南方医科大学学报 ›› 2024, Vol. 44 ›› Issue (9): 1776-1782.doi: 10.12122/j.issn.1673-4254.2024.09.18
收稿日期:2024-02-24
出版日期:2024-09-20
发布日期:2024-09-30
通讯作者:
张庆玲
E-mail:yemengnan3@163.com;zhangqingling@gdph.org.cn
作者简介:叶梦楠,硕士,E-mail: yemengnan3@163.com
基金资助:
Mengnan YE1,2( ), Hongmei WU2, Yan MEI2, Qingling ZHANG2(
), Hongmei WU2, Yan MEI2, Qingling ZHANG2( )
)
Received:2024-02-24
Online:2024-09-20
Published:2024-09-30
Contact:
Qingling ZHANG
E-mail:yemengnan3@163.com;zhangqingling@gdph.org.cn
Supported by:摘要:
目的 分析CREM在胃癌中的表达及其与临床预后的相关性。 方法 利用TCGA和GEO数据库分析胃癌和癌旁组织CREM mRNA表达差异,免疫组化染色(IHC)分析43例胃癌和配对癌旁组织的CREM蛋白表达差异,分析CREM表达与胃癌患者临床病理特征之间的关系;采用Kaplan-Meier生存分析探讨CREM表达水平与胃癌患者生存期之间的关系;利用LinkedOmics数据库注释CREM相关基因的GO功能和KEGG通路的富集情况。 结果 数据库分析显示,CREM在胃癌组织中高表达(P<0.05),与胃癌患者的不良预后正相关(P=0.01)。IHC结果显示,与癌旁组织相比,胃癌组织中CREM高表达(P<0.0001),CREM表达水平与T分期和N分期有关(P<0.05),CREM高表达的胃癌患者总生存期更短(RR=4.02,P=0.0046)。基因富集分析显示,CREM可能通过细胞黏附分子介导的信号通路促进胃癌发生发展。 结论 CREM在胃癌中高表达提示不良预后,CREM可作为候选的胃癌预后指标。
叶梦楠, 武鸿美, 梅琰, 张庆玲. CREM在胃癌中高表达并与患者的不良预后相关[J]. 南方医科大学学报, 2024, 44(9): 1776-1782.
Mengnan YE, Hongmei WU, Yan MEI, Qingling ZHANG. High expression of CREM is associated with poor prognosis in gastric cancer patients[J]. Journal of Southern Medical University, 2024, 44(9): 1776-1782.
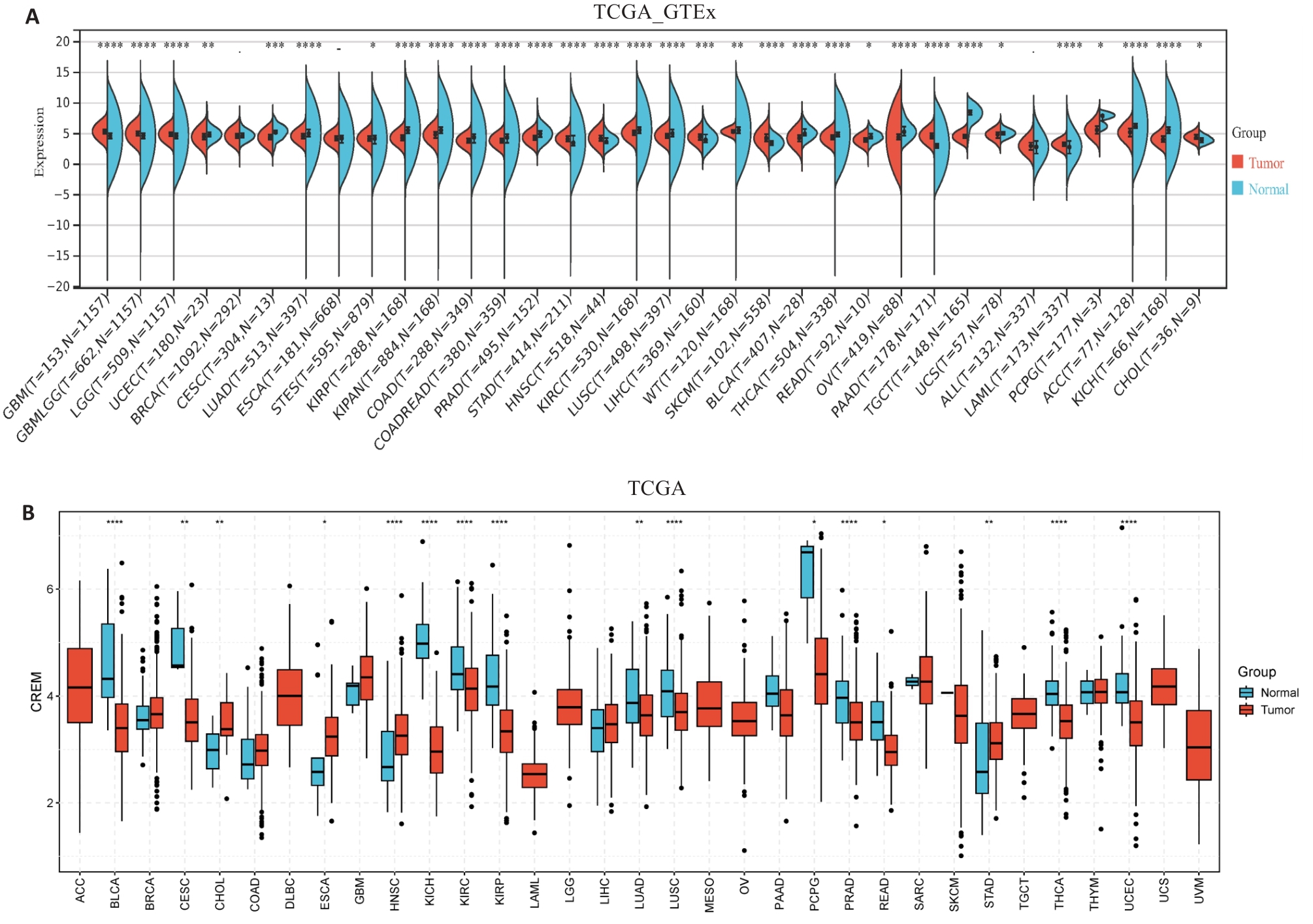
图1 CREM在多种恶性肿瘤中的差异性表达
Fig.1 Differential expression of CREM in different cancers based on data from TCGA-GTEx (A) and TCGA (B) databases. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
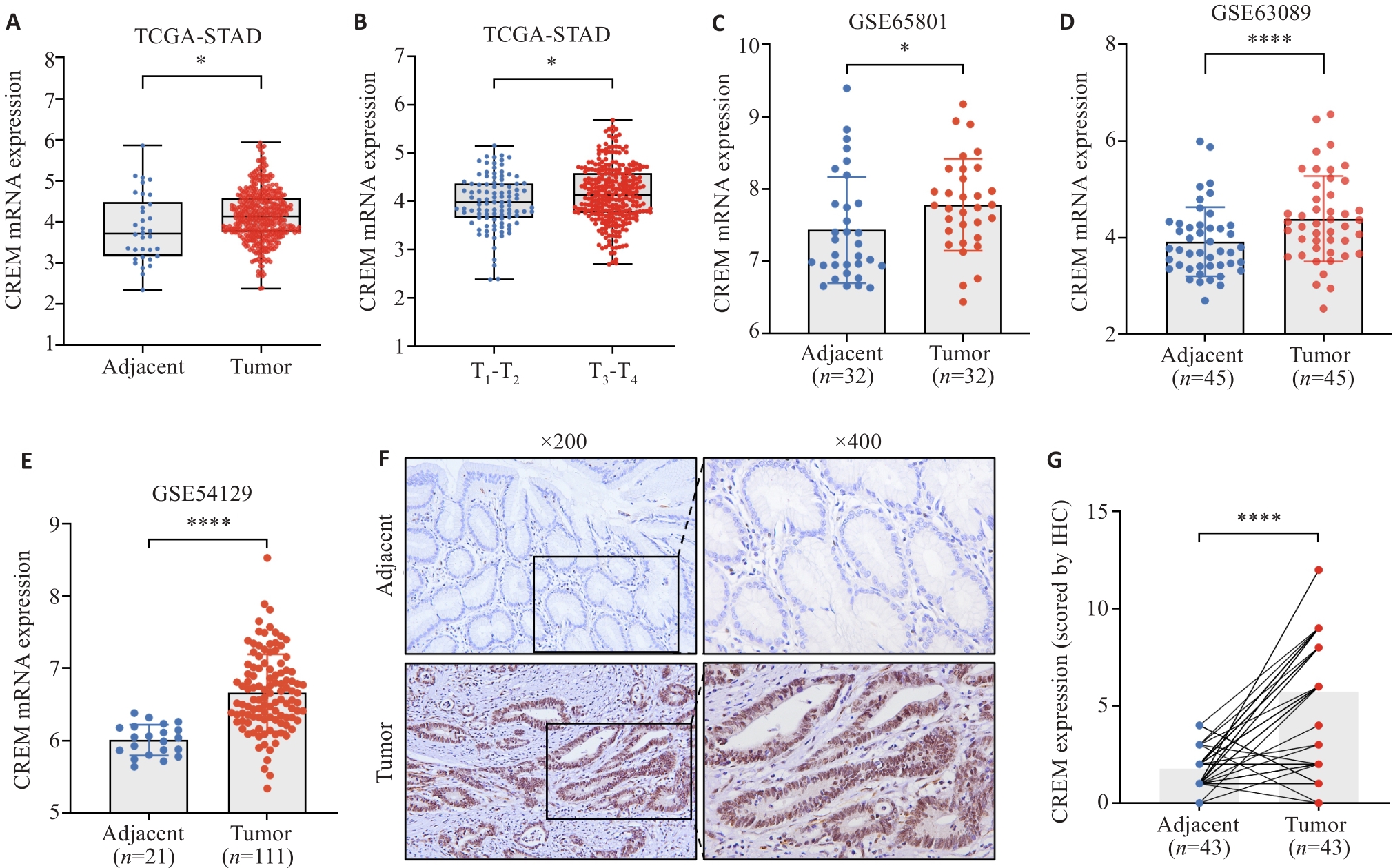
图3 CREM在胃癌组织中高表达
Fig.3 CREM is highly expressed in gastric cancer tissues. A: Expression of CREM in gastric cancer and adjacent tissues in TCGA database. B: Differential expression of CREM in tumors in different T-stages. C-E: Differential expression of CREM in gastric cancer tissues and adjacent tissues in GSE65801, GSE63089, and GSE54129 datasets. F, G: Immunohistochemical staining of CREM in gastric cancer and adjacent tissues and the immunohistochemical scores. *P<0.05, ****P<0.0001.
| Characteristic | n | CREM Expression | χ2 | P | |
|---|---|---|---|---|---|
| Low (n=11) | High (n=32) | ||||
| Age (year) | 1.815 | 0.178 | |||
| <60 | 15 | 2 (13.3) | 13 (86.7) | ||
| ≥60 | 28 | 9 (32.1) | 19 (67.9) | ||
| Gender | 1.120 | 0.290 | |||
| Male | 29 | 6 (20.7) | 23 (79.3) | ||
| Female | 14 | 5 (35.7) | 9 (64.3) | ||
| Ki-67(%) | 1.120 | 0.290 | |||
| <50 | 14 | 5 (35.7) | 9 (64.3) | ||
| ≥50 | 29 | 6 (20.7) | 23 (79.3) | ||
| Tumor size (cm) | 2.516 | 0.113 | |||
| <4 | 15 | 6 (40.0) | 9 (60.0) | ||
| ≥4 | 28 | 5 (17.9) | 23 (82.1) | ||
| T stage | 10.862 | 0.001 | |||
| T1-T2 | 14 | 8 (57.1) | 6 (42.9) | ||
| T3-T4 | 29 | 3 (10.3) | 26 (89.7) | ||
| N stage | 4.418 | 0.036 | |||
| N0 | 16 | 7 (43.8) | 9 (56.2) | ||
| N1-N3 | 27 | 4 (14.8) | 23 (85.2) | ||
| TNM staging | 3.376 | 0.066 | |||
| I-II | 21 | 8 (38.1) | 13 (61.9) | ||
| III-IV | 22 | 3 (13.6) | 19 (86.4) | ||
表1 43例胃癌患者CREM的表达与临床病理特征的关系
Tab. 1 Relationship between CREM expression and clinicopathological features of 43 patients with STAD
| Characteristic | n | CREM Expression | χ2 | P | |
|---|---|---|---|---|---|
| Low (n=11) | High (n=32) | ||||
| Age (year) | 1.815 | 0.178 | |||
| <60 | 15 | 2 (13.3) | 13 (86.7) | ||
| ≥60 | 28 | 9 (32.1) | 19 (67.9) | ||
| Gender | 1.120 | 0.290 | |||
| Male | 29 | 6 (20.7) | 23 (79.3) | ||
| Female | 14 | 5 (35.7) | 9 (64.3) | ||
| Ki-67(%) | 1.120 | 0.290 | |||
| <50 | 14 | 5 (35.7) | 9 (64.3) | ||
| ≥50 | 29 | 6 (20.7) | 23 (79.3) | ||
| Tumor size (cm) | 2.516 | 0.113 | |||
| <4 | 15 | 6 (40.0) | 9 (60.0) | ||
| ≥4 | 28 | 5 (17.9) | 23 (82.1) | ||
| T stage | 10.862 | 0.001 | |||
| T1-T2 | 14 | 8 (57.1) | 6 (42.9) | ||
| T3-T4 | 29 | 3 (10.3) | 26 (89.7) | ||
| N stage | 4.418 | 0.036 | |||
| N0 | 16 | 7 (43.8) | 9 (56.2) | ||
| N1-N3 | 27 | 4 (14.8) | 23 (85.2) | ||
| TNM staging | 3.376 | 0.066 | |||
| I-II | 21 | 8 (38.1) | 13 (61.9) | ||
| III-IV | 22 | 3 (13.6) | 19 (86.4) | ||
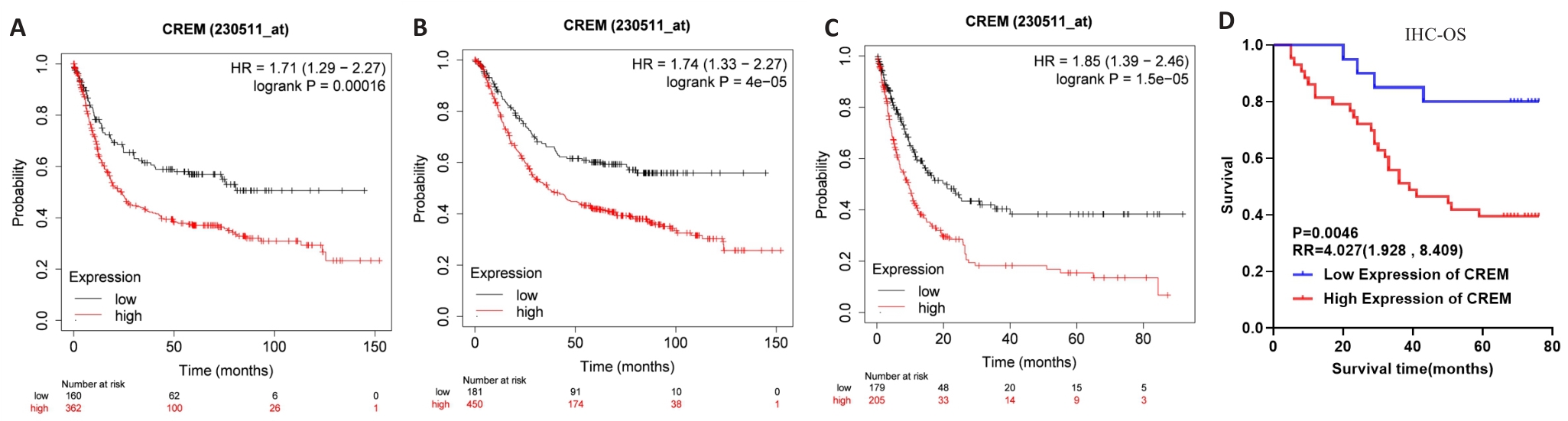
图4 CREM高表达与胃癌患者预后不良相关
Fig.4 High CREM expression is associated with poor prognosis in gastric cancer patients. A-C: FP, OS, and PPS curves of gastric cancer patients with different CREM mRNA expression levels in the Kaplan-Meier Plotter database. D: OS curves of patients with high and low CREM protein expression in clinical gastric cancer samples.
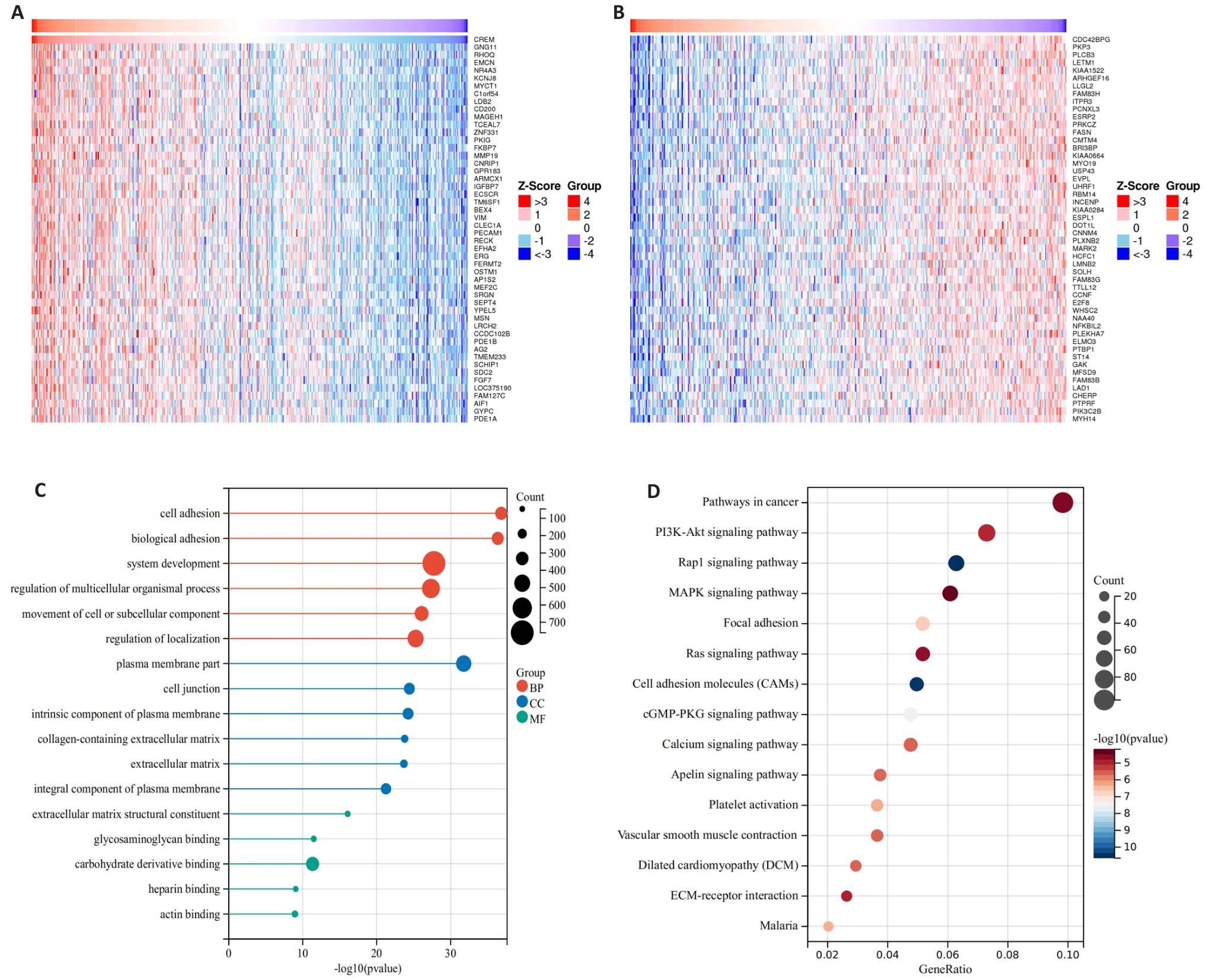
图5 胃癌中CREM相关基因分析及KEGG、GO富集分析结果
Fig.5 Analysis of CREM-related genes and results of KEGG and GO enrichment analysis in gastric cancer. A, B: LinkedOmics database analysis of genes positively and negatively correlated with CREM expression in the TCGA gastric cancer dataset. C: GO enrichment analysis to explore the role of CREM in gastric cancer, including the biological process, cellular composition, and molecular function. D: KEGG enrichment analysis to explore the relevant molecular pathways of CREM in gastric cancer development.
| 1 | Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-49. |
| 2 | Qiu HB, Cao SM, Xu RH. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020[J]. Cancer Commun, 2021, 41(10): 1037-48. |
| 3 | 赫 捷, 陈万青, 李兆申, 等. 中国胃癌筛查与早诊早治指南(2022,北京)[J]. 中华肿瘤杂志, 2022, 44(7): 634-66. |
| 4 | Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020[J]. Chin Med J, 2021, 134(7): 783-91. |
| 5 | Thrift AP, Wenker TN, El-Serag HB. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention[J]. Nat Rev Clin Oncol, 2023, 20(5): 338-49. |
| 6 | Shitara K, Chin K, Yoshikawa T, et al. Phase II study of adjuvant chemotherapy of S-1 plus oxaliplatin for patients with stage III gastric cancer after D2 gastrectomy[J]. Gastric Cancer, 2017, 20(1): 175-81. |
| 7 | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma[J]. Nature, 2014, 513(7517): 202-9. |
| 8 | Canale M, Casadei-Gardini A, Ulivi P, et al. Epigenetic mechanisms in gastric cancer: potential new therapeutic opportunities[J]. Int J Mol Sci, 2020, 21(15): 5500-9. |
| 9 | Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis[J]. Apoptosis, 2003, 8(3): 225-8. |
| 10 | Brindle PK, Montminy MR. The CREB family of transcription activators[J]. Curr Opin Genet Dev, 1992, 2(2): 199-204. |
| 11 | Sassone-Corsi P. Coupling gene expression to cAMP signalling: role of CREB and CREM[J]. Int J Biochem Cell Biol, 1998, 30(1): 27-38. |
| 12 | Sánchez-Jasso DE, López-Guzmán SF, Bermúdez-Cruz RM, et al. Novel aspects of cAMP-response element modulator (CREM) role in spermatogenesis and male fertility[J]. Int J Mol Sci, 2023, 24(16): 12558-67. |
| 13 | Wu XM, Jin W, Liu XJ, et al. Cyclic AMP response element modulator-1 (CREM-1) involves in neuronal apoptosis after traumatic brain injury[J]. J Mol Neurosci, 2012, 47(2): 357-67. |
| 14 | Kaprio H, Siddiqui A, Saustila L, et al. The oncogenic properties of the EWSR1: CREM fusion gene are associated with polyamine metabolism[J]. Sci Rep, 2023, 13(1): 4884-93. |
| 15 | Wang YC, Zhou SH, Yang XJ, et al. Low expression of cyclic AMP response element modulator-1 can increase the migration and invasion of esophageal squamous cell carcinoma[J]. Tumour Biol, 2013, 34(6): 3649-57. |
| 16 | Kaprio H, Heuser VD, Orte K, et al. Expression of transcription factor CREM in human tissues[J]. J Histochem Cytochem, 2021, 69(8): 495-509. |
| 17 | Shen WT, Song ZG, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform[J]. Imeta, 2022, 1(3): e36. |
| 18 | Hanahan D. Hallmarks of cancer: new dimensions[J]. Cancer Discov, 2022, 12(1): 31-46. |
| 19 | Alsina M, Arrazubi V, Diez M, et al. Current developments in gastric cancer: from molecular profiling to treatment strategy[J]. Nat Rev Gastroenterol Hepatol, 2023, 20(3): 155-70. |
| 20 | Sun JM, Shen L, Shah MA, et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study[J]. Lancet, 2021, 398(10302): 759-71. |
| 21 | Augustin JE, Soussan P, Bass AJ. Targeting the complexity of ERBB2 biology in gastroesophageal carcinoma[J]. Ann Oncol, 2022, 33(11): 1134-48. |
| 22 | Kawazoe A, Fukuoka S, Nakamura Y, et al. Lenvatinib plus pembrolizumab in patients with advanced gastric cancer in the first-line or second-line setting (EPOC1706): an open-label, single-arm, phase 2 trial[J]. Lancet Oncol, 2020, 21(8): 1057-65. |
| 23 | Wainberg ZA, Enzinger PC, Kang YK, et al. Bemarituzumab in patients with FGFR2b-selected gastric or gastro-oesophageal junction adenocarcinoma (FIGHT): a randomised, double-blind, placebo-controlled, phase 2 study[J]. Lancet Oncol, 2022, 23(11): 1430-40. |
| 24 | Qi CS, Gong JF, Li J, et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results[J]. Nat Med, 2022, 28(6): 1189-98. |
| 25 | Bardia A, Messersmith WA, Kio EA, et al. Sacituzumab govitecan, a Trop-2-directed antibody-drug conjugate, for patients with epithelial cancer: final safety and efficacy results from the phase I/II IMMU-132-01 basket trial[J]. Ann Oncol, 2021, 32(6): 746-56. |
| 26 | Servillo G, Della Fazia MA, Sassone-Corsi P. Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM[J]. Exp Cell Res, 2002, 275(2): 143-54. |
| 27 | Behr R, Weinbauer GF. cAMP response element modulator (CREM): an essential factor for spermatogenesis in Primates?[J]. Int J Androl, 2001, 24(3): 126-35. |
| 28 | Feng F, Wu JT, Gao ZL, et al. Screening the key microRNAs and transcription factors in prostate cancer based on microRNA functional synergistic relationships[J]. Medicine, 2017, 96(1): e5679. |
| 29 | Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: beyond the migration of single cells[J]. J Biol Chem, 2020, 295(8): 2495-505. |
| 30 | Chen JZ, Qin HY, Hao JZ, et al. Cardiac-specific overexpression of CREM-IbΔC-X via CRISPR/Cas9 in mice presents a new model of atrial cardiomyopathy with spontaneous atrial fibrillation[J]. Transl Res, 2024, 267: 54-66. |
| [1] | 陈孝华, 鲁辉, 王子良, 王炼, 夏勇生, 耿志军, 张小凤, 宋雪, 王月月, 李静, 胡建国, 左芦根. ABI2在胃癌进展和预后中的作用及其调控机制[J]. 南方医科大学学报, 2024, 44(9): 1653-1661. |
| [2] | 纪凯, 于冠宇, 周乐其, 张天帅, 凌潜龙, 满文江, 朱冰, 张卫. HNRNPA1基因在结直肠癌组织中高表达及其潜在的诊断和治疗价值[J]. 南方医科大学学报, 2024, 44(9): 1685-1695. |
| [3] | 耿志军, 杨晶晶, 牛民主, 刘馨悦, 施金冉, 刘亦珂, 姚新宇, 张雨路, 张小凤, 胡建国. 桑黄酮G通过调控PI3K/AKT/mTOR通路抑制胃癌细胞的生长、迁移和侵袭[J]. 南方医科大学学报, 2024, 44(8): 1476-1484. |
| [4] | 庞一丹, 刘雅, 陈思嫒, 张荆雷, 曾今, 潘元明, 安娟. SPAG5在胃癌细胞恶性增殖中的生物学作用[J]. 南方医科大学学报, 2024, 44(8): 1497-1507. |
| [5] | 陈莉莉, 吴天宇, 张铭, 丁子夏, 张妍, 杨依清, 郑佳倩, 张小楠. 类风湿关节炎的潜在生物标志物及其免疫调控机制:基于GEO数据库[J]. 南方医科大学学报, 2024, 44(6): 1098-1108. |
| [6] | 刘鹏程, 娄丽娟, 刘霞, 王建, 姜颖. M2巨噬细胞特征基因风险评分能准确预测HBV相关肝细胞癌患者的预后[J]. 南方医科大学学报, 2024, 44(5): 827-840. |
| [7] | 周伟, 聂军, 胡佳, 蒋艺枝, 张大发. 内质网应激相关基因在主动脉夹层疾病中的差异性表达及与免疫浸润的相关性[J]. 南方医科大学学报, 2024, 44(5): 859-866. |
| [8] | 夏勇生, 王炼, 陈孝华, 张雨路, 孙奥飞, 陈德利. 过表达TSR2通过下调PI3K/AKT信号通路抑制胃癌细胞的增殖和侵袭[J]. 南方医科大学学报, 2024, 44(5): 913-919. |
| [9] | 裴蓓, 张艺, 魏思源, 梅语, 宋标, 董港, 温子昂, 李学军. 基于转录组学测序及生物信息学方法鉴定肠上皮化生的潜在致病基因[J]. 南方医科大学学报, 2024, 44(5): 941-949. |
| [10] | 杨晶晶, 殷丽霞, 段婷, 牛民主, 何震东, 陈心蕊, 张小凤, 李静, 耿志军, 左芦根. 胃癌组织中高表达ATP5A1与患者术后的不良预后和肿瘤细胞的糖代谢有关[J]. 南方医科大学学报, 2024, 44(5): 974-980. |
| [11] | 王沁智, 宋冰, 郝诗睿, 肖志远, 金连辉, 郑通, 柴芳. 基于生物信息学分析CCND2在甲状腺乳头状癌中的表达及其对免疫浸润的影响[J]. 南方医科大学学报, 2024, 44(5): 981-988. |
| [12] | 高志强, 林 洁, 洪 鹏, 胡再宏, 董军君, 石秦林, 田小毛, 刘 丰, 魏光辉. 基于高通量 RNA 测序分析 Wilms 瘤中关键基因对预后及免疫应答的影响[J]. 南方医科大学学报, 2024, 44(4): 727-738. |
| [13] | 梁一豪, 赖颖君, 袁燕文, 袁 炜, 张锡波, 张拔山, 卢志锋. 基于GEO数据库筛选胃癌差异表达基因及其功能和通路富集分析[J]. 南方医科大学学报, 2024, 44(3): 605-616. |
| [14] | 沈梦迪, 赵 娜, 邓晓晶, 邓 敏. COX6B2在胃癌组织中高表达并影响患者的远期预后:基于抑制p53信号调控胃癌细胞的增殖及细胞周期[J]. 南方医科大学学报, 2024, 44(2): 289-297. |
| [15] | 张 诺, 张 震, 张雨路, 宋 雪, 张小凤, 李 静, 左芦根, 胡建国. PCID2在胃癌组织中高表达并通过调控细胞周期进程和增殖影响患者预后[J]. 南方医科大学学报, 2024, 44(2): 324-332. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||