 0.917 | 0.743-1.131 |
| Simple mode | 3 | 0.044 | 0.176 | 0.913 | 0.837-0.996 |
| Weighted mode | 3 | 0.041 | 0.144 | 0.909 | 0.839-0.985 |
Tab.2 Results of Mendelian randomization analysis for T Cells and colorectal cancer
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 3 | 0.032 | 0.035 | 0.935 | 0.878-0.995 |
| Weighted median | 3 | 0.031 | 0.006 | 0.919 | 0.866-0.976 |
| MR-Egger | 3 | 0.107 | 0.566 | 0.917 | 0.743-1.131 |
| Simple mode | 3 | 0.044 | 0.176 | 0.913 | 0.837-0.996 |
| Weighted mode | 3 | 0.041 | 0.144 | 0.909 | 0.839-0.985 |
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 25 | 0.056 | 0.195 | 1.076 | 0.963-1.201 |
| Weighted median | 25 | 0.079 | 0.142 | 1.123 | 0.962-1.312 |
| MR-Egger | 25 | 0.231 | 0.838 | 0.953 | 0.607-1.499 |
| Simple mode | 25 | 0.140 | 0.277 | 1.168 | 0.888-1.536 |
| Weighted mode | 25 | 0.124 | 0.222 | 1.168 | 0.916-1.489 |
Tab.3 Results of Mendelian randomization analysis for colorectal cancer and T cells
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 25 | 0.056 | 0.195 | 1.076 | 0.963-1.201 |
| Weighted median | 25 | 0.079 | 0.142 | 1.123 | 0.962-1.312 |
| MR-Egger | 25 | 0.231 | 0.838 | 0.953 | 0.607-1.499 |
| Simple mode | 25 | 0.140 | 0.277 | 1.168 | 0.888-1.536 |
| Weighted mode | 25 | 0.124 | 0.222 | 1.168 | 0.916-1.489 |
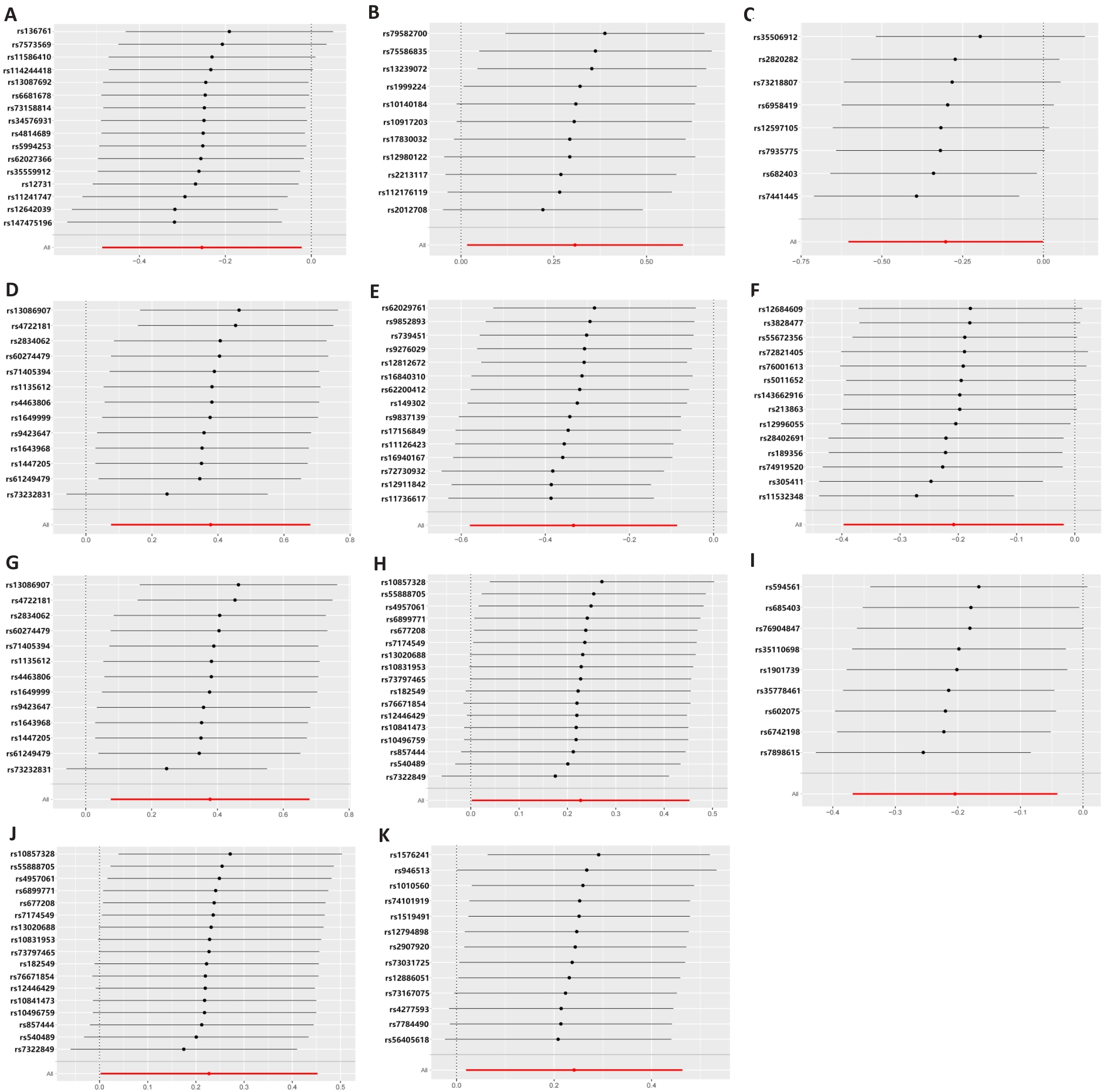
Fig.5 Leave-one-out plot of the association between gut microbiota and T cells under Mendelian randomization analysis.A: Ruminococcaceae NK4A214 group. B: Eubacterium xylanophilum group. C: Ruminococcaceae UCG010. D: Selenomonadales. E: XIII AD3011 group. F: Coprobacter. G: Negativicutes. H: Bifidobacteriaceae. I: Allisonella. J: Bifidobacteriales. K: Coprococcus1.
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Ruminococcaceae NK4A214 group | 5 | 0.028 | 0.947 | 0.998 | 0.945-1.054 |
| genus.Eubacterium xylanophilum group | 5 | 0.030 | 0.321 | 0.971 | 0.916-1.029 |
| genus.Ruminococcaceae UCG010 | 5 | 0.037 | 0.527 | 0.977 | 0.909-1.050 |
| order.Selenomonadales | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| genus.Family XIII AD3011 group | 5 | 0.029 | 0.762 | 1.009 | 0.953-1.067 |
| genus.Coprobacter | 5 | 0.069 | 0.843 | 1.014 | 0.885-1.162 |
| Class.Negativicutes | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| family.Bifidobacteriaceae | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Allisonella | 3 | 0.122 | 0.028 | 0.765 | 0.603-0.971 |
| order.Bifidobacteriales | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Coprococcus1 | 5 | 0.029 | 0.892 | 0.996 | 0.942-1.054 |
Tab.4 Results of inverse variance weighted method analysis in Mendelian randomization study on colorectal cancer and T cells
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Ruminococcaceae NK4A214 group | 5 | 0.028 | 0.947 | 0.998 | 0.945-1.054 |
| genus.Eubacterium xylanophilum group | 5 | 0.030 | 0.321 | 0.971 | 0.916-1.029 |
| genus.Ruminococcaceae UCG010 | 5 | 0.037 | 0.527 | 0.977 | 0.909-1.050 |
| order.Selenomonadales | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| genus.Family XIII AD3011 group | 5 | 0.029 | 0.762 | 1.009 | 0.953-1.067 |
| genus.Coprobacter | 5 | 0.069 | 0.843 | 1.014 | 0.885-1.162 |
| Class.Negativicutes | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| family.Bifidobacteriaceae | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Allisonella | 3 | 0.122 | 0.028 | 0.765 | 0.603-0.971 |
| order.Bifidobacteriales | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Coprococcus1 | 5 | 0.029 | 0.892 | 0.996 | 0.942-1.054 |
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| genus.Prevotella7 | CRC | MR-Rgger | 7.239 | 10 | 0.703 |
| genus.Prevotella7 | CRC | IVW | 9.043 | 11 | 0.618 |
| genus.Faecalibacterium | CRC | MR-Rgger | 13.261 | 11 | 0.277 |
| genus.Faecalibacterium | CRC | IVW | 13.262 | 12 | 0.350 |
| genus.Ruminococcaceae UCG011 | CRC | MR-Rgger | 6.409 | 6 | 0.379 |
| genus.Ruminococcaceae UCG011 | CRC | IVW | 6.820 | 7 | 0.448 |
| genus.Ruminococcaceae UCG004 | CRC | MR-Rgger | 10.434 | 10 | 0.403 |
| genus.Ruminococcaceae UCG004 | CRC | IVW | 11.115 | 11 | 0.434 |
| genus.Eubacterium brachy group | CRC | MR-Rgger | 9.150 | 9 | 0.424 |
| genus.Eubacterium brachy group | CRC | IVW | 9.264 | 10 | 0.507 |
| genus.Lachnospiraceae FCS020 group | CRC | MR-Rgger | 6.834 | 13 | 0.910 |
| genus.Lachnospiraceae FCS020 group | CRC | IVW | 6.848 | 14 | 0.940 |
| genus.Eubacterium xylanophilum group | CRC | MR-Rgger | 4.699 | 9 | 0.860 |
| genus.Eubacterium xylanophilum group | CRC | IVW | 7.284 | 10 | 0.698 |
| genus.Coprobacter | CRC | MR-Rgger | 15.480 | 12 | 0.216 |
| genus.Coprobacter | CRC | IVW | 15.643 | 13 | 0.269 |
| genus.Prevotella9 | CRC | MR-Rgger | 16.705 | 16 | 0.405 |
| genus.Prevotella9 | CRC | IVW | 16.715 | 17 | 0.474 |
| family.Enterobacteriaceae | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| family.Enterobacteriaceae | CRC | IVW | 9.399 | 10 | 0.495 |
| order.Enterobacteriales | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| order.Enterobacteriales | CRC | IVW | 9.399 | 10 | 0.495 |
| T Cells | CRC | MR-Rgger | 3.227 | 1 | 0.072 |
| T Cells | CRC | IVW | 3.356 | 2 | 0.187 |
| genus.Ruminococcaceae NK4A214 group | T Cells | MR-Rgger | 13.403 | 14 | 0.495 |
| genus.Ruminococcaceae NK4A214 group | T Cells | IVW | 14.064 | 15 | 0.521 |
| genus.Eubacterium xylanophilum group | T Cells | MR-Rgger | 12.167 | 9 | 0.204 |
| genus.Eubacterium xylanophilum group | T Cells | IVW | 12.770 | 10 | 0.237 |
| genus.Ruminococcaceae UCG010 | T Cells | MR-Rgger | 5.807 | 6 | 0.445 |
| genus.Ruminococcaceae UCG010 | T Cells | IVW | 6.325 | 7 | 0.502 |
| order.Selenomonadales | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| order.Selenomonadales | T Cells | IVW | 13.547 | 12 | 0.331 |
| genus.Family XIII AD3011 group | T Cells | MR-Rgger | 16.241 | 13 | 0.236 |
| genus.Family XIII AD3011 group | T Cells | IVW | 16.387 | 14 | 0.290 |
| genus.Coprobacter | T Cells | MR-Rgger | 17.482 | 12 | 0.132 |
| genus.Coprobacter | T Cells | IVW | 17.762 | 13 | 0.167 |
| Class.Negativicutes | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| Class.Negativicutes | T Cells | IVW | 13.547 | 12 | 0.331 |
| family.Bifidobacteriaceae | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| family.Bifidobacteriaceae | T Cells | IVW | 7.605 | 16 | 0.960 |
| genus.Allisonella | T Cells | MR-Rgger | 5.460 | 7 | 0.604 |
| genus.Allisonella | T Cells | IVW | 6.660 | 8 | 0.574 |
| order.Bifidobacteriales | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| order.Bifidobacteriales | T Cells | IVW | 7.605 | 16 | 0.960 |
Tab.5 Heterogeneity test for gut microbiota, T cells, and colorectal cancer
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| genus.Prevotella7 | CRC | MR-Rgger | 7.239 | 10 | 0.703 |
| genus.Prevotella7 | CRC | IVW | 9.043 | 11 | 0.618 |
| genus.Faecalibacterium | CRC | MR-Rgger | 13.261 | 11 | 0.277 |
| genus.Faecalibacterium | CRC | IVW | 13.262 | 12 | 0.350 |
| genus.Ruminococcaceae UCG011 | CRC | MR-Rgger | 6.409 | 6 | 0.379 |
| genus.Ruminococcaceae UCG011 | CRC | IVW | 6.820 | 7 | 0.448 |
| genus.Ruminococcaceae UCG004 | CRC | MR-Rgger | 10.434 | 10 | 0.403 |
| genus.Ruminococcaceae UCG004 | CRC | IVW | 11.115 | 11 | 0.434 |
| genus.Eubacterium brachy group | CRC | MR-Rgger | 9.150 | 9 | 0.424 |
| genus.Eubacterium brachy group | CRC | IVW | 9.264 | 10 | 0.507 |
| genus.Lachnospiraceae FCS020 group | CRC | MR-Rgger | 6.834 | 13 | 0.910 |
| genus.Lachnospiraceae FCS020 group | CRC | IVW | 6.848 | 14 | 0.940 |
| genus.Eubacterium xylanophilum group | CRC | MR-Rgger | 4.699 | 9 | 0.860 |
| genus.Eubacterium xylanophilum group | CRC | IVW | 7.284 | 10 | 0.698 |
| genus.Coprobacter | CRC | MR-Rgger | 15.480 | 12 | 0.216 |
| genus.Coprobacter | CRC | IVW | 15.643 | 13 | 0.269 |
| genus.Prevotella9 | CRC | MR-Rgger | 16.705 | 16 | 0.405 |
| genus.Prevotella9 | CRC | IVW | 16.715 | 17 | 0.474 |
| family.Enterobacteriaceae | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| family.Enterobacteriaceae | CRC | IVW | 9.399 | 10 | 0.495 |
| order.Enterobacteriales | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| order.Enterobacteriales | CRC | IVW | 9.399 | 10 | 0.495 |
| T Cells | CRC | MR-Rgger | 3.227 | 1 | 0.072 |
| T Cells | CRC | IVW | 3.356 | 2 | 0.187 |
| genus.Ruminococcaceae NK4A214 group | T Cells | MR-Rgger | 13.403 | 14 | 0.495 |
| genus.Ruminococcaceae NK4A214 group | T Cells | IVW | 14.064 | 15 | 0.521 |
| genus.Eubacterium xylanophilum group | T Cells | MR-Rgger | 12.167 | 9 | 0.204 |
| genus.Eubacterium xylanophilum group | T Cells | IVW | 12.770 | 10 | 0.237 |
| genus.Ruminococcaceae UCG010 | T Cells | MR-Rgger | 5.807 | 6 | 0.445 |
| genus.Ruminococcaceae UCG010 | T Cells | IVW | 6.325 | 7 | 0.502 |
| order.Selenomonadales | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| order.Selenomonadales | T Cells | IVW | 13.547 | 12 | 0.331 |
| genus.Family XIII AD3011 group | T Cells | MR-Rgger | 16.241 | 13 | 0.236 |
| genus.Family XIII AD3011 group | T Cells | IVW | 16.387 | 14 | 0.290 |
| genus.Coprobacter | T Cells | MR-Rgger | 17.482 | 12 | 0.132 |
| genus.Coprobacter | T Cells | IVW | 17.762 | 13 | 0.167 |
| Class.Negativicutes | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| Class.Negativicutes | T Cells | IVW | 13.547 | 12 | 0.331 |
| family.Bifidobacteriaceae | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| family.Bifidobacteriaceae | T Cells | IVW | 7.605 | 16 | 0.960 |
| genus.Allisonella | T Cells | MR-Rgger | 5.460 | 7 | 0.604 |
| genus.Allisonella | T Cells | IVW | 6.660 | 8 | 0.574 |
| order.Bifidobacteriales | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| order.Bifidobacteriales | T Cells | IVW | 7.605 | 16 | 0.960 |
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| CRC | genus.Prevotella7 | MR-Rgger | 33.667 | 20 | 0.028 |
| CRC | genus.Prevotella7 | IVW | 36.090 | 21 | 0.021 |
| CRC | genus.Faecalibacterium | MR-Rgger | 54.873 | 21 | <0.001 |
| CRC | genus.Faecalibacterium | IVW | 55.369 | 22 | <0.001 |
| CRC | genus.Ruminococcaceae UCG011 | MR-Rgger | 13.506 | 20 | 0.855 |
| CRC | genus.Ruminococcaceae UCG011 | IVW | 15.151 | 21 | 0.815 |
| CRC | genus.Ruminococcaceae UCG004 | MR-Rgger | 23.003 | 21 | 0.344 |
| CRC | genus.Ruminococcaceae UCG004 | IVW | 23.825 | 22 | 0.356 |
| CRC | genus.Eubacterium brachy group | MR-Rgger | 15.511 | 20 | 0.746 |
| CRC | genus.Eubacterium brachy group | IVW | 18.053 | 21 | 0.646 |
| CRC | genus.Lachnospiraceae FCS020 group | MR-Rgger | 29.009 | 21 | 0.114 |
| CRC | genus.Lachnospiraceae FCS020 group | IVW | 29.009 | 22 | 0.145 |
| CRC | genus.Eubacterium xylanophilum group | MR-Rgger | 30.851 | 21 | 0.076 |
| CRC | genus.Eubacterium xylanophilum group | IVW | 31.688 | 22 | 0.083 |
| CRC | genus.Coprobacter | MR-Rgger | 34.800 | 21 | 0.030 |
| CRC | genus.Coprobacter | IVW | 34.918 | 22 | 0.040 |
| CRC | genus.Prevotella9 | MR-Rgger | 19.765 | 21 | 0.536 |
| CRC | genus.Prevotella9 | IVW | 30.024 | 22 | 0.118 |
| CRC | family.Enterobacteriaceae | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | family.Enterobacteriaceae | IVW | 26.554 | 22 | 0.229 |
| CRC | order.Enterobacteriales | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | order.Enterobacteriales | IVW | 26.554 | 22 | 0.229 |
| CRC | T Cells | MR-Rgger | 17.594 | 23 | 0.779 |
| CRC | T Cells | IVW | 17.885 | 24 | 0.809 |
| T Cells | genus.Ruminococcaceae NK4A214 group | MR-Rgger | 1.310 | 3 | 0.727 |
| T Cells | genus.Ruminococcaceae NK4A214 group | IVW | 1.356 | 4 | 0.852 |
| T Cells | genus.Eubacterium xylanophilum group | MR-Rgger | 2.079 | 3 | 0.556 |
| T Cells | genus.Eubacterium xylanophilum group | IVW | 2.242 | 4 | 0.691 |
| T Cells | genus.Ruminococcaceae UCG010 | MR-Rgger | 0.939 | 3 | 0.816 |
| T Cells | genus.Ruminococcaceae UCG010 | IVW | 5.875 | 4 | 0.209 |
| T Cells | order.Selenomonadales | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | order.Selenomonadales | IVW | 8.155 | 4 | 0.086 |
| T Cells | genus.Family XIII AD3011 group | MR-Rgger | 1.403 | 3 | 0.705 |
| T Cells | genus.Family XIII AD3011 group | IVW | 1.482 | 4 | 0.830 |
| T Cells | genus.Coprobacter | MR-Rgger | 11.202 | 3 | 0.011 |
| T Cells | genus.Coprobacter | IVW | 11.278 | 4 | 0.024 |
| T Cells | Class.Negativicutes | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | Class.Negativicutes | IVW | 8.155 | 4 | 0.086 |
| T Cells | family.Bifidobacteriaceae | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | family.Bifidobacteriaceae | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Allisonella | MR-Rgger | 1.503 | 1 | 0.220 |
| T Cells | genus.Allisonella | IVW | 2.060 | 2 | 0.357 |
| T Cells | order.Bifidobacteriales | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | order.Bifidobacteriales | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Coprococcus1 | MR-Rgger | 4.329 | 3 | 0.228 |
| T Cells | genus.Coprococcus1 | IVW | 4.585 | 4 | 0.333 |
Tab.6 Heterogeneity test for gut microbiota, T cells, and colorectal cancer
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| CRC | genus.Prevotella7 | MR-Rgger | 33.667 | 20 | 0.028 |
| CRC | genus.Prevotella7 | IVW | 36.090 | 21 | 0.021 |
| CRC | genus.Faecalibacterium | MR-Rgger | 54.873 | 21 | <0.001 |
| CRC | genus.Faecalibacterium | IVW | 55.369 | 22 | <0.001 |
| CRC | genus.Ruminococcaceae UCG011 | MR-Rgger | 13.506 | 20 | 0.855 |
| CRC | genus.Ruminococcaceae UCG011 | IVW | 15.151 | 21 | 0.815 |
| CRC | genus.Ruminococcaceae UCG004 | MR-Rgger | 23.003 | 21 | 0.344 |
| CRC | genus.Ruminococcaceae UCG004 | IVW | 23.825 | 22 | 0.356 |
| CRC | genus.Eubacterium brachy group | MR-Rgger | 15.511 | 20 | 0.746 |
| CRC | genus.Eubacterium brachy group | IVW | 18.053 | 21 | 0.646 |
| CRC | genus.Lachnospiraceae FCS020 group | MR-Rgger | 29.009 | 21 | 0.114 |
| CRC | genus.Lachnospiraceae FCS020 group | IVW | 29.009 | 22 | 0.145 |
| CRC | genus.Eubacterium xylanophilum group | MR-Rgger | 30.851 | 21 | 0.076 |
| CRC | genus.Eubacterium xylanophilum group | IVW | 31.688 | 22 | 0.083 |
| CRC | genus.Coprobacter | MR-Rgger | 34.800 | 21 | 0.030 |
| CRC | genus.Coprobacter | IVW | 34.918 | 22 | 0.040 |
| CRC | genus.Prevotella9 | MR-Rgger | 19.765 | 21 | 0.536 |
| CRC | genus.Prevotella9 | IVW | 30.024 | 22 | 0.118 |
| CRC | family.Enterobacteriaceae | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | family.Enterobacteriaceae | IVW | 26.554 | 22 | 0.229 |
| CRC | order.Enterobacteriales | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | order.Enterobacteriales | IVW | 26.554 | 22 | 0.229 |
| CRC | T Cells | MR-Rgger | 17.594 | 23 | 0.779 |
| CRC | T Cells | IVW | 17.885 | 24 | 0.809 |
| T Cells | genus.Ruminococcaceae NK4A214 group | MR-Rgger | 1.310 | 3 | 0.727 |
| T Cells | genus.Ruminococcaceae NK4A214 group | IVW | 1.356 | 4 | 0.852 |
| T Cells | genus.Eubacterium xylanophilum group | MR-Rgger | 2.079 | 3 | 0.556 |
| T Cells | genus.Eubacterium xylanophilum group | IVW | 2.242 | 4 | 0.691 |
| T Cells | genus.Ruminococcaceae UCG010 | MR-Rgger | 0.939 | 3 | 0.816 |
| T Cells | genus.Ruminococcaceae UCG010 | IVW | 5.875 | 4 | 0.209 |
| T Cells | order.Selenomonadales | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | order.Selenomonadales | IVW | 8.155 | 4 | 0.086 |
| T Cells | genus.Family XIII AD3011 group | MR-Rgger | 1.403 | 3 | 0.705 |
| T Cells | genus.Family XIII AD3011 group | IVW | 1.482 | 4 | 0.830 |
| T Cells | genus.Coprobacter | MR-Rgger | 11.202 | 3 | 0.011 |
| T Cells | genus.Coprobacter | IVW | 11.278 | 4 | 0.024 |
| T Cells | Class.Negativicutes | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | Class.Negativicutes | IVW | 8.155 | 4 | 0.086 |
| T Cells | family.Bifidobacteriaceae | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | family.Bifidobacteriaceae | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Allisonella | MR-Rgger | 1.503 | 1 | 0.220 |
| T Cells | genus.Allisonella | IVW | 2.060 | 2 | 0.357 |
| T Cells | order.Bifidobacteriales | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | order.Bifidobacteriales | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Coprococcus1 | MR-Rgger | 4.329 | 3 | 0.228 |
| T Cells | genus.Coprococcus1 | IVW | 4.585 | 4 | 0.333 |
| [1] | Ionescu VA, Gheorghe G, Bacalbasa N, et al. Colorectal cancer: from risk factors to oncogenesis[J]. Medicina: Kaunas, 2023, 59(9): 1646. doi:10.3390/medicina59091646 |
| [2] | Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota[J]. Immunity, 2017, 46(4): 562-76. doi:10.1016/j.immuni.2017.04.008 |
| [3] | Hu ZJ, Zhu HR, Jin YJ, et al. Correlation between gut microbiota and tumor immune microenvironment: a bibliometric and visualized study[J]. World J Clin Oncol, 2025, 16(2): 101611. doi:10.5306/wjco.v16.i2.101611 |
| [4] | Schwabe RF, Jobin C. The microbiome and cancer[J]. Nat Rev Cancer, 2013, 13(11): 800-12. doi:10.1038/nrc3610 |
| [5] | Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis[J]. J Biomed Sci, 2018, 25(1): 79. doi:10.1186/s12929-018-0483-8 |
| [6] | Hou X, Zheng Z, Wei J, et al. Effects of gut microbiota on immune responses and immunotherapy in colorectal cancer[J]. Front Immunol, 2022, 13: 1030745. doi:10.3389/fimmu.2022.1030745 |
| [7] | Scarpellini E, Ianiro G, Attili F, et al. The human gut microbiota and virome: Potential therapeutic implications[J]. Dig Liver Dis, 2015, 47(12): 1007-12. doi:10.1016/j.dld.2015.07.008 |
| [8] | Quaglio AEV, Grillo TG, De Oliveira ECS, et al. Gut microbiota, inflammatory bowel disease and colorectal cancer[J]. World J Gastroenterol, 2022, 28(30): 4053-60. doi:10.3748/wjg.v28.i30.4053 |
| [9] | Shim JA, Ryu JH, Jo Y, et al. The role of gut microbiota in T cell immunity and immune mediated disorders[J]. Int J Biol Sci, 2023, 19(4): 1178-91. doi:10.7150/ijbs.79430 |
| [10] | Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life[J]. Immunity, 2018, 48(2): 202-13. doi:10.1016/j.immuni.2018.01.007 |
| [11] | Ni JJ, Li XS, Zhang H, et al. Mendelian randomization study of causal link from gut microbiota to colorectal cancer[J]. BMC Cancer, 2022, 22(1): 1371. doi:10.1186/s12885-022-10483-w |
| [12] | Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis[J]. Proc Natl Acad Sci USA, 2011, 108(37): 15354-9. doi:10.1073/pnas.1010203108 |
| [13] | Brennan CA, Clay SL, et al. Fusobacterium nucleatumdrives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression[J]. Gut Microbes, 2021, 13: 1987780. doi:10.1080/19490976.2021.1987780 |
| [14] | Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome[J]. Science, 2006, 313(5795): 1960-4. doi:10.1126/science.1129139 |
| [15] | Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer[J]. J Clin Oncol, 2009, 27(2): 186-92. doi:10.1200/jco.2008.18.7229 |
| [16] | Zheng Z, Wieder T, Mauerer B, et al. T cells in colorectal cancer: unravelling the function of different T cell subsets in the tumor microenvironment[J]. Int J Mol Sci, 2023, 24(14): 11673. doi:10.3390/ijms241411673 |
| [17] | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data[J]. Genet Epidemiol, 2013, 37(7): 658-65. doi:10.1002/gepi.21758 |
| [18] | Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer[J]. Oncogene, 2020, 39(26): 4925-43. doi:10.1038/s41388-020-1341-1 |
| [19] | Yuan H, Gui R, Wang Z, et al. Gut microbiota: a novel and potential target for radioimmunotherapy in colorectal cancer[J]. Front Immunol, 2023, 14: 1128774. doi:10.3389/fimmu.2023.1128774 |
| [20] | Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental deter-minants of disease[J]? Int J Epidemiol, 2003, 32(1): 1-22. doi:10.1093/ije/dyg070 |
| [21] | 胡旭焘.肠道菌群,循环代谢物与胆石症:一项孟德尔随机化研究[D].吉林大学, 2024. 003850. |
| [22] | Shan J, Hu X, Chen T, et al. COVID-19 vaccination and the risk of autoimmune diseases: a Mendelian randomization study[J]. Front Public Health, 2024, 12: 1322140. doi:10.3389/fpubh.2024.1322140 |
| [23] | Burgess S, Bowden J, Fall T, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants[J]. Epidemiology, 2017, 28(1): 30-42. doi:10.1097/ede.0000000000000559 |
| [24] | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression[J]. Int J Epidemiol, 2015, 44(2): 512-25. doi:10.1093/ije/dyv080 |
| [25] | Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted Median estimator[J]. Genet Epidemiol, 2016, 40(4): 304-14. doi:10.1002/gepi.21965 |
| [26] | Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption[J]. Int J Epidemiol, 2017, 46(6): 1985-98. doi:10.1093/ije/dyx102 |
| [27] | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method[J]. Eur J Epidemiol, 2017, 32(5): 377-89. doi:10.1007/s10654-017-0255-x |
| [28] | Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases[J]. Nat Genet, 2018, 50(5): 693-8. doi:10.1038/s41588-018-0099-7 |
| [29] | Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies[J]. Hum Mol Genet, 2018, 27(r2): R195-208. doi:10.1093/hmg/ddy163 |
| [30] | Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data[J]. PLoS Genet, 2017, 13(11): e1007081. doi:10.1371/journal.pgen.1007081 |
| [31] | Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians[J]. BMJ, 2018, 362: k601. doi:10.1136/bmj.k601 |
| [32] | Teng NMY, Kiu R, Evans R, et al. Allocoprobacillus halotolerans gen. nov., sp. nov and Coprobacter tertius sp. nov., isolated from human gut microbiota[J]. Int J Syst Evol Microbiol, 2023, 73(7): 5950-8. doi:10.1099/ijsem.0.005950 |
| [33] | Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer[J]. Gut, 2017, 66(1): 70-8. doi:10.1136/gutjnl-2015-309800 |
| [34] | Silva M, Brunner V, Tschurtschenthaler M. Microbiota and colorectal cancer: from gut to bedside[J]. Front Pharmacol, 2021, 12: 760280. doi:10.3389/fphar.2021.760280 |
| [35] | Wang Y, Wan X, Wu X, et al. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis[J]. Gut Pathog, 2021, 13(1): 2. doi:10.1186/s13099-020-00396-z |
| [36] | Li HY, Wang Y, Shao SM, et al. Rabdosia serra alleviates dextran sulfate sodium salt-induced colitis in mice through anti-inflammation, regulating Th17/Treg balance, maintaining intestinal barrier integrity, and modulating gut microbiota[J]. J Pharm Anal, 2022, 12(6): 824-38. doi:10.1016/j.jpha.2022.08.001 |
| [37] | Park JS, Gazzaniga FS, Wu M, et al. Targeting PD-L2-RGMb overcomes microbiome-related immunotherapy resistance[J]. Nature, 2023, 617(7960): 377-85. doi:10.1038/s41586-023-06026-3 |
| [1] | Rongmao HE, Zeyang FANG, Yunyun ZHANG, Youliang WU, Shixiu LIANG, Tao JI, Kequan CHEN, Siqi WANG. Diagnostic and predictive value of ferroptosis-related genes in patients with ulcerative colitis [J]. Journal of Southern Medical University, 2025, 45(9): 1927-1937. |
| [2] | Chuyu DENG, Xueying WANG, Lixiang GAN, Dayu WANG, Xiaoyan ZHENG, Chunzhi TANG. Electroacupuncture at Zusanli improves blood lipid disorders in hyperlipidemic mice by improving gut microbiota structure [J]. Journal of Southern Medical University, 2025, 45(8): 1633-1642. |
| [3] | Hongbo ZHANG, Mengyu YAN, Jiandong ZHANG, Peiwang SUN, Rui WANG, Yuanyuan GUO. Pirfenidone inhibits bladder cancer xenograft growth in mice by regulating regulatory T cells [J]. Journal of Southern Medical University, 2025, 45(7): 1513-1518. |
| [4] | Kang WANG, Haibin LI, Jing YU, Yuan MENG, Hongli ZHANG. High expression of ELFN1 is a prognostic biomarker and promotes proliferation and metastasis of colorectal cancer cells [J]. Journal of Southern Medical University, 2025, 45(7): 1543-1553. |
| [5] | Yuexuan ZHU, Zhangrui ZHU, Peng WU. Pentosan polysulfate alleviates cyclophosphamide-induced interstitial cystitis/bladder pain syndrome in mice by modulating gut microbiota and bile acid metabolism [J]. Journal of Southern Medical University, 2025, 45(6): 1270-1279. |
| [6] | Nuozhou WENG, Bin TAN, Wentao ZENG, Jiayu GU, Lianji WENG, Kehong ZHENG. RGL1 overexpression promotes metastasis of colorectal cancer by upregulating motile focal adhesion assembly via activating the CDC42/RAC1 complex [J]. Journal of Southern Medical University, 2025, 45(5): 1031-1038. |
| [7] | Anbang ZHANG, Xiuqi SUN, Bo PANG, Yuanhua WU, Jingyu SHI, Ning ZHANG, Tao YE. Electroacupuncture pretreatment alleviates cerebral ischemia-reperfusion injury in rats by inhibiting ferroptosis through the gut-brain axis and the Nrf2/HO-1 signaling pathway [J]. Journal of Southern Medical University, 2025, 45(5): 911-920. |
| [8] | Zhennan MA, Fuquan LIU, Xuefeng ZHAO, Xiaowei ZHANG. High expression of DTX2 promotes proliferation, invasion and epithelial-mesenchymal transition of oxaliplatin-resistant colorectal cancer cells [J]. Journal of Southern Medical University, 2025, 45(4): 829-836. |
| [9] | Wenjie LI, Yaonan HONG, Rui HUANG, Yuchen LI, Ying ZHANG, Yun ZHANG, Dijiong WU. Causal relationship between autoimmune diseases and aplastic anemia: A Mendelian randomization study [J]. Journal of Southern Medical University, 2025, 45(4): 871-879. |
| [10] | Shunjie QING, Zhiyong SHEN. High expression of hexokinase 2 promotes proliferation, migration and invasion of colorectal cancer cells by activating the JAK/STAT pathway and regulating tumor immune microenvironment [J]. Journal of Southern Medical University, 2025, 45(3): 542-553. |
| [11] | Jiachun LUO, Sodnomjamts Batzaya, Xuefeng GAO, Jingyu CHEN, Zhengying YU, Shasha XIONG, Hong CAO. Akkermansia muciniphila gavage improves gut-brain interaction disorders in gp120 transgenic mice [J]. Journal of Southern Medical University, 2025, 45(3): 554-565. |
| [12] | Lixia YIN, Minzhu NIU, Keni ZHANG, Zhijun GENG, Jianguo HU, Jiangyan LI, Jing LI. Cimifugin ameliorates Crohn's disease-like colitis in mice by modulating Th-cell immune balance via inhibiting the MAPK pathway [J]. Journal of Southern Medical University, 2025, 45(3): 595-602. |
| [13] | Junjie GAO, Kai YE, Jing WU. Quercetin inhibits proliferation and migration of clear cell renal cell carcinoma cells by regulating TP53 gene [J]. Journal of Southern Medical University, 2025, 45(2): 313-321. |
| [14] | Xingxu PAN, Bingqi ZHANG, Zhihua ZHANG, Qiushi CAO. Reduced intestinal abundance of Gordonibacter increases risk of kidney stones: a Mendelian randomization study and evidence from rat models [J]. Journal of Southern Medical University, 2025, 45(11): 2405-2415. |
| [15] | Huihua MA, Kuipo YAN, Gang LIU, Yazhou XU, Lei ZHANG, Yizhuo LI. Epidemiology of atrial fibrillation/atrial flutter and its risk factors from 1990 to 2021: a systematic analysis and Mendelian randomization study based on the China and Global Burden of Disease Study 2021 [J]. Journal of Southern Medical University, 2025, 45(10): 2182-2190. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||