Journal of Southern Medical University ›› 2026, Vol. 46 ›› Issue (1): 208-218.doi: 10.12122/j.issn.1673-4254.2026.01.23
Xueqi LU1( ), Huayuan CHEN2(
), Huayuan CHEN2( ), Qiucen WU1, Yaoqi WEN1, Guoguang LIU3(
), Qiucen WU1, Yaoqi WEN1, Guoguang LIU3( ), Chaomin CHEN1(
), Chaomin CHEN1( )
)
Received:2025-06-24
Online:2026-01-20
Published:2026-01-16
Contact:
Guoguang LIU, Chaomin CHEN
E-mail:415081161@qq.com;67936668@qq.com;571611621@qq.com
Xueqi LU, Huayuan CHEN, Qiucen WU, Yaoqi WEN, Guoguang LIU, Chaomin CHEN. Evaluation of an interpretable 12-lead ECG automatic diagnosis model based on deep feature fusion[J]. Journal of Southern Medical University, 2026, 46(1): 208-218.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2026.01.23
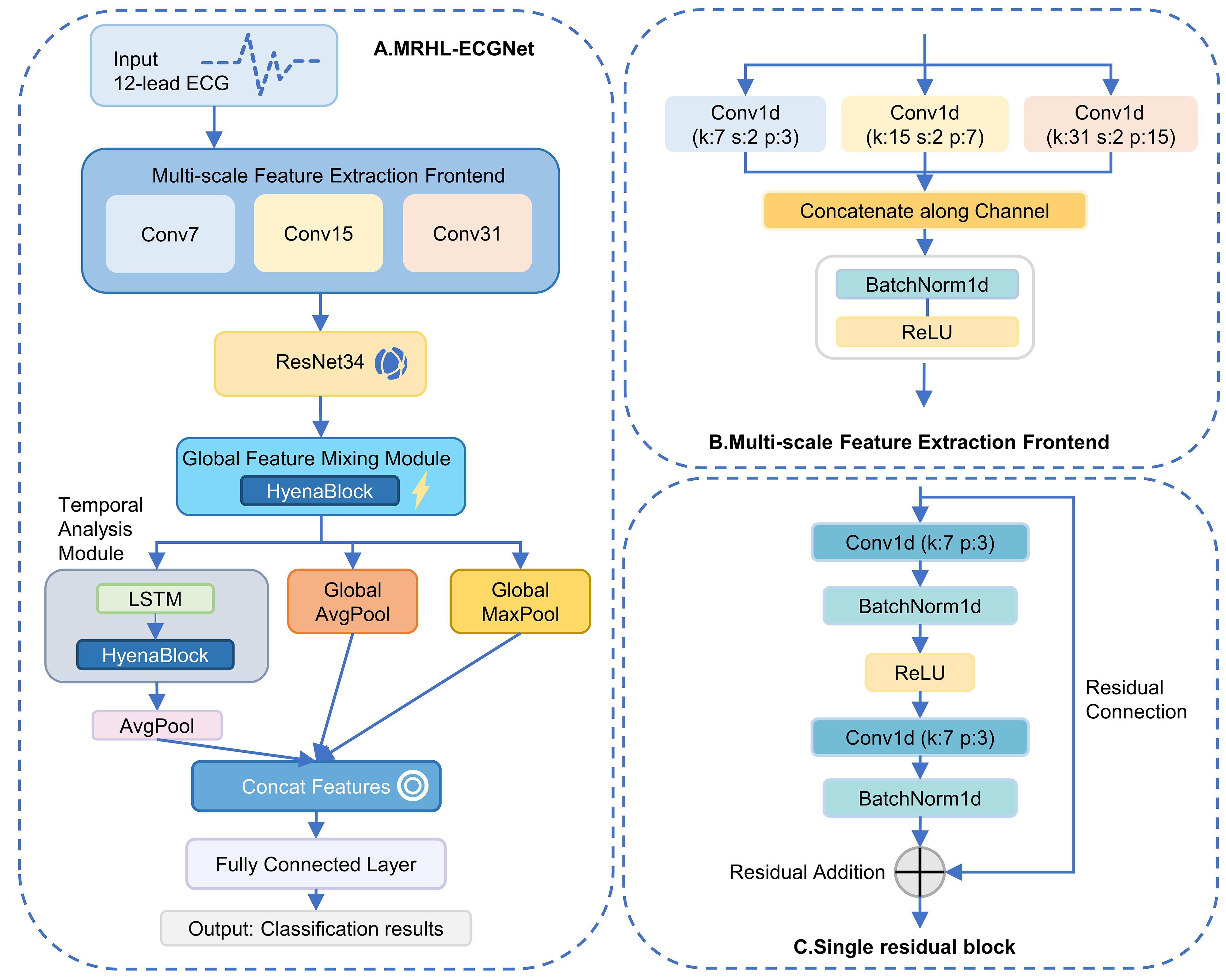
Fig.2 Schematic diagram of the model structure 1. A: Overall structure diagram of MRHL-ECGNet. B: Structure diagram of the multi-scale feature extraction front-end. C: Structure diagram of the single residual block.
| Module | In/Out & K and Other Parameters |
|---|---|
| Input Layer | In: 12×T |
| Multi-scale feature extraction frontend | In: 12×T; Out: 64×T/2; K: 7, 15, 31; Stride: 2 |
| ResNet-34 Layer1 | In: 64; Out: 64; K: 7; BasicBlock1d: 3; Downsample: No |
| ResNet-34 Layer2 | In: 64; Out: 128; K: 7; BasicBlock1d: 4; Downsample: Yes |
| ResNet-34 Layer3 | In: 128; Out: 256; K: 7; BasicBlock1d: 6; Downsample: Yes |
| ResNet-34 Layer4 | In: 256; Out: 512; K: 7; BasicBlock1d: 3; Downsample: Yes |
| Global feature mixing module | In: 512; Out: 512; K: 31 |
| Temporal analysis module (LSTM) | In: 512; Hidden state: 64 |
| Temporal analysis module (Hyena) | In: 64; Out: 64; K: 15 |
| Feature fusion and classifier | Concatenated in: 512×2+64; Fully connected layer Output: Classification results |
Tab.1 Parameter settings of the MRHL-ECGNet model
| Module | In/Out & K and Other Parameters |
|---|---|
| Input Layer | In: 12×T |
| Multi-scale feature extraction frontend | In: 12×T; Out: 64×T/2; K: 7, 15, 31; Stride: 2 |
| ResNet-34 Layer1 | In: 64; Out: 64; K: 7; BasicBlock1d: 3; Downsample: No |
| ResNet-34 Layer2 | In: 64; Out: 128; K: 7; BasicBlock1d: 4; Downsample: Yes |
| ResNet-34 Layer3 | In: 128; Out: 256; K: 7; BasicBlock1d: 6; Downsample: Yes |
| ResNet-34 Layer4 | In: 256; Out: 512; K: 7; BasicBlock1d: 3; Downsample: Yes |
| Global feature mixing module | In: 512; Out: 512; K: 31 |
| Temporal analysis module (LSTM) | In: 512; Hidden state: 64 |
| Temporal analysis module (Hyena) | In: 64; Out: 64; K: 15 |
| Feature fusion and classifier | Concatenated in: 512×2+64; Fully connected layer Output: Classification results |
| Item | Configuration |
|---|---|
| Operation system | Windows 11 |
| CPU | Intel Core i5-13490F |
| GPU | NVIDIA GeForce RTX 4060Ti |
| RAM | 16GB |
| Cuda version | 12.7 |
| Pytorch version | 2.3.1 |
| Anaconda version | 24.9.2 |
Tab.2 Experimental environment for model testing
| Item | Configuration |
|---|---|
| Operation system | Windows 11 |
| CPU | Intel Core i5-13490F |
| GPU | NVIDIA GeForce RTX 4060Ti |
| RAM | 16GB |
| Cuda version | 12.7 |
| Pytorch version | 2.3.1 |
| Anaconda version | 24.9.2 |
| Method | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|
| Zhang D et al.[ | 0.963 | 0.961 | 0.823 | 0.826 | 0.821 |
| Reddy L et al.[ | 0.895 | 0.913 | 0.810 | 0.812 | 0.809 |
| Hwang S et al.[ | 0.905 | 0.928 | 0.797 | 0.808 | 0.787 |
| Strodthoff N et al.[ | 0.901 | 0.941 | 0.813 | 0.836 | 0.792 |
| MRHL-ECGNet | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
Tab.3 Comparison results of the performance of the 12-lead ECG automatic diagnosis model
| Method | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|
| Zhang D et al.[ | 0.963 | 0.961 | 0.823 | 0.826 | 0.821 |
| Reddy L et al.[ | 0.895 | 0.913 | 0.810 | 0.812 | 0.809 |
| Hwang S et al.[ | 0.905 | 0.928 | 0.797 | 0.808 | 0.787 |
| Strodthoff N et al.[ | 0.901 | 0.941 | 0.813 | 0.836 | 0.792 |
| MRHL-ECGNet | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
| Diagnosis category | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|
| Normal | 0.963 | 0.980 | 0.819 | 0.837 | 0.802 |
| AF | 0.977 | 0.995 | 0.927 | 0.944 | 0.911 |
| I-AVB | 0.975 | 0.975 | 0.896 | 0.912 | 0.882 |
| LBBB | 0.994 | 0.998 | 0.920 | 0.986 | 0.862 |
| RBBB | 0.961 | 0.989 | 0.925 | 0.954 | 0.898 |
| PAC | 0.966 | 0.952 | 0.739 | 0.683 | 0.804 |
| PVC | 0.965 | 0.982 | 0.836 | 0.803 | 0.872 |
| STD | 0.951 | 0.976 | 0.793 | 0.844 | 0.747 |
| STE | 0.996 | 0.998 | 0.914 | 0.889 | 0.941 |
| AVG | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
Tab.4 Performance of MRHL-ECGNet model on 9 different diagnostic categories of ECG
| Diagnosis category | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|
| Normal | 0.963 | 0.980 | 0.819 | 0.837 | 0.802 |
| AF | 0.977 | 0.995 | 0.927 | 0.944 | 0.911 |
| I-AVB | 0.975 | 0.975 | 0.896 | 0.912 | 0.882 |
| LBBB | 0.994 | 0.998 | 0.920 | 0.986 | 0.862 |
| RBBB | 0.961 | 0.989 | 0.925 | 0.954 | 0.898 |
| PAC | 0.966 | 0.952 | 0.739 | 0.683 | 0.804 |
| PVC | 0.965 | 0.982 | 0.836 | 0.803 | 0.872 |
| STD | 0.951 | 0.976 | 0.793 | 0.844 | 0.747 |
| STE | 0.996 | 0.998 | 0.914 | 0.889 | 0.941 |
| AVG | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
| Experiment ID | Configuration | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|---|
| 1 | Remove multi-scale feature extraction frontend | 0.951 | 0.968 | 0.846 | 0.844 | 0.851 |
| 2 | Remove resnet-34 | 0.898 | 0.901 | 0.803 | 0.796 | 0.812 |
| 3 | Remove temporal analysis module | 0.912 | 0.919 | 0.820 | 0.821 | 0.819 |
| 4 | Remove global avgpool | 0.956 | 0.971 | 0.837 | 0.835 | 0.840 |
| 5 | Remove global maxpool | 0.967 | 0.973 | 0.833 | 0.829 | 0.838 |
| 6 | Remove all hyenablock | 0.905 | 0.909 | 0.799 | 0.798 | 0.802 |
| 7 | Replace hyenablock with transformerblock | 0.970 | 0.985 | 0.847 | 0.836 | 0.859 |
| 8 | Full MRHL-ECGNet | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
Tab.5 Results of the ablation experiment and the mechanism replacement experiment
| Experiment ID | Configuration | Accuracy | AUC | F1s | Precision | Recall |
|---|---|---|---|---|---|---|
| 1 | Remove multi-scale feature extraction frontend | 0.951 | 0.968 | 0.846 | 0.844 | 0.851 |
| 2 | Remove resnet-34 | 0.898 | 0.901 | 0.803 | 0.796 | 0.812 |
| 3 | Remove temporal analysis module | 0.912 | 0.919 | 0.820 | 0.821 | 0.819 |
| 4 | Remove global avgpool | 0.956 | 0.971 | 0.837 | 0.835 | 0.840 |
| 5 | Remove global maxpool | 0.967 | 0.973 | 0.833 | 0.829 | 0.838 |
| 6 | Remove all hyenablock | 0.905 | 0.909 | 0.799 | 0.798 | 0.802 |
| 7 | Replace hyenablock with transformerblock | 0.970 | 0.985 | 0.847 | 0.836 | 0.859 |
| 8 | Full MRHL-ECGNet | 0.972 | 0.983 | 0.864 | 0.873 | 0.857 |
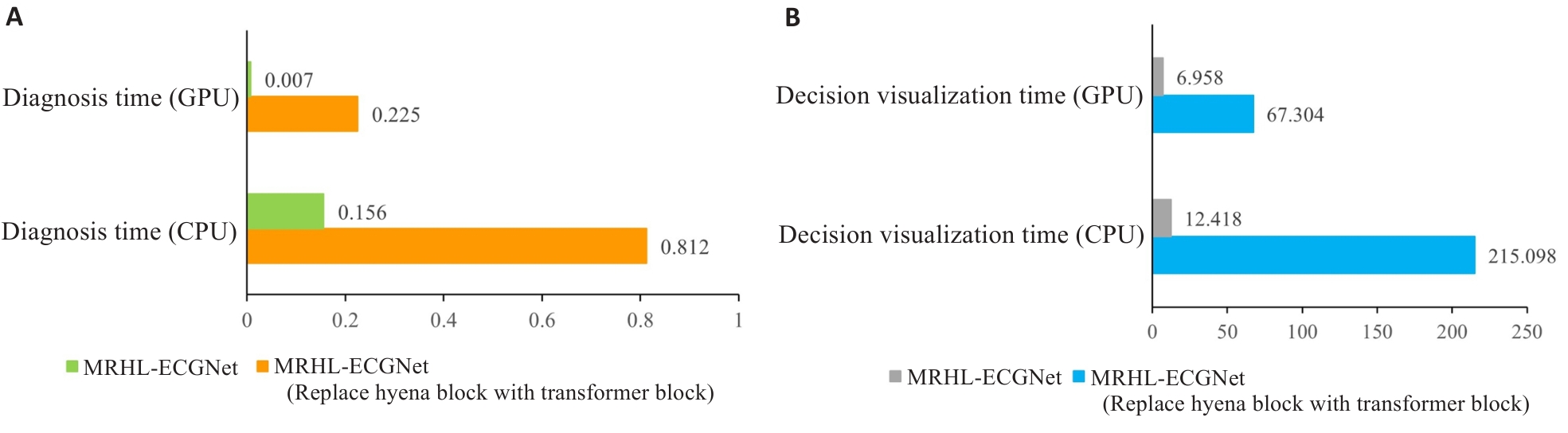
Fig.5 Model running time. A: Time required for diagnostic results output under different conditions. B: Time for generating the decision-making basis diagram under different conditions.
| Module | Parameters | Memory (MB) |
|---|---|---|
| Multi-scale feature extraction frontend | 13920 | 0.053 |
| ResNet-34 | 16589312 | 63.283 |
| Global feature mixing module | 295424 | 1.127 |
| Temporal analysis module | 154176 | 0.588 |
| Hyenablock | 8256 | 0.031 |
| Mrhl-ecgnet | 17615017 | 67.196 |
| Mrhl-ecgnet (replace hyenablock with transformerblock) | 20746921 | 80.226 |
Tab.6 Model parameter quantity and memory usage
| Module | Parameters | Memory (MB) |
|---|---|---|
| Multi-scale feature extraction frontend | 13920 | 0.053 |
| ResNet-34 | 16589312 | 63.283 |
| Global feature mixing module | 295424 | 1.127 |
| Temporal analysis module | 154176 | 0.588 |
| Hyenablock | 8256 | 0.031 |
| Mrhl-ecgnet | 17615017 | 67.196 |
| Mrhl-ecgnet (replace hyenablock with transformerblock) | 20746921 | 80.226 |
| [1] | Berkaya SK, Uysal AK, Gunal ES, et al. A survey on ECG analysis[J]. Biomed Sig Proc Control, 2018, 43: 216-35. doi:10.1016/j.bspc.2018.03.003 |
| [2] | Chou's Electrocardiography in Clinical Practice[M]. Elsevier Science Health Science div,2008 . doi:10.1016/b978-141603774-3.10027-9 |
| [3] | Hong SD, Zhou YX, Shang JY, et al. Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review[J]. Comput Biol Med, 2020, 122: 103801. doi:10.1016/j.compbiomed.2020.103801 |
| [4] | Priori SG, Blomström-Lundqvist C, Mazzanti A, et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC)[J]. Eur Heart J, 2015, 36(41): 2793-867. doi:10.1093/eurheartj/ehv316 |
| [5] | Hannun AY, Rajpurkar P, Haghpanahi M, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electro-cardiograms using a deep neural network[J]. Nat Med, 2019, 25(1): 65-9. doi:10.1038/s41591-018-0268-3 |
| [6] | Yang XZ, Zhang XY, Yang MY, et al. 12-Lead ECG arrhythmia classification using cascaded convolutional neural network and expert feature[J]. J Electrocardiol, 2021, 67: 56-62. doi:10.1016/j.jelectrocard.2021.04.016 |
| [7] | Vaswani A, Shazeer NM, Parmar N, et al. Attention is all you need[C]//Neural Information Processing Systems., 2017 |
| [8] | Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: Transformers for image recognition at scale[J]. arxiv preprint arxiv: 2010. 11929, 2020. |
| [9] | Zhou HY, Zhang SH, Peng JQ, et al. Informer: beyond efficient transformer for long sequence time-series forecasting[J]. Proc AAAI Conf Artif Intell, 2021, 35(12): 11106-15. doi:10.1609/aaai.v35i12.17325 |
| [10] | Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining[J]. Bioinformatics, 2020, 36(4): 1234-40. doi:10.1093/bioinformatics/btz682 |
| [11] | Zhang SY, Lian C, Xu BR, et al. A token selection-based multi-scale dual-branch CNN-transformer network for 12-lead ECG signal classification[J]. Knowl Based Syst, 2023, 280: 111006. doi:10.1016/j.knosys.2023.111006 |
| [12] | Zheng JW, Zhang JM, Danioko S, et al. A 12-lead electrocardiogram database for arrhythmia research covering more than 10, 000 patients[J]. Sci Data, 2020, 7(1): 48. doi:10.1038/s41597-020-0386-x |
| [13] | Petch J, Di S, Nelson W. Opening the black box: the promise and limitations of explainable machine learning in cardiology[J]. Can J Cardiol, 2022, 38(2): 204-13. doi:10.1016/j.cjca.2021.09.004 |
| [14] | Zhang DD, Yang S, Yuan XH, et al. Interpretable deep learning for automatic diagnosis of 12-lead electrocardiogram[J]. iScience, 2021, 24(4): 102373. doi:10.1016/j.isci.2021.102373 |
| [15] | Lundberg SM, Lee SI. A unified approach to interpreting model predictions[C]//Neural Information Processing Systems., 2017 |
| [16] | Reddy L, Talwar V, Alle S, et al. IMLE-net: an interpretable multi-level multi-channel model for ECG classification[C]//2021 IEEE International Conference on Systems, Man, and Cybernetics (SMC). October 17-20, 2021. Melbourne, Australia. IEEE, 2021: 1068-1074. doi:10.1109/smc52423.2021.9658706 |
| [17] | Liu FF, Liu CY, Zhao LN, et al. An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection[J]. J Med Imaging Hlth Inform, 2018, 8(7): 1368-73. doi:10.1166/jmihi.2018.2442 |
| [18] | Zhang HZ, Zhao W, Liu S. SE-ECGNet: a multi-scale deep residual network with squeeze-and-excitation module for ECG signal classification[C]//2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). December 16-19, 2020. Seoul, Korea. IEEE, 2020: 2685-2691. doi:10.1109/bibm49941.2020.9313548 |
| [19] | He KM, Zhang XY, Ren SQ, et al. Deep residual learning for image recognition[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). June 27-30, 2016, Las Vegas, NV, USA. IEEE, 2016: 770-8. doi:10.1109/cvpr.2016.90 |
| [20] | Poli M, Massaroli S, Nguyen E, et al. Hyena hierarchy: Towards larger convolutional language models[C]//International Conference on Machine Learning. PMLR, 2023: 28043-78. |
| [21] | Yang W, Deo R, Guo WS. Functional feature extraction and validation from twelve-lead electrocardiograms to identify atrial fibrillation[J]. Commun Med (Lond), 2025, 5(1): 32. doi:10.1038/s43856-025-00749-2 |
| [22] | Hochreiter S, Schmidhuber J. Long short-term memory[J]. Neural Comput, 1997, 9(8): 1735-80. doi:10.1162/neco.1997.9.8.1735 |
| [23] | Kingma D P, Ba J. Adam: A method for stochastic optimization[J]. arxiv preprint arxiv:, 2014. |
| [24] | Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI[J]. Inf Fusion, 2020, 58: 82-115. doi:10.1016/j.inffus.2019.12.012 |
| [25] | Sundararajan M, Taly A, Yan Q. Gradients of counterfactuals[J]. arxiv preprint arxiv:, 2016. |
| [26] | Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks[J]. Inf Process Manag, 2009, 45(4): 427-37. doi:10.1016/j.ipm.2009.03.002 |
| [27] | Hwang S, Cha J, Heo J, et al. Multi-label abnormality classification from 12-lead ECG using a 2D residual U-net[C]//ICASSP 2024 - 2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). April 14-19, 2024, Seoul, Korea, Republic of. IEEE, 2024: 2265-9. doi:10.1109/icassp48485.2024.10448259 |
| [28] | Strodthoff N, Wagner P, Schaeffter T, et al. Deep learning for ECG analysis: benchmarks and insights from PTB-XL[J]. IEEE J Biomed Health Inform, 2021, 25(5): 1519-28. doi:10.1109/jbhi.2020.3022989 |
| [29] | Conen D, Adam M, Roche F, et al. Premature atrial contractions in the general population: frequency and risk factors[J]. Circulation, 2012, 126(19): 2302-8. doi:10.1161/circulationaha.112.112300 |
| [30] | Joglar JA, Chung MK, Armbruster AL, et al. 2023 ACC/AHA/ACCP/HRS guideline for the diagnosis and management of atrial fibrillation: a report of the American college of cardiology/American heart association joint committee on clinical practice guidelines[J]. Circulation, 2024, 149(1): e1-156. doi:10.1161/cir.0000000000001207 |
| [31] | Ikeda T. Right bundle branch block: current considerations[J]. Curr Cardiol Rev, 2021, 17(1): 24-30. doi:10.2174/1573403x16666200708111553 |
| [1] | Qiucen WU, Xueqi LU, Yaoqi WEN, Yong HONG, Yuliang WU, Chaomin CHEN. A myocardial infarction detection and localization model based on multi-scale field residual blocks fusion with modified channel attention [J]. Journal of Southern Medical University, 2025, 45(8): 1777-1790. |
| [2] | Ziyu ZHENG, Xiaying YANG, Shengjie WU, Shijie ZHANG, Guorong LYU, Peizhong LIU, Jun WANG, Shaozheng HE. A multi-feature fusion-based model for fetal orientation classification from intrapartum ultrasound videos [J]. Journal of Southern Medical University, 2025, 45(7): 1563-1570. |
| [3] | Meng QU, Rong FU. ResLSTM-TemporalSE: an automated classification model for multi-lead ECG signals [J]. Journal of Southern Medical University, 2025, 45(12): 2708-2717. |
| [4] | Huirong XIE, Chaobin HU, Guohua LIANG, Hongzhe HAN, Mu HUANG, Qianjin FENG. Design and validation of a multimodal model integrating text and imaging data for intelligent assessment of psychological stress in college students [J]. Journal of Southern Medical University, 2025, 45(11): 2504-2510. |
| [5] | Yadi HE, Xuanru ZHOU, Jinhui JIN, Ting SONG. PE-CycleGAN network based CBCT-sCT generation for nasopharyngeal carsinoma adaptive radiotherapy [J]. Journal of Southern Medical University, 2025, 45(1): 179-186. |
| [6] | Weiyang FANG, Hui XIAO, Shuang WANG, Xiaoming LIN, Chaomin CHEN. A deep learning model based on magnetic resonance imaging and clinical feature fusion for predicting preoperative cytokeratin 19 status in hepatocellular carcinoma [J]. Journal of Southern Medical University, 2024, 44(9): 1738-1751. |
| [7] | Jiazhi OU, Chang'an ZHAN, Feng YANG. An autoencoder model based on one-dimensional neural network for epileptic EEG anomaly detection [J]. Journal of Southern Medical University, 2024, 44(9): 1796-1804. |
| [8] | Chen WANG, Mingqiang MENG, Mingqiang LI, Yongbo WANG, Dong ZENG, Zhaoying BIAN, Jianhua MA. Reconstruction from CT truncated data based on dual-domain transformer coupled feature learning [J]. Journal of Southern Medical University, 2024, 44(5): 950-959. |
| [9] | LONG Kaixing, WENG Danyi, GENG Jian, LU Yanmeng, ZHOU Zhitao, CAO Lei. Automatic classification of immune-mediated glomerular diseases based on multi-modal multi-instance learning [J]. Journal of Southern Medical University, 2024, 44(3): 585-593. |
| [10] | XIAO Hui, FANG Weiyang, LIN Mingjun, ZHOU Zhenzhong, FEI Hongwen, CHEN Chaomin. A multiscale carotid plaque detection method based on two-stage analysis [J]. Journal of Southern Medical University, 2024, 44(2): 387-396. |
| [11] | Caolin LIU, Qingqing ZOU, Menghong WANG, Qinmei YANG, Liwen SONG, Zixiao LU, Qianjin FENG, Yinghua ZHAO. Identification of osteoid and chondroid matrix mineralization in primary bone tumors using a deep learning fusion model based on CT and clinical features: a multi-center retrospective study [J]. Journal of Southern Medical University, 2024, 44(12): 2412-2420. |
| [12] | MI Jia, ZHOU Yujia, FENG Qianjin. A 3D/2D registration method based on reconstruction of orthogonal-view Xray images [J]. Journal of Southern Medical University, 2023, 43(9): 1636-1643. |
| [13] | CHU Zhiqin, QU Yaoming, ZHONG Tao, LIANG Shujun, WEN Zhibo, ZHANG Yu. A Dual-Aware deep learning framework for identification of glioma isocitrate dehydrogenase genotype using magnetic resonance amide proton transfer modalities [J]. Journal of Southern Medical University, 2023, 43(8): 1379-1387. |
| [14] | YU Jiahong, ZHANG Kunpeng, JIN Shuang, SU Zhe, XU Xiaotong, ZHANG Hua. Sinogram interpolation combined with unsupervised image-to-image translation network for CT metal artifact correction [J]. Journal of Southern Medical University, 2023, 43(7): 1214-1223. |
| [15] | ZHOU Hao, ZENG Dong, BIAN Zhaoying, MA Jianhua. A semi-supervised network-based tissue-aware contrast enhancement method for CT images [J]. Journal of Southern Medical University, 2023, 43(6): 985-993. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||