Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (11): 2518-2526.doi: 10.12122/j.issn.1673-4254.2025.11.25
Xinyang LI1( ), Guixiao XU4,5,6,7, Jiehong LIU1, Yanqiu FENG1,2,3(
), Guixiao XU4,5,6,7, Jiehong LIU1, Yanqiu FENG1,2,3( )
)
Received:2025-09-01
Online:2025-11-20
Published:2025-11-28
Contact:
Yanqiu FENG
E-mail:lixinyang91@163.com;foree@smu.edu.cn
Supported by:Xinyang LI, Guixiao XU, Jiehong LIU, Yanqiu FENG. Effect of AI-assisted compressed sensing acceleration on MRI radiomic feature extraction and staging model performance for nasopharyngeal carcinoma[J]. Journal of Southern Medical University, 2025, 45(11): 2518-2526.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.11.25
| Characteristics | Dataset values |
|---|---|
| Number of patients (%) | 64 |
| Age (year, Mean±SD) | 44.2±11.3 (18-68) |
| Gender | |
| Male | 42(65.6%) |
| Female | 22(34.4%) |
| Histological type | |
| NKUC | 64 (100%) |
| T stage | |
| T1 | 7 (10.9%) |
| T2 | 6 (9.4%) |
| T3 | 40 (62.5%) |
| T4 | 11 (17.2%) |
| N stage | |
| N0 | 1 (1.6%) |
| N1 | 32 (50%) |
| N2 | 15 (23.4%) |
| N3 | 16 (25%) |
| M stage | |
| M0 | 59 (92.2%) |
| M1 | 5 (7.8%) |
Tab.1 Characteristics of the patients with NPC
| Characteristics | Dataset values |
|---|---|
| Number of patients (%) | 64 |
| Age (year, Mean±SD) | 44.2±11.3 (18-68) |
| Gender | |
| Male | 42(65.6%) |
| Female | 22(34.4%) |
| Histological type | |
| NKUC | 64 (100%) |
| T stage | |
| T1 | 7 (10.9%) |
| T2 | 6 (9.4%) |
| T3 | 40 (62.5%) |
| T4 | 11 (17.2%) |
| N stage | |
| N0 | 1 (1.6%) |
| N1 | 32 (50%) |
| N2 | 15 (23.4%) |
| N3 | 16 (25%) |
| M stage | |
| M0 | 59 (92.2%) |
| M1 | 5 (7.8%) |
| Sequences parameter | AX T2WI FSE ACS | AX T2WI FSE PI | AX T1WI FSE ACS | AX T1WI FSE PI | Post-contrast AX T1WI FSE ACS | Post-contrast AX T1WI FSE PI |
|---|---|---|---|---|---|---|
| Fov (mm) | 240×240 | 240×240 | 240×240 | 240×240 | 240×240 | 240×240 |
| TR/TE (ms) | 4800/120 | 4800/120 | 662/8.16 | 662/8.16 | 789/8.12 | 789/8.12 |
| Matrix | 384×269 | 384×269 | 384×307 | 384×307 | 384×307 | 384×307 |
| ETL | 28 | 28 | 2 | 2 | 2 | 2 |
| Bandwidth (Hz) | 260 | 260 | 280 | 280 | 250 | 250 |
| Average | 1 | 1 | 1 | 1 | 1 | 1 |
| Number slices | 40 | 40 | 40 | 40 | 40 | 40 |
| Spatial resolution | 0.89×0.63×5 | 0.89×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 |
| Acquisition | ACS | PI | ACS | PI | ACS | PI |
| Accelerating factors | 2.25 | 2 | 2.25 | 2 | 2.25 | 2 |
| Sequence acquisition time (s) | 59 | 82 | 93 | 140 | 75 | 90 |
Tab.2 MRI sequences and parameters
| Sequences parameter | AX T2WI FSE ACS | AX T2WI FSE PI | AX T1WI FSE ACS | AX T1WI FSE PI | Post-contrast AX T1WI FSE ACS | Post-contrast AX T1WI FSE PI |
|---|---|---|---|---|---|---|
| Fov (mm) | 240×240 | 240×240 | 240×240 | 240×240 | 240×240 | 240×240 |
| TR/TE (ms) | 4800/120 | 4800/120 | 662/8.16 | 662/8.16 | 789/8.12 | 789/8.12 |
| Matrix | 384×269 | 384×269 | 384×307 | 384×307 | 384×307 | 384×307 |
| ETL | 28 | 28 | 2 | 2 | 2 | 2 |
| Bandwidth (Hz) | 260 | 260 | 280 | 280 | 250 | 250 |
| Average | 1 | 1 | 1 | 1 | 1 | 1 |
| Number slices | 40 | 40 | 40 | 40 | 40 | 40 |
| Spatial resolution | 0.89×0.63×5 | 0.89×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 | 0.78×0.63×5 |
| Acquisition | ACS | PI | ACS | PI | ACS | PI |
| Accelerating factors | 2.25 | 2 | 2.25 | 2 | 2.25 | 2 |
| Sequence acquisition time (s) | 59 | 82 | 93 | 140 | 75 | 90 |
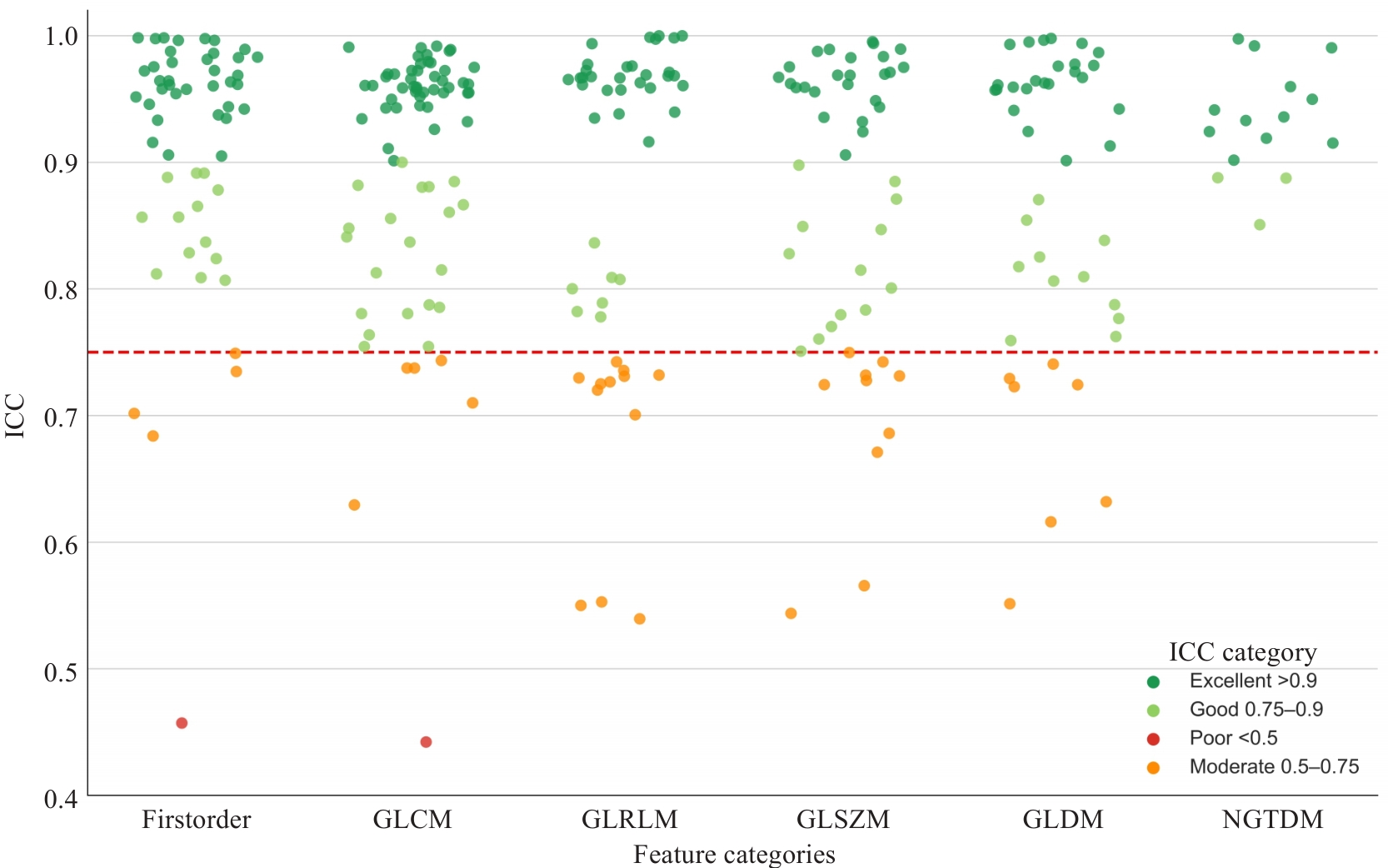
Fig.2 Scatter plot of intraclass correlation coefficients (ICCs) for all radiomic features extracted from T1WI, T2WI, and contrast-enhanced T1WI sequences, categorized by consistency levels. The red dashed line represents the ICC threshold of 0.75.
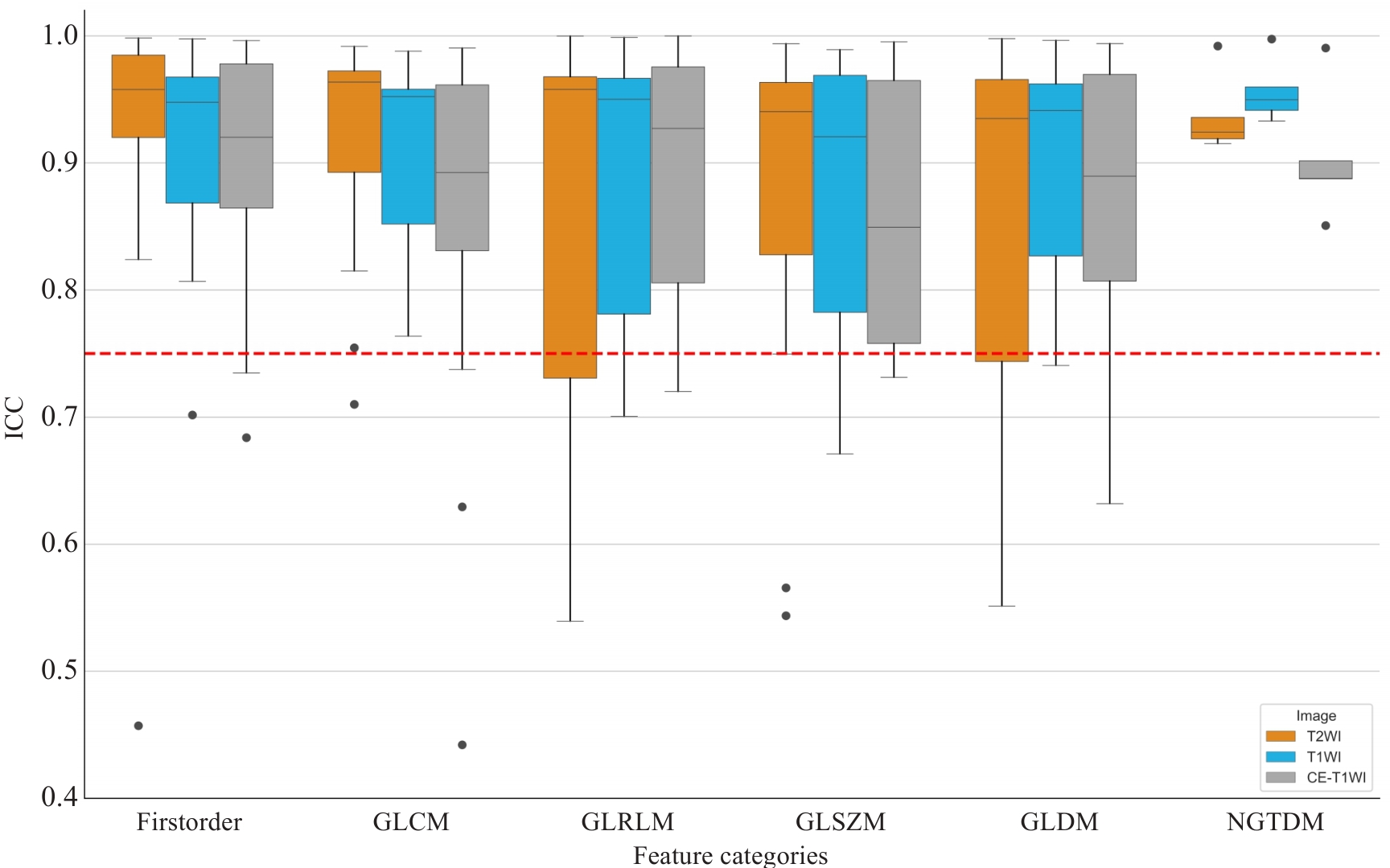
Fig.3 Boxplots of ICCs for different categories of radiomic features extracted from T1WI, T2WI, and contrast-enhanced T1WI (CE-T1WI) sequences. Each boxplot represents the distribution of ICCs within a specific feature category. The red dashed line indicates the reproducibility threshold at ICC=0.75, above which features are considered to have good agreement.
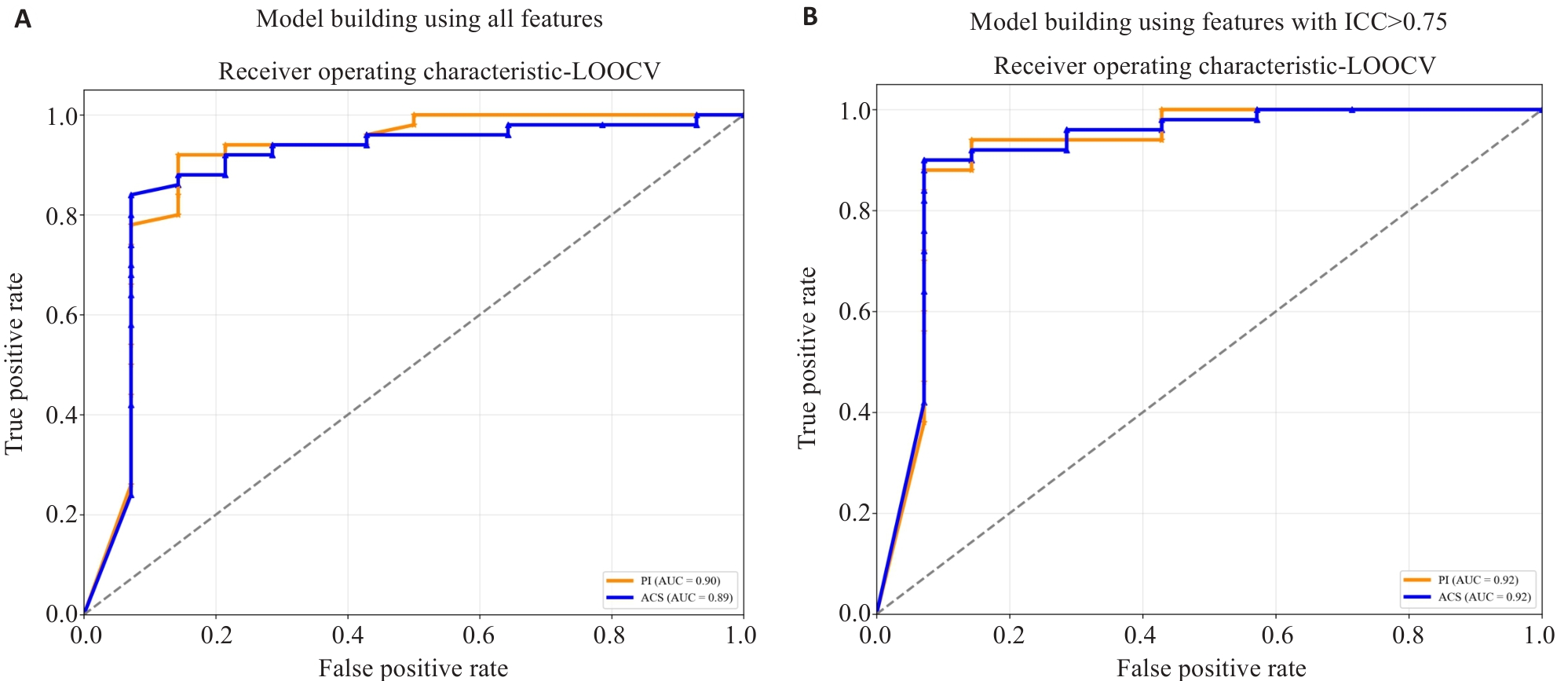
Fig.4 ROC curves of radiomics models constructed using PI- and ACS-accelerated images. The plots show the ROC curves of models built using all extracted features (A) and models built using only features with ICC>0.75 (B). The orange curve represents the model based on PI-accelerated images, and the blue curve represents the model based on ACS-accelerated images.
| Feature type | Method | Accuracy | AUC | F1-Score | Sensitivity | Specificity | P |
|---|---|---|---|---|---|---|---|
| All Features | PI | 0.891 | 0.889 | 0.931 | 0.940 | 0.714 | 0.991 |
| ACS | 0.891 | 0.902 | 0.931 | 0.940 | 0.714 | ||
| Features ICC>0.75 | PI | 0.891 | 0.916 | 0.931 | 0.940 | 0.714 | 0.998 |
| ACS | 0.906 | 0.916 | 0.939 | 0.920 | 0.857 |
Tab.3 Predictive performance of different acceleration sequences
| Feature type | Method | Accuracy | AUC | F1-Score | Sensitivity | Specificity | P |
|---|---|---|---|---|---|---|---|
| All Features | PI | 0.891 | 0.889 | 0.931 | 0.940 | 0.714 | 0.991 |
| ACS | 0.891 | 0.902 | 0.931 | 0.940 | 0.714 | ||
| Features ICC>0.75 | PI | 0.891 | 0.916 | 0.931 | 0.940 | 0.714 | 0.998 |
| ACS | 0.906 | 0.916 | 0.939 | 0.920 | 0.857 |
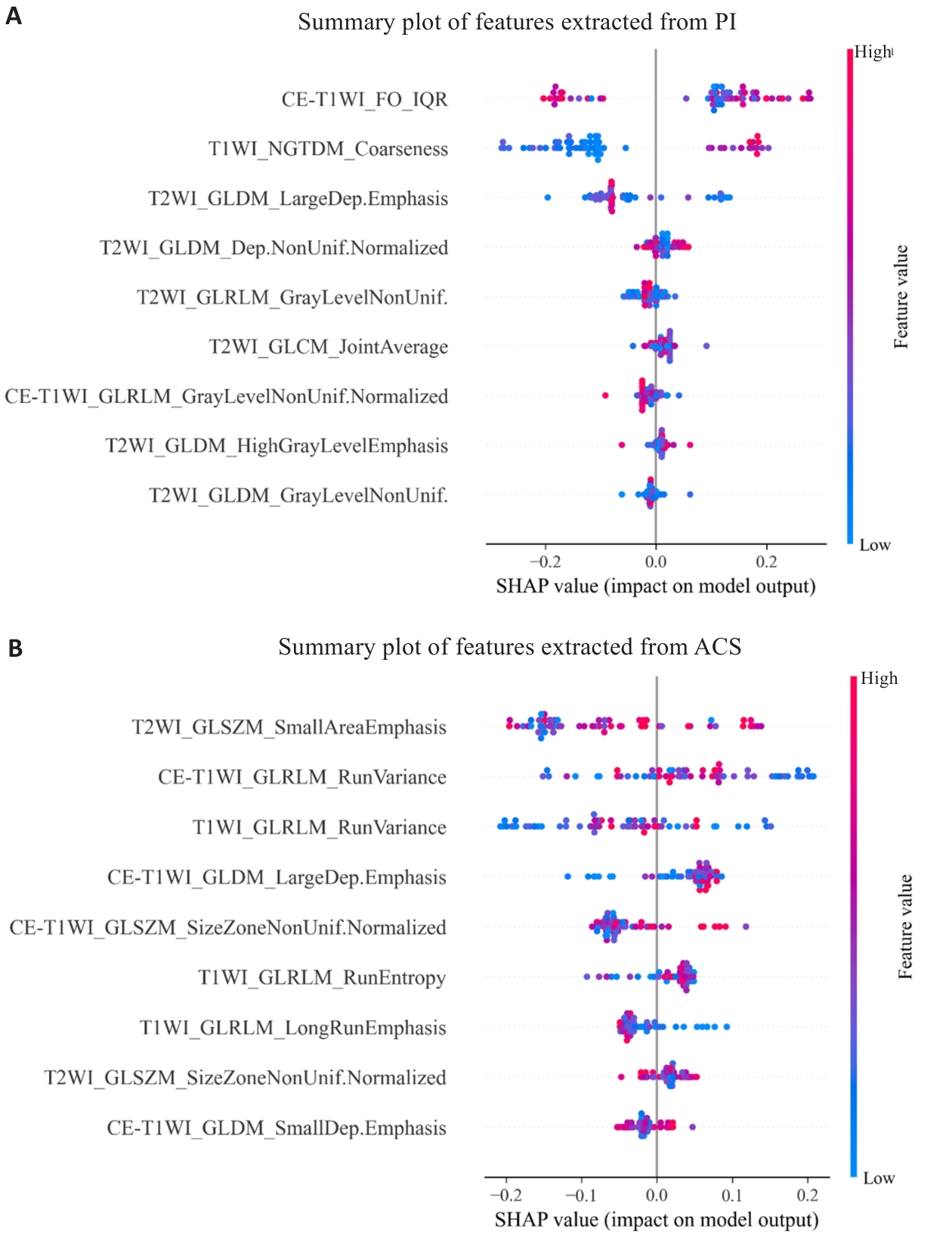
Fig.5 SHAP summary plots of feature contributions in random forest models. The plots show feature contributions for models constructed using features extracted from PI-accelerated images (A) and ACS-accelerated images (B).
| [1] | Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma[J]. Lancet, 2019, 394(10192): 64-80. doi:10.1016/s0140-6736(19)30956-0 |
| [2] | Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview[J]. Int J Cancer, 2021, 149(4): 778-89. doi:10.1002/ijc.33588 |
| [3] | Liao XB, Mao YP, Liu LZ, et al. How does magnetic resonance imaging influence staging according to AJCC staging system for nasopharyngeal carcinoma compared with computed tomography [J]? Int J Radiat Oncol Biol Phys, 2008, 72(5): 1368-77. doi:10.1016/j.ijrobp.2008.03.017 |
| [4] | Wang F, Chen G, Zhang Z, et al. The Chinese society of clinical oncology (CSCO): clinical guidelines for the diagnosis and treatment of colorectal cancer, 2024 update[J]. Cancer Commun: Lond, 2025, 45(3): 332-79. doi:10.1002/cac2.12639 |
| [5] | Deshmane A, Gulani V, Griswold MA, et al. Parallel MR imaging[J]. J Magn Reson Imaging, 2012, 36(1): 55-72. doi:10.1002/jmri.23639 |
| [6] | Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging[J]. Magn Reson Med, 2007, 58(6): 1182-95. doi:10.1002/mrm.21391 |
| [7] | Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA)[J]. Magn Reson Med, 2002, 47(6): 1202-10. doi:10.1002/mrm.10171 |
| [8] | Pruessmann KP, Weiger M, Scheidegger MB, et al. SENSE: Sensitivity encoding for fast MRI[J], Magn Reson Med, 1999;42(5): 952-62. doi:10.1002/(sici)1522-2594(199911)42:5<952::aid-mrm16>3.3.co;2-j |
| [9] | Fransen SJ, Roest C, Simonis FFJ, et al. The scientific evidence of commercial AI products for MRI acceleration: a systematic review[J]. Eur Radiol, 2025, 35(8): 4736-46. doi:10.1007/s00330-025-11423-5 |
| [10] | Lin DJ, Johnson PM, Knoll F, et al. Artificial intelligence for MR image reconstruction: an overview for clinicians[J]. J Magn Reson Imaging, 2021, 53(4): 1015-28. doi:10.1002/jmri.27078 |
| [11] | Ghodrati V, Shao J, Bydder M, et al. MR image reconstruction using deep learning: evaluation of network structure and loss functions[J]. Quant Imaging Med Surg, 2019, 9(9): 1516-27. doi:10.21037/qims.2019.08.10 |
| [12] | Sheng RF, Zheng LY, Jin KP, et al. Single-breath-hold T2WI liver MRI with deep learning-based reconstruction: a clinical feasibility study in comparison to conventional multi-breath-hold T2WI liver MRI[J]. Magn Reson Imag, 2021, 81: 75-81. doi:10.1016/j.mri.2021.06.014 |
| [13] | Liu HB, Deng DL, Zeng WL, et al. AI-assisted compressed sensing and parallel imaging sequences for MRI of patients with nasopharyngeal carcinoma: comparison of their capabilities in terms of examination time and image quality[J]. Eur Radiol, 2023, 33(11): 7686-96. doi:10.1007/s00330-023-09742-6 |
| [14] | Wang QZ, Zhao WL, Xing XY, et al. Feasibility of AI-assisted compressed sensing protocols in knee MR imaging: a prospective multi-reader study[J]. Eur Radiol, 2023, 33(12): 8585-96. doi:10.1007/s00330-023-09823-6 |
| [15] | 王思敏, 李 彦, 严福华, 等. 不同医用磁共振成像设备在中枢神经系统的临床图像质量评价与比较研究[J]. 磁共振成像, 2019(2): 83-8. |
| [16] | Ni M, He M, Yang YX, et al. Application research of AI-assisted compressed sensing technology in MRI scanning of the knee joint: 3D-MRI perspective[J]. Eur Radiol, 2024, 34(5): 3046-58. doi:10.1007/s00330-023-10368-x |
| [17] | Zhao Q, Xu J, Yang YX, et al. AI-assisted accelerated MRI of the ankle: clinical practice assessment[J]. Eur Radiol Exp, 2023, 7(1): 62. doi:10.1186/s41747-023-00374-5 |
| [18] | Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine[J]. Nat Rev Clin Oncol, 2017, 14(12): 749-62. doi:10.1038/nrclinonc.2017.141 |
| [19] | Liu Z, Wang S, Dong D, et al. The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges[J]. Theranostics, 2019, 9(5): 1303-22. doi:10.7150/thno.30309 |
| [20] | Kim MJ, Choi Y, Sung YE, et al. Early risk-assessment of patients with nasopharyngeal carcinoma: the added prognostic value of MR-based radiomics[J]. Transl Oncol, 2021, 14(10): 101180. doi:10.1016/j.tranon.2021.101180 |
| [21] | Zhang L, Dong D, Li HL, et al. Development and validation of a magnetic resonance imaging-based model for the prediction of distant metastasis before initial treatment of nasopharyngeal carcinoma: a retrospective cohort study[J]. EBioMedicine, 2019, 40: 327-35. doi:10.1016/j.ebiom.2019.01.013 |
| [22] | Zhang B, Tian J, Dong D, et al. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma[J]. Clin Cancer Res, 2017, 23(15): 4259-69. doi:10.1158/1078-0432.ccr-16-2910 |
| [23] | Zhang LL, Huang MY, Li Y, et al. Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 nasopharyngeal carcinoma[J]. EBioMedicine, 2019, 42: 270-80. doi:10.1016/j.ebiom.2019.03.050 |
| [24] | 张 璐. 基于磁共振影像组学的鼻咽癌远处转移风险评估模型构建及验证[D]. 南方医科大学, 2019. |
| [25] | Teng X, Wang Y, Nicol AJ, et al. Enhancing the clinical utility of radiomics: addressing the challenges of repeatability and repro-ducibility in CT and MRI[J]. Diagnostics: Basel, 2024, 14(16): 1835. doi:10.3390/diagnostics14161835 |
| [26] | Li S, Deng YQ, Zhu ZL, et al. A comprehensive review on radiomics and deep learning for nasopharyngeal carcinoma imaging[J]. Diagnostics: Basel, 2021, 11(9): 1523. doi:10.3390/diagnostics11091523 |
| [27] | Joo L, Jung SC, Lee H, et al. Stability of MRI radiomic features according to various imaging parameters in fast scanned T2-FLAIR for acute ischemic stroke patients[J]. Sci Rep, 2021, 11(1): 17143. doi:10.1038/s41598-021-96621-z |
| [28] | Rai R, Barton MB, Chlap P, et al. Repeatability and reproducibility of magnetic resonance imaging-based radiomic features in rectal cancer[J]. J Med Imaging: Bellingham, 2022, 9(4): 044005. doi:10.1117/1.jmi.9.4.044005 |
| [29] | Klontzas ME. Radiomics feature reproducibility: The elephant in the room[J]. Eur J Radiol, 2024, 175: 111430. doi:10.1016/j.ejrad.2024.111430 |
| [30] | Reschke P, Koch V, Gruenewald LD, et al. Deep learning-accelerated prostate MRI: improving speed, accuracy, and sustainability[J]. Acad Radiol, 2025, 14: S1076-6332(25)00571-9. doi:10.1016/j.acra.2025.06.022 |
| [31] | Schwier M, van Griethuysen J, Vangel MG, et al. Repeatability of multiparametric prostate MRI radiomics features[J]. Sci Rep, 2019, 9(1): 9441. doi:10.1038/s41598-019-45766-z |
| [32] | Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping[J]. Radiology, 2020, 295(2): 328-38. doi:10.1148/radiol.2020191145 |
| [33] | Granzier RWY, Ibrahim A, Primakov S, et al. Test-retest data for the assessment of breast MRI radiomic feature repeatability[J]. J Magn Reson Imaging, 2022, 56(2): 592-604. doi:10.1002/jmri.28027 |
| [34] | Lu H, Parra NA, Qi J, et al. Repeatability of quantitative imaging features in prostate magnetic resonance imaging[J]. Front Oncol, 2020, 10: 551. doi:10.3389/fonc.2020.00551 |
| [35] | Scalco E, Belfatto A, Mastropietro A, et al. T2w-MRI signal normalization affects radiomics features reproducibility[J]. Med Phys, 2020, 47(4): 1680-91. doi:10.1002/mp.14038 |
| [36] | Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research[J]. J Chiropr Med, 2016, 15(2): 155-63. doi:10.1016/j.jcm.2016.02.012 |
| [37] | Bi Y, Xiang DX, Ge ZY, et al. An interpretable prediction model for identifying N7-methylguanosine sites based on XGBoost and SHAP[J]. Mol Ther Nucleic Acids, 2020, 22: 362-72. doi:10.1016/j.omtn.2020.08.022 |
| [38] | Shukla-Dave A, Obuchowski NA, Chenevert TL, et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials[J]. J Magn Reson Imaging, 2019, 49(7): e101-21. doi:10.1002/jmri.26805 |
| [39] | Salluzzi M, McCreary CR, Gobbi DG, et al. Short-term repeatability and long-term reproducibility of quantitative MR imaging biomarkers in a single centre longitudinal study[J]. NeuroImage, 2022, 260: 119488. doi:10.1016/j.neuroimage.2022.119488 |
| [40] | Ye S, Lim JY, Huang W. Statistical considerations for repeatability and reproducibility of quantitative imaging biomarkers[J]. BJR Open, 2022, 4(1): 20210083. doi:10.1259/bjro.20210083 |
| [41] | Orlhac F, Lecler A, Savatovski J, et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure[J]. Eur Radiol, 2021, 31(4): 2272-80. doi:10.1007/s00330-020-07284-9 |
| [42] | Carré A, Battistella E, Niyoteka S, et al. AutoComBat: a generic method for harmonizing MRI-based radiomic features[J]. Sci Rep, 2022, 12(1): 12762. doi:10.1038/s41598-022-16609-1 |
| [43] | Zhao W, Hu Z, Kazerooni AF, et al. Physics-informed discretization for reproducible and robust radiomic feature extraction using quantitative MRI[J]. Invest Radiol, 2024, 59(5): 359-71. doi:10.1097/rli.0000000000001026 |
| [44] | 胡丽霞, 朱 进, 周阳泱, 等. 鼻咽癌动态增强MRI参数与临床病理特征的关系[J]. 中国医学影像技术, 2014, 30(4): 514-7. |
| [45] | Carbonell G, Kennedy P, Bane O, et al. Precision of MRI radiomics features in the liver and hepatocellular carcinoma[J]. Eur Radiol, 2022, 32(3): 2030-40. doi:10.1007/s00330-021-08282-1 |
| [46] | Perrin T, Midya A, Yamashita R, et al. Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging[J]. Abdom Radiol, 2018, 43(12): 3271-8. doi:10.1007/s00261-018-1600-6 |
| [47] | Ibrahim A, Guha S, Lu L, et al. The reproducibility and predictivity of radiomic features extracted from dynamic contrast-enhanced computed tomography of hepatocellular carcinoma[J]. PLoS One, 2024, 19(9): e0310486. doi:10.1371/journal.pone.0310486 |
| [1] | Liying ZHANG, Tongzhen ZHANG, Xin ZHAO. Diagnostic value of morphological features of breast lesions on DWI and T2WI assessed using Breast Imaging Reporting and Data System lexicon descriptors [J]. Journal of Southern Medical University, 2025, 45(9): 1809-1817. |
| [2] | Qizhi HUANG, Daipeng XIE, Lintong YAO, Qiaxuan LI, Shaowei WU, Haiyu ZHOU. Tumor microenvironment-specific CT radiomics signature for predicting immunotherapy response in non-small cell lung cancer [J]. Journal of Southern Medical University, 2025, 45(9): 1903-1918. |
| [3] | Zhenxiang DONG, Yihao GUO, Qiang LIU, Yizhe ZHANG, Qianyi QIU, Xiaodong ZHANG, Yanqiu FENG. A single repetition time quantitative magnetic susceptibility imaging method for the lumbar spine using bipolar readout gradient [J]. Journal of Southern Medical University, 2025, 45(6): 1336-1342. |
| [4] | Yumei ZENG, Jike LI, Zhongxi HUANG, Yibo ZHOU. Villin-like protein VILL suppresses proliferation of nasopharyngeal carcinoma cells by interacting with LMO7 protein [J]. Journal of Southern Medical University, 2025, 45(5): 954-961. |
| [5] | Lu TAO, Zhuoli WEI, Yueyue WANG, Ping XIANG. CEACAM6 inhibits proliferation and migration of nasopharyngeal carcinoma cells by suppressing epithelial-mesenchymal transition [J]. Journal of Southern Medical University, 2025, 45(3): 566-576. |
| [6] | Jinyu LIU, Shujun LIANG, Yu ZHANG. A multi-scale supervision and residual feedback optimization algorithm for improving optic chiasm and optic nerve segmentation accuracy in nasopharyngeal carcinoma CT images [J]. Journal of Southern Medical University, 2025, 45(3): 632-642. |
| [7] | Xiaoyuan LI, Yiyue ZHANG, Yucheng GU, Nihong CHEN, Xinyu QIAN, Pengjun ZHANG, Jiaxin HAO, Feng WANG. Association between Tau protein deposition and brain metabolites: N-acetylaspartate and creatine as potential biomarkers for advanced Alzheimer's disease [J]. Journal of Southern Medical University, 2025, 45(11): 2350-2357. |
| [8] | Weiyang FANG, Hui XIAO, Shuang WANG, Xiaoming LIN, Chaomin CHEN. A deep learning model based on magnetic resonance imaging and clinical feature fusion for predicting preoperative cytokeratin 19 status in hepatocellular carcinoma [J]. Journal of Southern Medical University, 2024, 44(9): 1738-1751. |
| [9] | Xiaofan CONG, Teng CHEN, Shuo LI, Yuanyuan WANG, Longyun ZHOU, Xiaolong LI, Pei ZHANG, Xiaojin SUN, Surong ZHAO. Dihydroartemisinin enhances sensitivity of nasopharyngeal carcinoma HNE1/DDP cells to cisplatin-induced apoptosis by promoting ROS production [J]. Journal of Southern Medical University, 2024, 44(8): 1553-1560. |
| [10] | Yuanyuan WANG, Teng CHEN, Xiaofan CONG, Yiran LI, Rui CHEN, Pei ZHANG, Xiaojin SUN, Surong ZHAO. Pristimerin enhances cisplatin-induced apoptosis in nasopharyngeal carcinoma cells via ROS-mediated deactivation of the PI3K/AKT signaling pathway [J]. Journal of Southern Medical University, 2024, 44(5): 904-912. |
| [11] | LIU Yunze, LI Chengrun, GUO Juntang, LIU Yang. A clinical-radiomics nomogram for differentiating focal organizing pneumonia and lung adenocarcinoma [J]. Journal of Southern Medical University, 2024, 44(2): 397-404. |
| [12] | Yue HU, Yu ZENG, Linjing WANG, Zhiwei LIAO, Jianming TAN, Yanhao KUANG, Pan GONG, Bin QI, Xin ZHEN. Performance of multi-modality and multi-classifier fusion models for predicting radiation-induced oral mucositis in patients with nasopharyngeal carcinoma [J]. Journal of Southern Medical University, 2024, 44(12): 2434-2442. |
| [13] | Xiaoyin HUANG, Fenglian CHEN, Yu ZHANG, Shujun LIANG. A predictive model for survival outcomes of glioma patients based on multi-parametric, multi-regional MRI radiomics features and clinical features [J]. Journal of Southern Medical University, 2024, 44(10): 2004-2014. |
| [14] | HE Huishan, GUO Erjia, MENG Wenyi, WANG Yu, WANG Wen, HE Wenle, WU Yuankui, YANG Wei. Predicting cerebral glioma enhancement pattern using a machine learning-based magnetic resonance imaging radiomics model [J]. Journal of Southern Medical University, 2024, 44(1): 194-200. |
| [15] | LIU Yuxuan, CHU Zhiqin, ZHANG Yu. Physical model-based cascaded generative adversarial networks for accelerating quantitative multi-parametric magnetic resonance imaging [J]. Journal of Southern Medical University, 2023, 43(8): 1402-1409. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||