Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (2): 313-321.doi: 10.12122/j.issn.1673-4254.2025.02.12
Previous Articles Next Articles
Junjie GAO( ), Kai YE, Jing WU(
), Kai YE, Jing WU( )
)
Received:2024-09-03
Online:2025-02-20
Published:2025-03-03
Contact:
Jing WU
E-mail:631517383@qq.com;wjwjwj1973@126.com
Supported by:Junjie GAO, Kai YE, Jing WU. Quercetin inhibits proliferation and migration of clear cell renal cell carcinoma cells by regulating TP53 gene[J]. Journal of Southern Medical University, 2025, 45(2): 313-321.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.02.12
| Exposure | Outcome | Gen | Method | Nsnp | b | Se | P |
|---|---|---|---|---|---|---|---|
| 9816_37_ISOC1_ISOC1 | KIRC | ISOC1 | IVW | 8 | 0.736746 | 0.171048 | 1.65E-05 |
| 6354_13_HEPHL1_HPHL1 | KIRC | HEPHL1 | IVW | 5 | -1.28851 | 0.310833 | 3.39E-05 |
| 17742_2_RRAS_RRAS | KIRC | RRAS | IVW | 5 | -1.33358 | 0.325935 | 4.29E-05 |
| 12621_55_PPP2R1A_PP2AAA | KIRC | PPP2R1A | IVW | 4 | -0.9648 | 0.243754 | 7.56E-05 |
| 5345_51_CKAP2_CKAP2 | KIRC | CKAP2 | IVW | 15 | -0.66094 | 0.169567 | 9.71E-05 |
| 12807_89_ARHGAP30_RHG30 | KIRC | ARHGAP30 | IVW | 3 | 1.721247 | 0.446005 | 0.000114 |
| 6123_69_TP53_p53 | KIRC | TP53 | IVW | 7 | 1.201512 | 0.311593 | 0.000115 |
| 9511_61_NXPH2_NXPH2 | KIRC | NXPH2 | IVW | 18 | -0.39638 | 0.103952 | 0.000137 |
| 6388_21_CCDC126_CC126 | KIRC | CCDC126 | IVW | 18 | -0.46547 | 0.124775 | 0.000191 |
| 13673_21_CCT7_TCPH | KIRC | CCT7 | IVW | 4 | -1.0039 | 0.269598 | 0.000196 |
| 7083_74_MATN4_MATN4 | KIRC | MATN4 | IVW | 7 | 0.634937 | 0.174364 | 0.000271 |
| 4568_17_SLITRK5_SLIK5 | KIRC | SLITRK5 | IVW | 12 | 0.821988 | 0.226306 | 0.000281 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | IVW | 3 | -1.75547 | 0.495302 | 0.000394 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | IVW | 8 | -0.76964 | 0.218311 | 0.000423 |
| 8040_9_LEMD1_LEMD1 | KIRC | LEMD1 | IVW | 3 | -2.26505 | 0.647584 | 0.000469 |
| 6578_29_TNMD_TNMD | KIRC | TNMD | IVW | 3 | -1.94913 | 0.558892 | 0.000488 |
| 9012_1_PRPF6_PRP6 | KIRC | PRPF6 | IVW | 2 | -2.88761 | 0.828756 | 0.000493 |
| 8535_102_DMKN_Dermokine | KIRC | DMKN | IVW | 2 | -1.63853 | 0.479425 | 0.000632 |
| 8748_45_CD74_HG2A | KIRC | CD74 | IVW | 3 | -2.22233 | 0.652226 | 0.000656 |
| 10085_25_STAR_STAR | KIRC | STAR | IVW | 5 | -1.39378 | 0.418791 | 0.000874 |
| 18198_51_HBQ1_HBAT | KIRC | HBQ1 | IVW | 8 | -0.86326 | 0.261832 | 0.000977 |
| 18380_78_ALB_Albumin | KIRC | ALB | IVW | 11 | -0.41249 | 0.13172 | 0.001739 |
| 12587_65_ARL2_ARL2 | KIRC | ARL2 | IVW | 12 | -0.47391 | 0.152486 | 0.001884 |
| 12041_33_HSPA1L_HS71L | KIRC | HSPA1L | IVW | 11 | -0.55241 | 0.178001 | 0.001913 |
| 10842_7_GCNT4_GCNT4 | KIRC | GCNT4 | IVW | 4 | -1.6224 | 0.541404 | 0.00273 |
| 11160_56_RNF122_RN122 | KIRC | RNF122 | IVW | 6 | -0.87475 | 0.299384 | 0.00348 |
Tab.1 Result of Mendelian randomization between plasma proteins and ccRCC
| Exposure | Outcome | Gen | Method | Nsnp | b | Se | P |
|---|---|---|---|---|---|---|---|
| 9816_37_ISOC1_ISOC1 | KIRC | ISOC1 | IVW | 8 | 0.736746 | 0.171048 | 1.65E-05 |
| 6354_13_HEPHL1_HPHL1 | KIRC | HEPHL1 | IVW | 5 | -1.28851 | 0.310833 | 3.39E-05 |
| 17742_2_RRAS_RRAS | KIRC | RRAS | IVW | 5 | -1.33358 | 0.325935 | 4.29E-05 |
| 12621_55_PPP2R1A_PP2AAA | KIRC | PPP2R1A | IVW | 4 | -0.9648 | 0.243754 | 7.56E-05 |
| 5345_51_CKAP2_CKAP2 | KIRC | CKAP2 | IVW | 15 | -0.66094 | 0.169567 | 9.71E-05 |
| 12807_89_ARHGAP30_RHG30 | KIRC | ARHGAP30 | IVW | 3 | 1.721247 | 0.446005 | 0.000114 |
| 6123_69_TP53_p53 | KIRC | TP53 | IVW | 7 | 1.201512 | 0.311593 | 0.000115 |
| 9511_61_NXPH2_NXPH2 | KIRC | NXPH2 | IVW | 18 | -0.39638 | 0.103952 | 0.000137 |
| 6388_21_CCDC126_CC126 | KIRC | CCDC126 | IVW | 18 | -0.46547 | 0.124775 | 0.000191 |
| 13673_21_CCT7_TCPH | KIRC | CCT7 | IVW | 4 | -1.0039 | 0.269598 | 0.000196 |
| 7083_74_MATN4_MATN4 | KIRC | MATN4 | IVW | 7 | 0.634937 | 0.174364 | 0.000271 |
| 4568_17_SLITRK5_SLIK5 | KIRC | SLITRK5 | IVW | 12 | 0.821988 | 0.226306 | 0.000281 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | IVW | 3 | -1.75547 | 0.495302 | 0.000394 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | IVW | 8 | -0.76964 | 0.218311 | 0.000423 |
| 8040_9_LEMD1_LEMD1 | KIRC | LEMD1 | IVW | 3 | -2.26505 | 0.647584 | 0.000469 |
| 6578_29_TNMD_TNMD | KIRC | TNMD | IVW | 3 | -1.94913 | 0.558892 | 0.000488 |
| 9012_1_PRPF6_PRP6 | KIRC | PRPF6 | IVW | 2 | -2.88761 | 0.828756 | 0.000493 |
| 8535_102_DMKN_Dermokine | KIRC | DMKN | IVW | 2 | -1.63853 | 0.479425 | 0.000632 |
| 8748_45_CD74_HG2A | KIRC | CD74 | IVW | 3 | -2.22233 | 0.652226 | 0.000656 |
| 10085_25_STAR_STAR | KIRC | STAR | IVW | 5 | -1.39378 | 0.418791 | 0.000874 |
| 18198_51_HBQ1_HBAT | KIRC | HBQ1 | IVW | 8 | -0.86326 | 0.261832 | 0.000977 |
| 18380_78_ALB_Albumin | KIRC | ALB | IVW | 11 | -0.41249 | 0.13172 | 0.001739 |
| 12587_65_ARL2_ARL2 | KIRC | ARL2 | IVW | 12 | -0.47391 | 0.152486 | 0.001884 |
| 12041_33_HSPA1L_HS71L | KIRC | HSPA1L | IVW | 11 | -0.55241 | 0.178001 | 0.001913 |
| 10842_7_GCNT4_GCNT4 | KIRC | GCNT4 | IVW | 4 | -1.6224 | 0.541404 | 0.00273 |
| 11160_56_RNF122_RN122 | KIRC | RNF122 | IVW | 6 | -0.87475 | 0.299384 | 0.00348 |
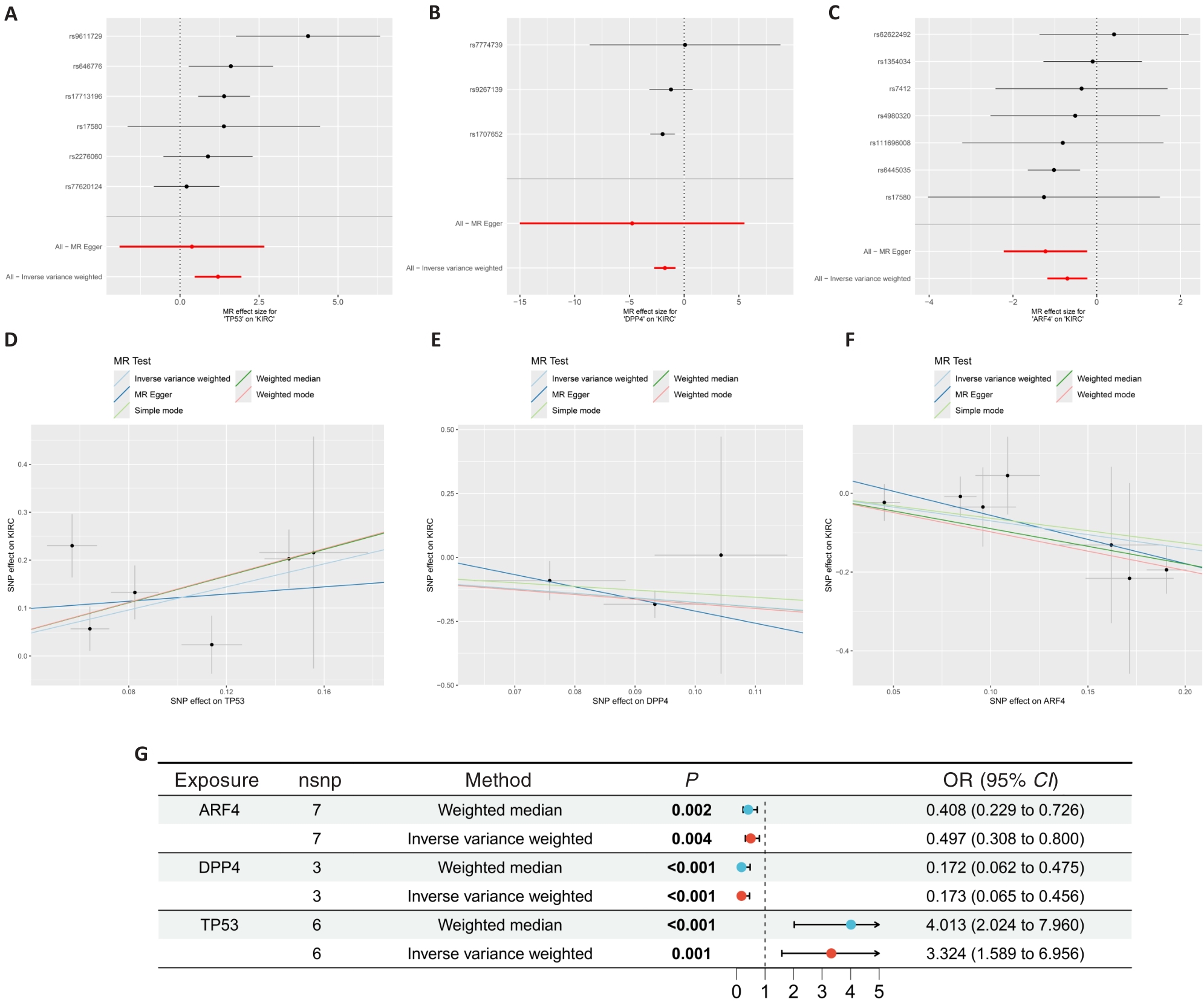
Fig.3 Mendelian randomization results on ccRCC. A-C: Forest plot showing the results of Mendelian randomization of the core genes. D-F: Scatter plots of Mendelian randomization results on core targets and ccRCC. G: Forest plot showing Mendelian randomization results of the core genes.
| Id.exposure | Outcome | Exposure | Method | Q | Q_pval |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | MR Egger | 2.46079 | 0.782388313 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | Inverse variance weighted | 3.836625 | 0.698774273 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | MR Egger | 0.27862 | 0.597607084 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | Inverse variance weighted | 0.608893 | 0.737531351 |
| 6123_69_TP53_p53 | KIRC | TP53 | MR Egger | 9.021383 | 0.060567227 |
| 6123_69_TP53_p53 | KIRC | TP53 | Inverse variance weighted | 10.29088 | 0.06740046 |
Tab. 2 Heterogeneity analysis of Mendelian randomization between plasma proteins and ccRCC
| Id.exposure | Outcome | Exposure | Method | Q | Q_pval |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | MR Egger | 2.46079 | 0.782388313 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | Inverse variance weighted | 3.836625 | 0.698774273 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | MR Egger | 0.27862 | 0.597607084 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | Inverse variance weighted | 0.608893 | 0.737531351 |
| 6123_69_TP53_p53 | KIRC | TP53 | MR Egger | 9.021383 | 0.060567227 |
| 6123_69_TP53_p53 | KIRC | TP53 | Inverse variance weighted | 10.29088 | 0.06740046 |
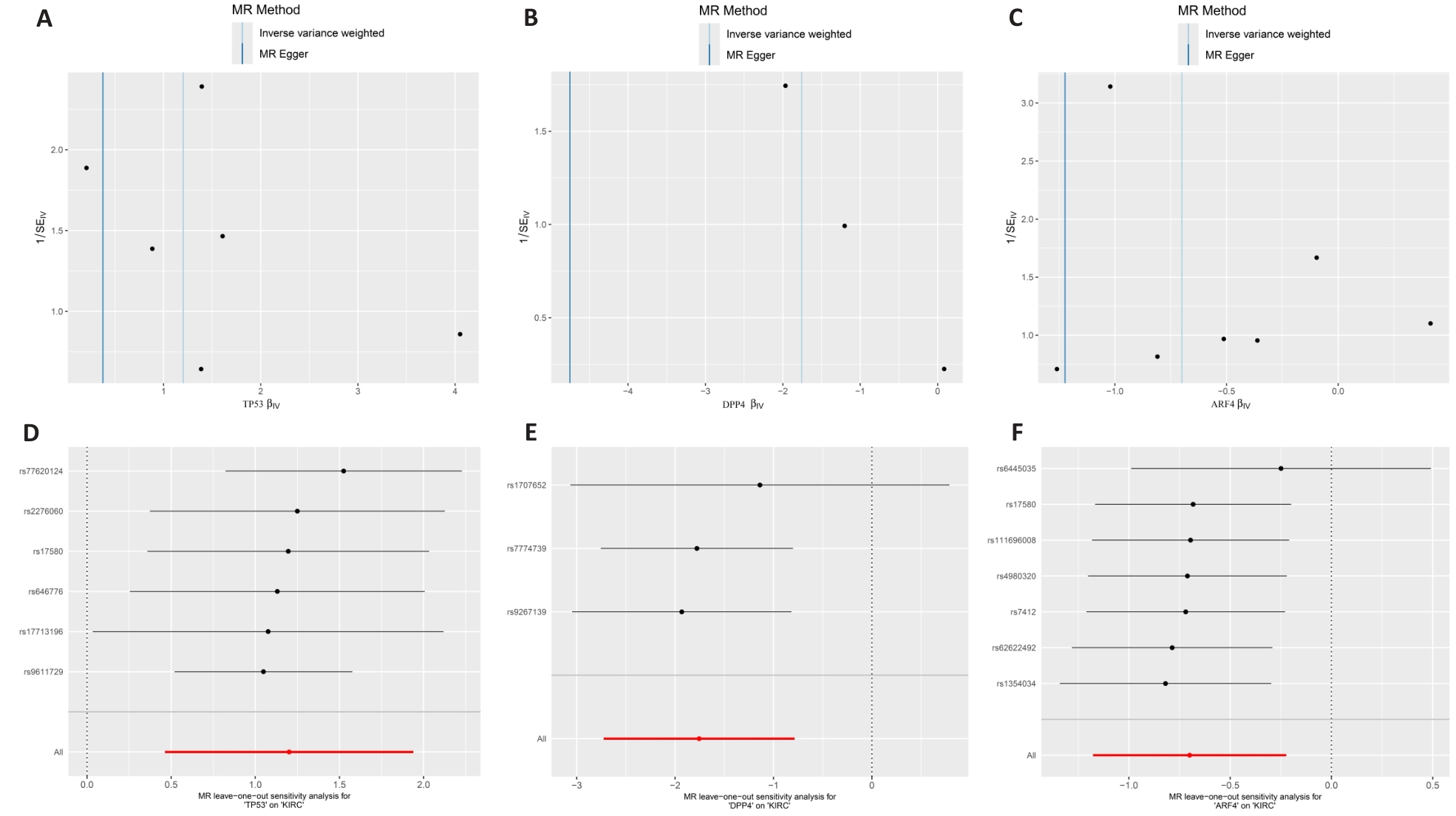
Fig.4 Sensitivity analysis of Mendelian randomization. A-C: MR funnel plot for core targets and ccRCC. D-F: Leave-one-out sensitivity analysis for potential core genes and ccRCC.
| Id.exposure | Outcome | Exposure | Egger_intercept | Se | P |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | 0.066322526 | 0.056543 | 0.293645 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | 0.264971682 | 0.461066 | 0.667936 |
| 6123_69_TP53_p53 | KIRC | TP53 | 0.08422687 | 0.112264 | 0.494821 |
Tab.3 Pleiotropy analysis of Mendelian randomization between plasma proteins and ccRCC
| Id.exposure | Outcome | Exposure | Egger_intercept | Se | P |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | 0.066322526 | 0.056543 | 0.293645 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | 0.264971682 | 0.461066 | 0.667936 |
| 6123_69_TP53_p53 | KIRC | TP53 | 0.08422687 | 0.112264 | 0.494821 |
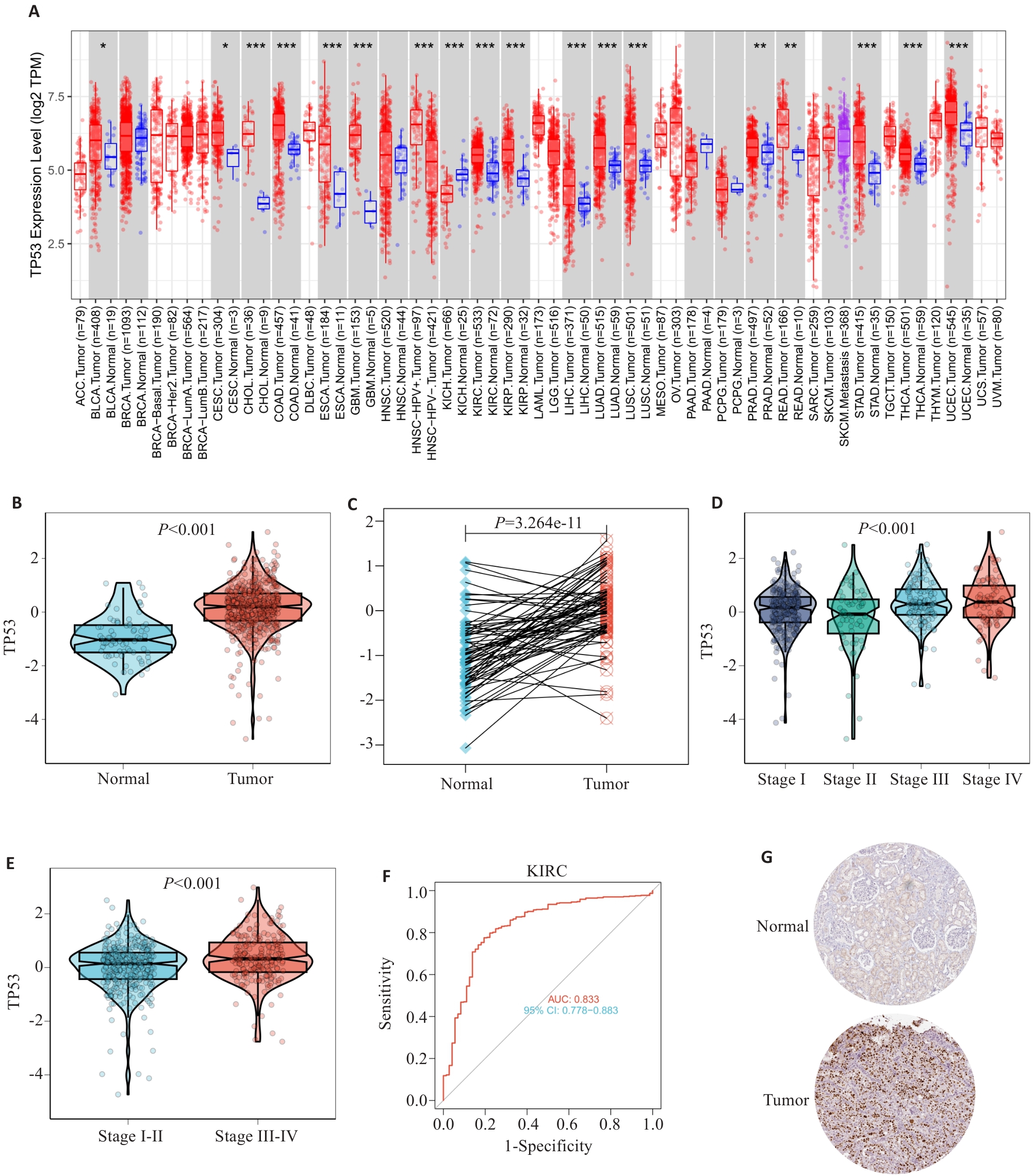
Fig.5 TP53 expression in ccRCC. A: TP53 mRNA evpression in different tumor samples. B: TP53 mRNA expression in ccRCC samples and normal tissues. C: TP53 mRNA expression in ccRCC and paired adjacent normal tissues. D,E: TP53 mRNA expression levels in different ccRCC clinical stages. F: Receiver operating characteristic analysis (ROC) of TP53 in patients with ccRCC (AUC=0.833). G: Immunohistochemical staining of TP53 in renal cancer tissues and normal renal tissues (Original magnification: ×20).
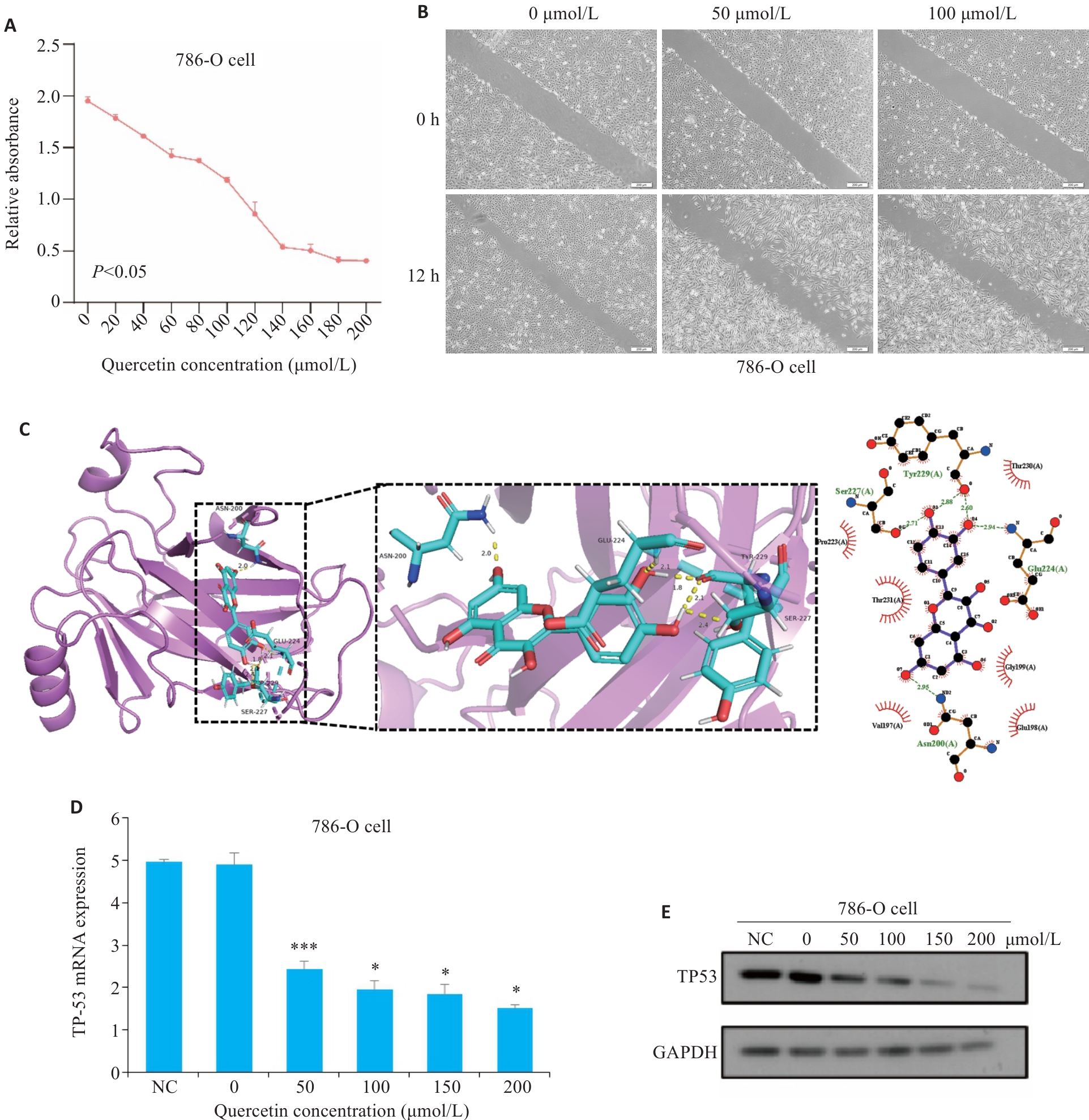
Fig.6 Cell experiments and molecular docking validation. A: Viability of 786-O cells measured using CCK-8 assay after 24 h of quercetin treatment. B: Wound healing assay for assessing migration capacity of 786-O cells after quercetin treatment for 12 h (scale bar=200 μm). C: Docking of quercetin with TP53 molecule. D: Effect of different concentrations of quercetin on TP53 mRNA expression in 786-O cells detected using RT-qPCR (*P<0.05,***P<0.001 vs 0 μmol/L). E: Effect of different concentrations of quercetin on TP53 protein expression in 786-O cells detected using Western blotting.
| 1 | Marston Linehan W, Schmidt LS, Crooks DR, et al. The metabolic basis of kidney cancer[J]. Cancer Discov, 2019, 9(8): 1006-21. |
| 2 | Singh D, Vignat J, Lorenzoni V, et al. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative[J]. Lancet Glob Health, 2023, 11(2): e197-206. |
| 3 | Capitanio U, Montorsi F. Renal cancer[J]. Lancet, 2016, 387(10021): 894-906. |
| 4 | Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European association of urology guidelines on renal cell carcinoma: the 2022 update[J]. Eur Urol, 2022, 82(4): 399-410. |
| 5 | Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma[J]. N Engl J Med, 2019, 380(12): 1103-15. |
| 6 | Panossian A, Wikman G. Pharmacology of Schisandra chinensis bail.: an overview of Russian research and uses in medicine[J]. J Ethnopharmacol, 2008, 118(2): 183-212. |
| 7 | Li Y, Yao JY, Han CY, et al. Quercetin, inflammation and immunity[J]. Nutrients, 2016, 8(3): 167. |
| 8 | Lim H, Min DS, Yun HE, et al. Impressic acid from Acanthopanax koreanum, possesses matrix metalloproteinase-13 down-regulating capacity and protects cartilage destruction[J]. J Ethnopharmacol, 2017, 209: 73-81. |
| 9 | Wong CK. Simplifying electrocardiographic assessment in STEMI reperfusion management: Pros and cons[J]. Int J Cardiol, 2017, 227: 30-6. |
| 10 | Vandemoortele A, Babat P, Yakubu M, et al. Reactivity of free malondialdehyde during in vitro simulated gastrointestinal digestion[J]. J Agric Food Chem, 2017, 65(10): 2198-204. |
| 11 | Ru JL, Li P, Wang JN, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines[J]. J Cheminform, 2014, 6: 13. |
| 12 | Wang X, Shen YH, Wang SW, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database[J]. Nucleic Acids Res, 2017, 45(W1): W356-60. |
| 13 | Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules[J]. Nucleic Acids Res, 2019, 47(W1): W357-64. |
| 14 | Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018[J]. Nucleic Acids Res, 2018, 46(D1): D1074-82. |
| 15 | Gilson MK, Liu TQ, Baitaluk M, et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology[J]. Nucleic Acids Res, 2016, 44(D1): D1045-53. |
| 16 | Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility[J]. J Comput Chem, 2009, 30(16): 2785-91. |
| 17 | Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina[J]. J Comput Aided Mol Des, 2010, 24(5): 417-22. |
| 18 | Larson AJ, David Symons J, Jalili T. Therapeutic potential of quercetin to decrease blood pressure: review of efficacy and mechanisms[J]. Adv Nutr, 2012, 3(1): 39-46. |
| 19 | Russo M, Spagnuolo C, Tedesco I, et al. Phytochemicals in cancer prevention and therapy: truth or dare[J]? Toxins, 2010, 2(4): 517-51. |
| 20 | Tang SM, Deng XT, Zhou J, et al. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects[J]. Biomed Pharmacother, 2020, 121: 109604. |
| 21 | Levine AJ. p53: 800 million years of evolution and 40 years of discovery[J]. Nat Rev Cancer, 2020, 20(8): 471-80. |
| 22 | Network CGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma[J]. Nature, 2013, 499(7456): 43-9. |
| 23 | Wang B, Song Q, Wei YA, et al. Comprehensive investigation into cuproptosis in the characterization of clinical features, molecular characteristics, and immune situations of clear cell renal cell carcinoma[J]. Front Immunol, 2022, 13: 948042. |
| 24 | Paul JY, Harding R, Tushemereirwe W, et al. Banana21: from gene discovery to deregulated golden bananas[J]. Front Plant Sci, 2018, 9: 558. |
| 25 | D’Andrea G. Quercetin: a flavonol with multifaceted therapeutic applications[J]? Fitoterapia, 2015, 106: 256-71. |
| 26 | Li FZ, Aljahdali IAM, Zhang RY, et al. Kidney cancer biomarkers and targets for therapeutics: survivin (BIRC5), XIAP, MCL-1, HIF1α, HIF2α, NRF2, MDM2, MDM4, p53, KRAS and AKT in renal cell carcinoma[J]. J Exp Clin Cancer Res, 2021, 40(1): 254. |
| 27 | Uhlman DL, Nguyen PL, Manivel JC, et al. Association of immunohistochemical staining for p53 with metastatic progression and poor survival in patients with renal cell carcinoma[J]. J Natl Cancer Inst, 1994, 86(19): 1470-5. |
| 28 | Shvarts O, Seligson D, Lam J, et al. p53 is an independent predictor of tumor recurrence and progression after nephrectomy in patients with localized renal cell carcinoma[J]. J Urol, 2005, 173(3): 725-8. |
| 29 | Noon AP, Vlatković N, Polański R, et al. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets[J]? Cancer, 2010, 116(4): 780-90. |
| [1] | Xinyuan CHEN, Chengting WU, Ruidi LI, Xueqin PAN, Yaodan ZHANG, Junyu TAO, Caizhi LIN. Shuangshu Decoction inhibits growth of gastric cancer cell xenografts by promoting cell ferroptosis via the P53/SLC7A11/GPX4 axis [J]. Journal of Southern Medical University, 2025, 45(7): 1363-1371. |
| [2] | Liming WANG, Hongrui CHEN, Yan DU, Peng ZHAO, Yujie WANG, Yange TIAN, Xinguang LIU, Jiansheng LI. Yiqi Zishen Formula ameliorates inflammation in mice with chronic obstructive pulmonary disease by inhibiting the PI3K/Akt/NF-κB signaling pathway [J]. Journal of Southern Medical University, 2025, 45(7): 1409-1422. |
| [3] | Yinfu ZHU, Yiran LI, Yi WANG, Yinger HUANG, Kunxiang GONG, Wenbo HAO, Lingling SUN. Therapeutic mechanism of hederagenin, an active component in Guizhi Fuling Pellets, against cervical cancer in nude mice [J]. Journal of Southern Medical University, 2025, 45(7): 1423-1433. |
| [4] | Lijun HE, Xiaofei CHEN, Chenxin YAN, Lin SHI. Inhibitory effect of Fuzheng Huaji Decoction against non-small cell lung cancer cells in vitro and the possible molecular mechanism [J]. Journal of Southern Medical University, 2025, 45(6): 1143-1152. |
| [5] | Guoyong LI, Renling LI, Yiting LIU, Hongxia KE, Jing LI, Xinhua WANG. Therapeutic mechanism of Arctium lappa extract for post-viral pneumonia pulmonary fibrosis: a metabolomics, network pharmacology analysis and experimental verification [J]. Journal of Southern Medical University, 2025, 45(6): 1185-1199. |
| [6] | Liping GUAN, Yan YAN, Xinyi LU, Zhifeng LI, Hui GAO, Dong CAO, Chenxi HOU, Jingyu ZENG, Xinyi LI, Yang ZHAO, Junjie WANG, Huilong FANG. Compound Centella asiatica formula alleviates Schistosoma japonicum-induced liver fibrosis in mice by inhibiting the inflammation-fibrosis cascade via regulating the TLR4/MyD88 pathway [J]. Journal of Southern Medical University, 2025, 45(6): 1307-1316. |
| [7] | Peipei TANG, Yong TAN, Yanyun YIN, Xiaowei NIE, Jingyu HUANG, Wenting ZUO, Yuling LI. Tiaozhou Ziyin recipe for treatment of premature ovarian insufficiency: efficacy, safety and mechanism [J]. Journal of Southern Medical University, 2025, 45(5): 929-941. |
| [8] | Xiaotao LIANG, Yifan XIONG, Xueqi LIU, Xiaoshan LIANG, Xiaoyu ZHU, Wei XIE. Huoxue Shufeng Granule alleviates central sensitization in chronic migraine mice via TLR4/NF-κB inflammatory pathway [J]. Journal of Southern Medical University, 2025, 45(5): 986-994. |
| [9] | Niandong RAN, Jie LIU, Jian XU, Yongping ZHANG, Jiangtao GUO. n-butanol fraction of ethanol extract of Periploca forrestii Schltr.: its active components, targets and pathways for treating Alcheimer's disease in rats [J]. Journal of Southern Medical University, 2025, 45(4): 785-798. |
| [10] | Wenjie LI, Yaonan HONG, Rui HUANG, Yuchen LI, Ying ZHANG, Yun ZHANG, Dijiong WU. Causal relationship between autoimmune diseases and aplastic anemia: A Mendelian randomization study [J]. Journal of Southern Medical University, 2025, 45(4): 871-879. |
| [11] | Hongyan SUN, Guoqing LU, Chengwen FU, Mengwen XU, Xiaoyi ZHU, Guoquan XING, Leqiang LIU, Yufei KE, Lemei CUI, Ruiyang CHEN, Lei WANG, Pinfang KANG, Bi TANG. Quercetin ameliorates myocardial injury in diabetic rats by regulating L-type calcium channels [J]. Journal of Southern Medical University, 2025, 45(3): 531-541. |
| [12] | Haonan¹ XU, Fang³ ZHANG, Yuying² HUANG, Qisheng⁴ YAO, Yueqin⁴ GUAN, Hao CHEN. Thesium chinense Turcz. alleviates antibiotic-associated diarrhea in mice by modulating gut microbiota structure and regulating the EGFR/PI3K/Akt signaling pathway [J]. Journal of Southern Medical University, 2025, 45(2): 285-295. |
| [13] | Pengwei HUANG, Jie CHEN, Jinhu ZOU, Xuefeng GAO, Hong CAO. Quercetin mitigates HIV-1 gp120-induced rat astrocyte neurotoxicity via promoting G3BP1 disassembly in stress granules [J]. Journal of Southern Medical University, 2025, 45(2): 304-312. |
| [14] | Ying LIU, Borui LI, Yongcai LI, Lubo CHANG, Jiao WANG, Lin YANG, Yonggang YAN, Kai QV, Jiping LIU, Gang ZHANG, Xia SHEN. Jiawei Xiaoyao Pills improves depression-like behavior in rats by regulating neurotransmitters, inhibiting inflammation and oxidation and modulating intestinal flora [J]. Journal of Southern Medical University, 2025, 45(2): 347-358. |
| [15] | Xiaorui CHEN, Qingzheng WEI, Zongliang ZHANG, Jiangshui YUAN, Weiqing SONG. Overexpression of CHMP2B suppresses proliferation of renal clear cell carcinoma cells [J]. Journal of Southern Medical University, 2025, 45(1): 126-136. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||