Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (3): 632-642.doi: 10.12122/j.issn.1673-4254.2025.03.21
Jinyu LIU( ), Shujun LIANG(
), Shujun LIANG( ), Yu ZHANG(
), Yu ZHANG( )
)
Received:2024-07-06
Online:2025-03-20
Published:2025-03-28
Contact:
Shujun LIANG, Yu ZHANG
E-mail:1377981055@qq.com;390611257@qq.com;yuzhang@smu.edu.cn
Supported by:Jinyu LIU, Shujun LIANG, Yu ZHANG. A multi-scale supervision and residual feedback optimization algorithm for improving optic chiasm and optic nerve segmentation accuracy in nasopharyngeal carcinoma CT images[J]. Journal of Southern Medical University, 2025, 45(3): 632-642.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.03.21
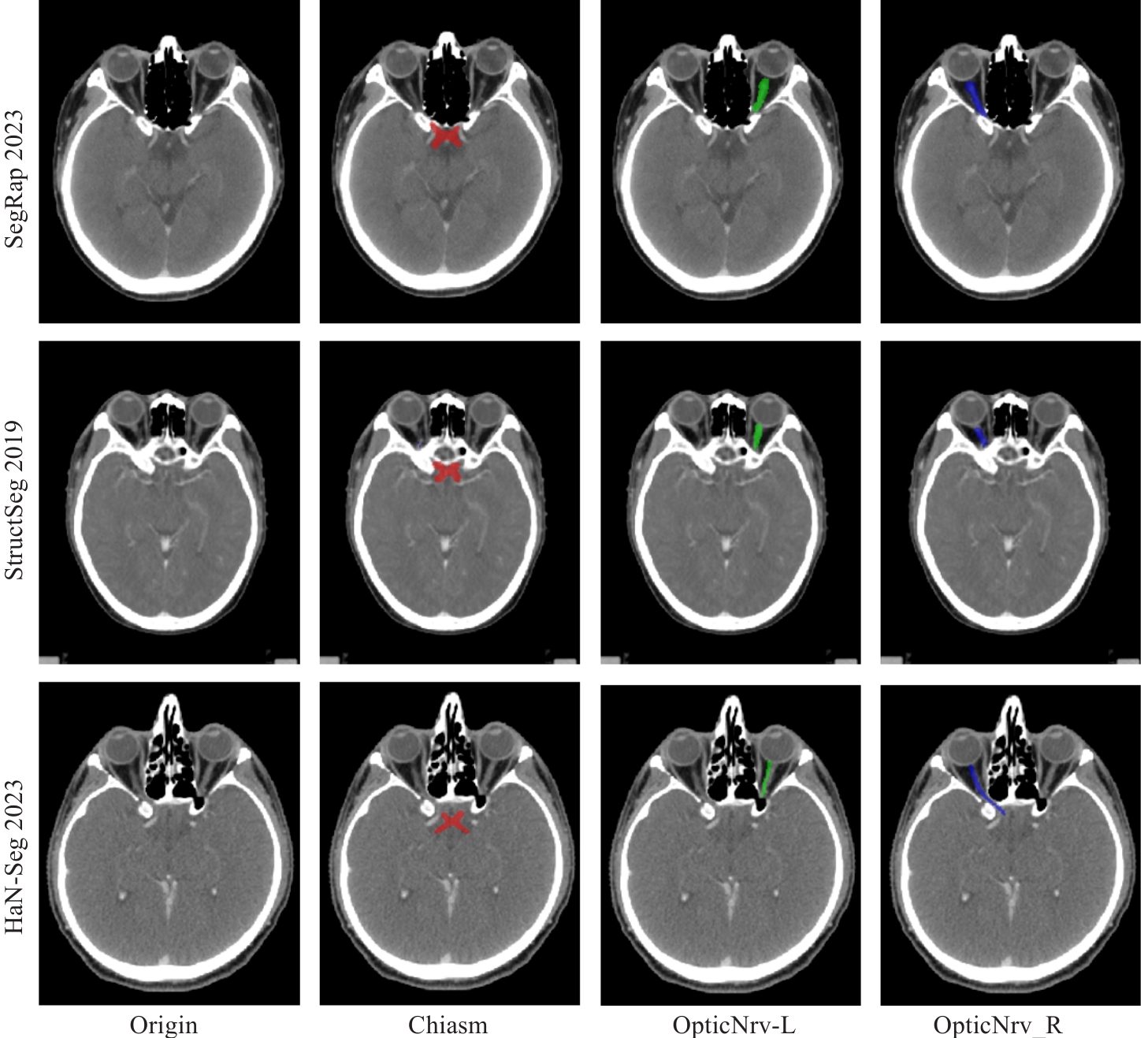
Fig.1 CT images and ground truth labels for segmentation of the optic chiasm and optic nerve in CT images from 3 datasets. The optic chiasm is shown in red, the left optic nerve in green, and the right optic nerve in blue.
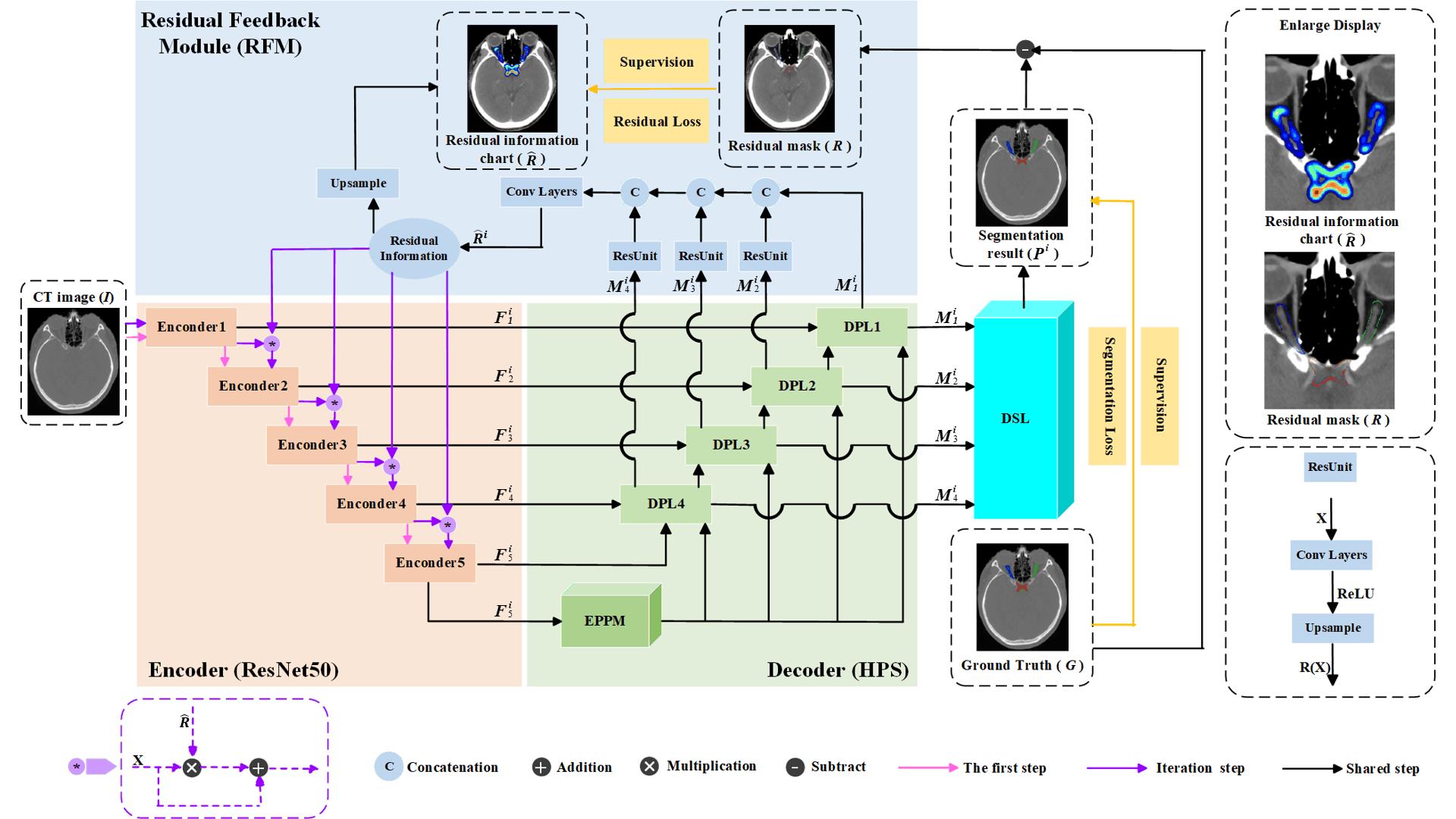
Fig.2 Deep supervision residual feedback network framework. The orange components represent the encoder, the green components represent the decoder based on the Hybrid Pooling Strategy (HPS), the blue components represent the Residual Feedback Module (RFM), and the cyan components represent the Multi-scale Deep Supervision Layers (DSL). The top-right corner shows a magnified view of the residual information map and residual mask. The bottom-right corner illustrates the structure of the residual unit.
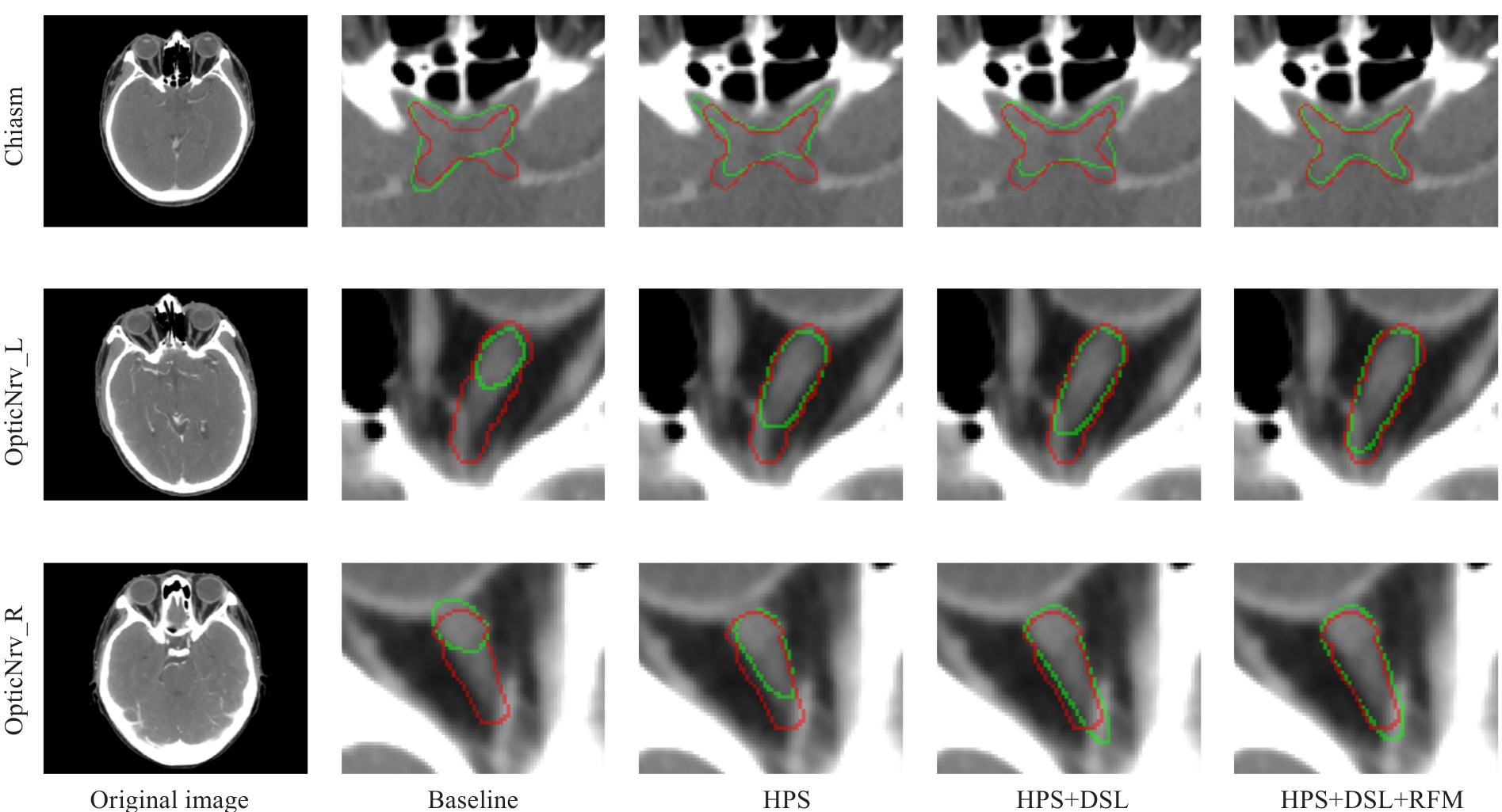
Fig.5 Visualized results of the ablation experiments on the internal test set for DSRF. The ground truth labels are shown in red, and the segmentation results of the model are shown in green.
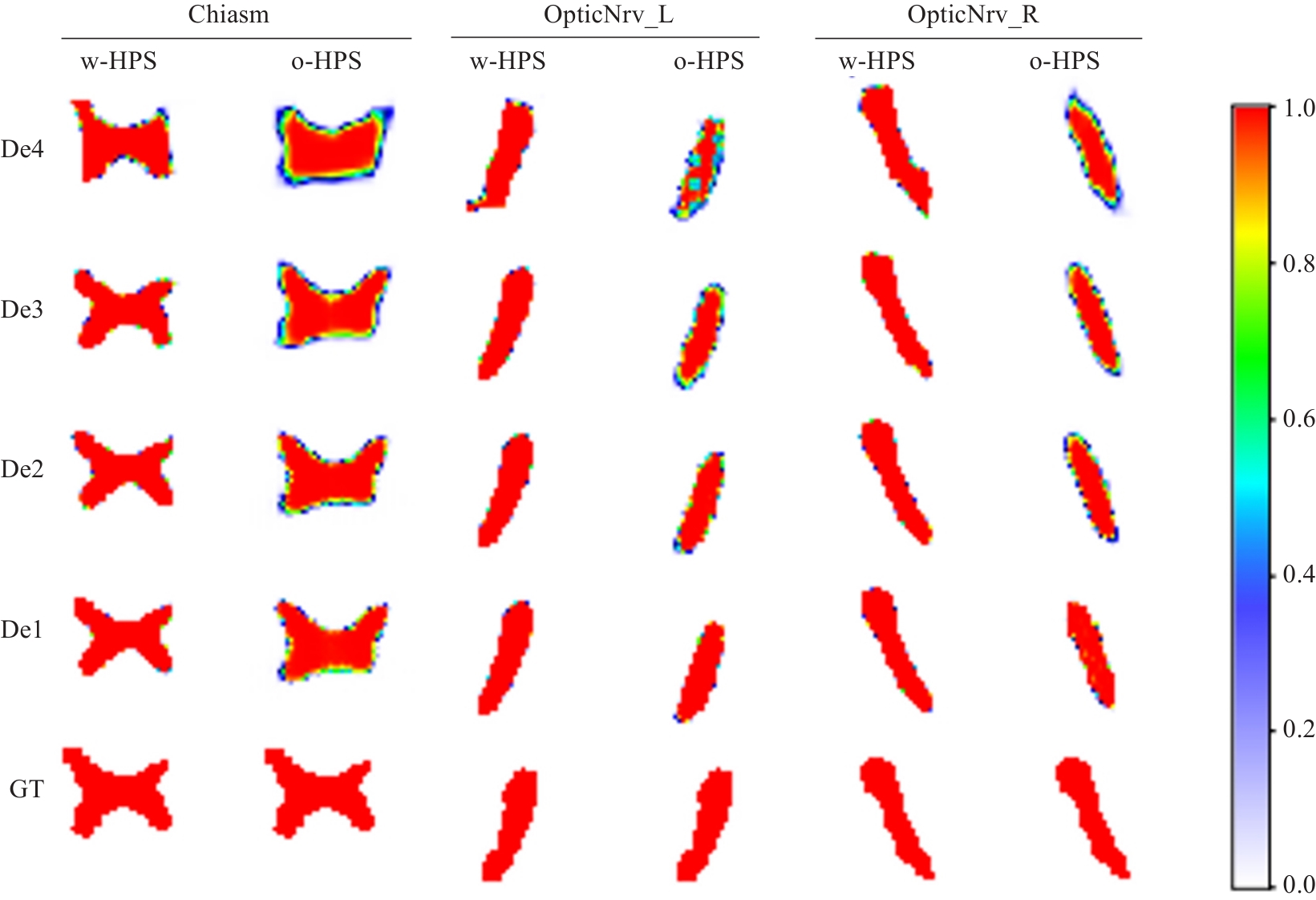
Fig.6 Examples of semantic feature heatmaps in the decoder blocks. w-HPS represents the DSRF model, o-HPS represents the DSL+RFM model, and De4 to De1 correspond to decoder blocks from deep to shallow layers. GT denotes the ground truth labels. The heatmap colors range from white to red, indicating semantic feature values from low probability to high probability.
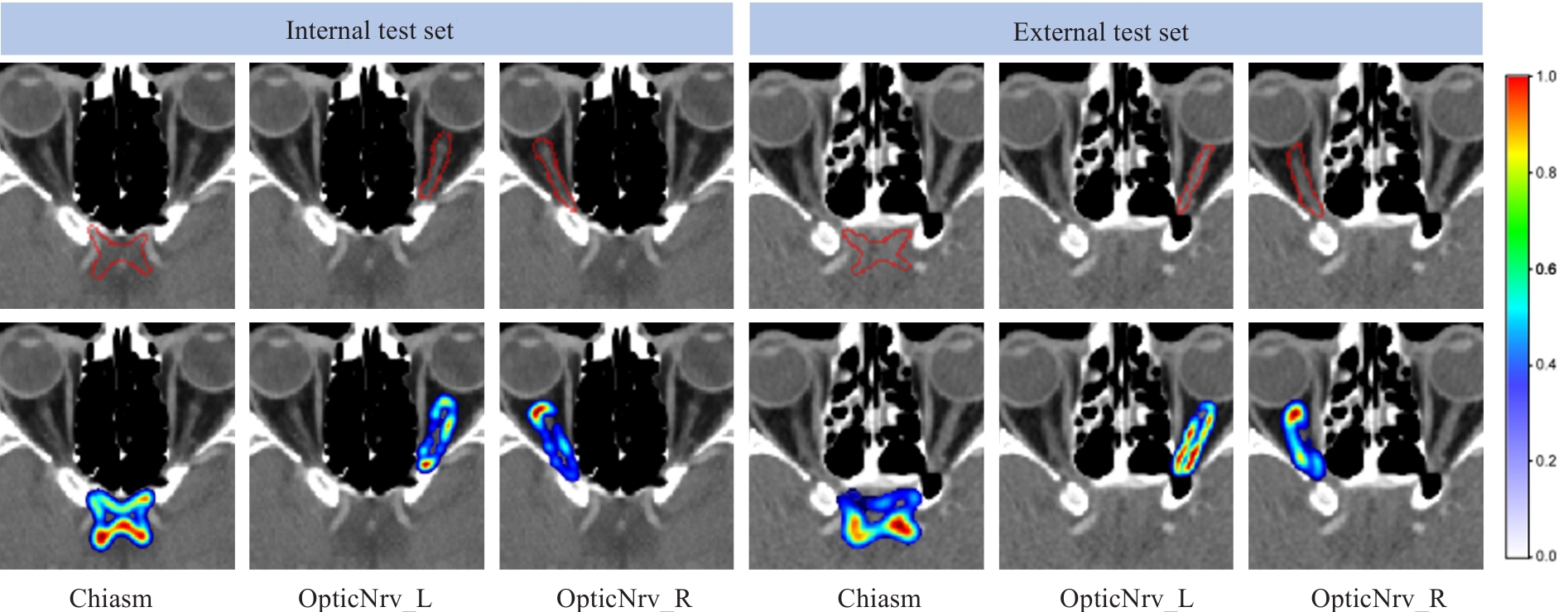
Fig.7 Two examples of residual expression. The red contour in the first row represents the edge of the ground truth label. In the second row, the residual information color changes (from blue to red) indicate low to high probabilities.
| HPS | DSL | RFM | Internal Test | External Test | ||||
|---|---|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||||
| 0.689 | 0.789 | 0.793 | 0.537 | 0.711 | 0.706 | |||
| √ | 0.729 | 0.836 | 0.838 | 0.659 | 0.768 | 0.773 | ||
| √ | 0.706 | 0.808 | 0.801 | 0.594 | 0.730 | 0.736 | ||
| √ | 0.701 | 0.796 | 0.796 | 0.564 | 0.718 | 0.717 | ||
| √ | √ | 0.712 | 0.818 | 0.821 | 0.601 | 0.747 | 0.751 | |
| √ | √ | 0.743 | 0.852 | 0.858 | 0.672 | 0.788 | 0.789 | |
| √ | √ | 0.752 | 0.858 | 0.857 | 0.688 | 0.801 | 0.813 | |
| √ | √ | √ | 0.764 | 0.872 | 0.874 | 0.708 | 0.819 | 0.828 |
Tab.1 Average dice similarity coefficient of DSRF in ablation experiments on the internal and external test sets
| HPS | DSL | RFM | Internal Test | External Test | ||||
|---|---|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||||
| 0.689 | 0.789 | 0.793 | 0.537 | 0.711 | 0.706 | |||
| √ | 0.729 | 0.836 | 0.838 | 0.659 | 0.768 | 0.773 | ||
| √ | 0.706 | 0.808 | 0.801 | 0.594 | 0.730 | 0.736 | ||
| √ | 0.701 | 0.796 | 0.796 | 0.564 | 0.718 | 0.717 | ||
| √ | √ | 0.712 | 0.818 | 0.821 | 0.601 | 0.747 | 0.751 | |
| √ | √ | 0.743 | 0.852 | 0.858 | 0.672 | 0.788 | 0.789 | |
| √ | √ | 0.752 | 0.858 | 0.857 | 0.688 | 0.801 | 0.813 | |
| √ | √ | √ | 0.764 | 0.872 | 0.874 | 0.708 | 0.819 | 0.828 |
| HPS | DSL | RFM | Internal Test | External Test | ||||
|---|---|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||||
| 0.875 | 0.450 | 0.451 | 1.386 | 0.728 | 0.783 | |||
| √ | 0.668 | 0.326 | 0.343 | 0.925 | 0.537 | 0.541 | ||
| √ | 0.838 | 0.437 | 0.447 | 1.168 | 0.626 | 0.646 | ||
| √ | 0.867 | 0.448 | 0.452 | 1.327 | 0.658 | 0.679 | ||
| √ | √ | 0.768 | 0.366 | 0.414 | 1.068 | 0.568 | 0.614 | |
| √ | √ | 0.657 | 0.279 | 0.270 | 0.859 | 0.447 | 0.440 | |
| √ | √ | 0.640 | 0.247 | 0.270 | 0.690 | 0.374 | 0.388 | |
| √ | √ | √ | 0.601 | 0.220 | 0.232 | 0.615 | 0.334 | 0.333 |
Tab.2 Average symmetric surface distance of DSRF in ablation experiments on the internal and external test sets
| HPS | DSL | RFM | Internal Test | External Test | ||||
|---|---|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||||
| 0.875 | 0.450 | 0.451 | 1.386 | 0.728 | 0.783 | |||
| √ | 0.668 | 0.326 | 0.343 | 0.925 | 0.537 | 0.541 | ||
| √ | 0.838 | 0.437 | 0.447 | 1.168 | 0.626 | 0.646 | ||
| √ | 0.867 | 0.448 | 0.452 | 1.327 | 0.658 | 0.679 | ||
| √ | √ | 0.768 | 0.366 | 0.414 | 1.068 | 0.568 | 0.614 | |
| √ | √ | 0.657 | 0.279 | 0.270 | 0.859 | 0.447 | 0.440 | |
| √ | √ | 0.640 | 0.247 | 0.270 | 0.690 | 0.374 | 0.388 | |
| √ | √ | √ | 0.601 | 0.220 | 0.232 | 0.615 | 0.334 | 0.333 |
| Method | DSC | ASSD | ||||
|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||
| DSRF0 | 0.752 | 0.858 | 0.857 | 0.640 | 0.247 | 0.270 |
| DSRF1 | 0.764 | 0.872 | 0.874 | 0.601 | 0.220 | 0.232 |
| DSRF2 | 0.759 | 0.875 | 0.876 | 0.598 | 0.214 | 0.220 |
Tab.3 Quantitative results of different iteration counts for DSRF
| Method | DSC | ASSD | ||||
|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||
| DSRF0 | 0.752 | 0.858 | 0.857 | 0.640 | 0.247 | 0.270 |
| DSRF1 | 0.764 | 0.872 | 0.874 | 0.601 | 0.220 | 0.232 |
| DSRF2 | 0.759 | 0.875 | 0.876 | 0.598 | 0.214 | 0.220 |
| Method | DSC | ASSD | ||||
|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||
| nnU-Net[ | 0.721 | 0.809 | 0.824 | 0.632 | 0.354 | 0.363 |
| PoolNet[ | 0.722 | 0.838 | 0.834 | 0.676 | 0.330 | 0.353 |
| STU-Net[ | 0.732 | 0.802 | 0.828 | 0.640 | 0.422 | 0.368 |
| UMamba[ | 0.733 | 0.816 | 0.814 | 0.769 | 0.371 | 0.409 |
| RF-Net[ | 0.735 | 0.863 | 0.869 | 0.807 | 0.246 | 0.242 |
| DSRF | 0.764 | 0.872 | 0.874 | 0.601 | 0.220 | 0.232 |
Tab.4 Segmentation results of DSRF and other methods on the internal test set
| Method | DSC | ASSD | ||||
|---|---|---|---|---|---|---|
| OpticNrv_L | OpticNrv_R | OpticNrv_L | OpticNrv_R | |||
| nnU-Net[ | 0.721 | 0.809 | 0.824 | 0.632 | 0.354 | 0.363 |
| PoolNet[ | 0.722 | 0.838 | 0.834 | 0.676 | 0.330 | 0.353 |
| STU-Net[ | 0.732 | 0.802 | 0.828 | 0.640 | 0.422 | 0.368 |
| UMamba[ | 0.733 | 0.816 | 0.814 | 0.769 | 0.371 | 0.409 |
| RF-Net[ | 0.735 | 0.863 | 0.869 | 0.807 | 0.246 | 0.242 |
| DSRF | 0.764 | 0.872 | 0.874 | 0.601 | 0.220 | 0.232 |
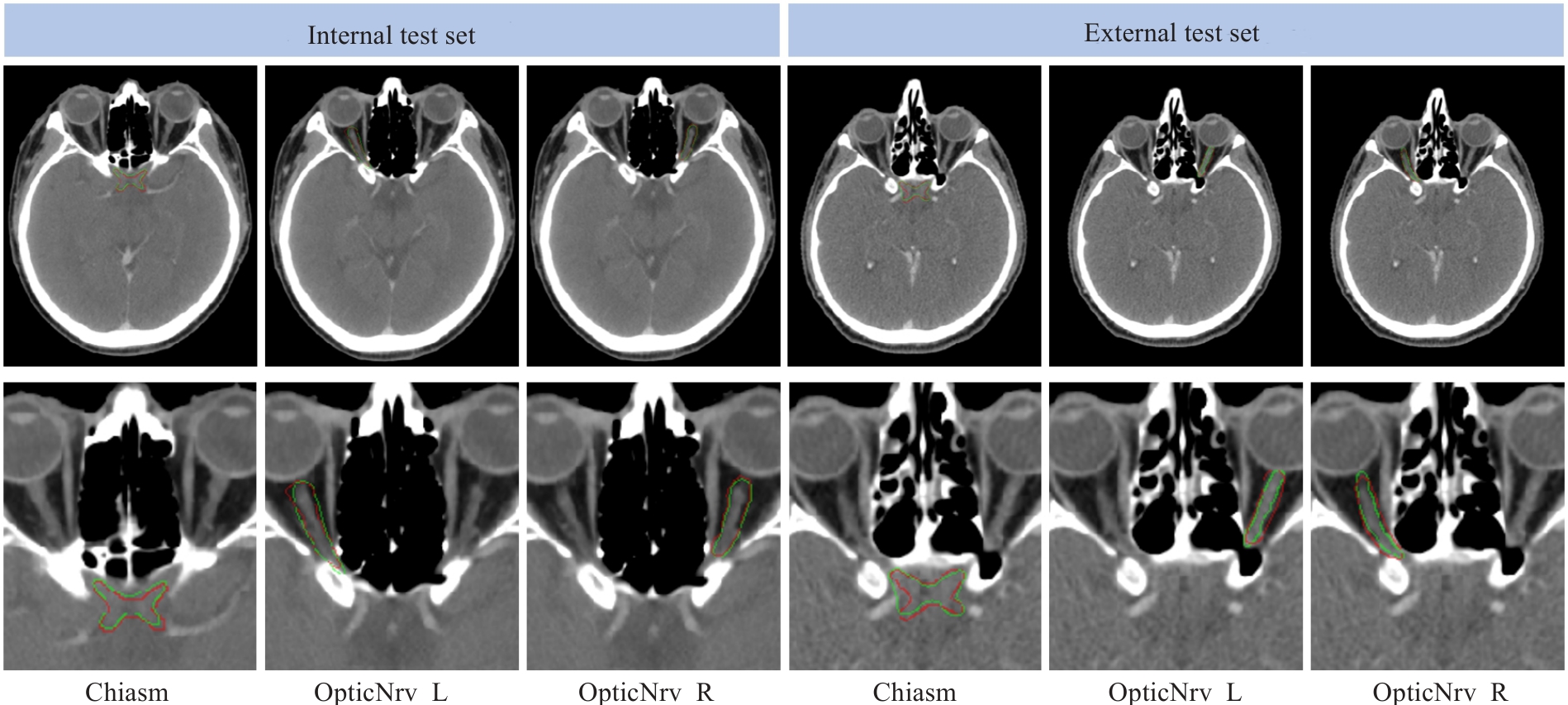
Fig.8 Visual comparison of the segmentation results between the ground truth (red) and the proposed framework (green) for two randomly selected testing subjects from the internal and external test sets.
| 1 | Du XJ, Wang GY, Zhu XD, et al. Refining the 8th edition TNM classification for EBV related nasopharyngeal carcinoma[J]. Cancer Cell, 2024, 42(3): 464-73.e3. |
| 2 | 陈美宁, 刘懿梅, 彭应林, 等. 不同级别肿瘤中心医师对鼻咽癌调强放疗靶区和危及器官勾画差异比较[J]. 中国医学物理学杂志, 2024, 41(3): 265-72. |
| 3 | Guo DZ, Jin DK, Zhu Z, et al. Organ at risk segmentation for head and neck cancer using stratified learning and neural architecture search[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, 2020: 4223-32. |
| 4 | Guo H, Wang J, Xia X, et al. The dosimetric impact of deep learning-based auto-segmentation of organs at risk on nasopharyngeal and rectal cancer[J]. Radiat Oncol, 2021, 16(1): 113. |
| 5 | Costea M, Zlate A, Durand M, et al. Comparison of atlas-based and deep learning methods for organs at risk delineation on head-and-neck CT images using an automated treatment planning system[J]. Radiother Oncol, 2022, 177: 61-70. |
| 6 | Peng YL, Liu YM, Shen GZ, et al. Improved accuracy of auto-segmentation of organs at risk in radiotherapy planning for nasopharyngeal carcinoma based on fully convolutional neural network deep learning[J]. Oral Oncol, 2023, 136: 106261. |
| 7 | Luan S, Wei C, Ding Y, et al. PCG-net: feature adaptive deep learning for automated head and neck organs-at-risk segmentation[J]. Front Oncol, 2023, 13: 1177788. |
| 8 | Liu P, Sun Y, Zhao X, et al. Deep learning algorithm performance in contouring head and neck organs at risk: a systematic review and single-arm meta-analysis[J]. Biomed Eng Online, 2023, 22(1): 104. |
| 9 | Wang K, Liang SJ, Zhang Y. Residual feedback network for breast lesion segmentation in ultrasound image[M]//Medical Image Computing and Computer Assisted Intervention – MICCAI 2021. Cham: Springer International Publishing, 2021: 471-81. |
| 10 | Luo XD, Fu J, Zhong YX, et al. SegRap2023: a benchmark of organs-at-risk and gross tumor volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma[J]. Med Image Anal, 2025, 101: 103447. |
| 11 | Automatic Structure Segmentation for Radiotherapy Planning Challenge 2019. The MICCAI 2019 Challenge[OL]. Retrieved from |
| 12 | Podobnik G, Strojan P, Peterlin P, et al. HaN-Seg: The head and neck organ-at-risk CT and MR segmentation dataset[J]. Med Phys, 2023, 50(3): 1917-27. |
| 13 | Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation[J]. Nat Meth, 2021, 18: 203-11. |
| 14 | He KM, Zhang XY, Ren SQ, et al. Deep residual learning for image recognition[C]//2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). June 27-30, 2016. Las Vegas, NV, USA. IEEE, 2016: 770-778. |
| 15 | Kingma DP, Ba J. Adam: A method for stochastic optimization [J]. arXiv preprint arXiv:, 2014. |
| 16 | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks[J]. Commun ACM, 2017, 60(6): 84-90. |
| 17 | Liu JJ, Hou QB, Cheng MM, et al. A simple pooling-based design for real-time salient object detection[C]//2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). June 15-20, 2019. Long Beach, CA, USA. IEEE, 2019: 3917-3926. |
| 18 | Huang ZY, Ye J, Wang HY, et al. Evaluating STU-net for brain tumor segmentation[M]//Brain Tumor Segmentation, and Cross-Modality Domain Adaptation for Medical Image Segmentation. Cham: Springer Nature Switzerland, 2024: 140-51. |
| 19 | Ma J, Li FF, Wang B. U-mamba: enhancing long-range dependency for biomedical image segmentation[EB/OL]. 2024: 2401.04722. . |
| 20 | Wenderott K, Krups J, Zaruchas F, et al. Effects of artificial intelligence implementation on efficiency in medical imaging: a systematic literature review and meta-analysis[J]. NPJ Digit Med, 2024, 7: 265. |
| 21 | 赖建军, 陈丽婷, 胡海丽, 等. 基于深度学习自动勾画在鼻咽癌调强放射治疗计划中的系统性评价研究[J]. 中国现代医药杂志, 2023, 25(10): 24-30. |
| 22 | 黄 新, 王新卓, 薛 涛, 等. 鼻咽癌放射治疗危及器官自动勾画的几何和剂量学分析[J]. 生物医学工程与临床, 2024, 28(1): 26-34. |
| 23 | Su ZY, Siak PY, Lwin YY, et al. Epidemiology of nasopharyngeal carcinoma: current insights and future outlook[J]. Cancer Metastasis Rev, 2024, 43(3): 919-39. |
| 24 | Azad R, Aghdam EK, Rauland A, et al. Medical image segmentation review: the success of U-net[J]. IEEE Trans Pattern Anal Mach Intell, 2024, 46(12): 10076-95. |
| 25 | Liu JJ, Hou Q, Liu ZA, et al. PoolNet+: exploring the potential of pooling for salient object detection[J]. IEEE Trans Pattern Anal Mach Intell, 2023, 45(1): 887-904. |
| 26 | Hu K, Chen W, Sun Y, et al. PPNet: Pyramid pooling based network for polyp segmentation[J]. Comput Biol Med, 2023, 160: 107028. |
| 27 | Wu YH, Liu Y, Zhan X, et al. P2T: pyramid pooling transformer for scene understanding[J]. IEEE Trans Pattern Anal Mach Intell, 2023, 45(11): 12760-71. |
| 28 | Wang LW, Lee CY, Tu ZW, et al. Training deeper convolutional networks with deep supervision[EB/OL]. 2015: 1505.02496. . |
| 29 | Zhang LF, Chen X, Zhang JB, et al. Contrastive deep supervision[M]//Computer Vision-ECCV 2022. Cham: Springer Nature Switzerland, 2022: 1-19. |
| 30 | Ahmad S, Ullah Z, Gwak J. Multi-teacher cross-modal distillation with cooperative deep supervision fusion learning for unimodal segmentation[J]. Knowl Based Syst, 2024, 297: 111854. |
| 31 | Umer MJ, Sharif MI, Kim J. Breast cancer segmentation from ultrasound images using multiscale cascaded convolution with residual attention-based double decoder network[J]. IEEE Access, 2024, 12: 107888-902. |
| 32 | Wang J, Liang J, Xiao Y, et al. TaiChiNet: negative-positive cross-attention network for breast lesion segmentation in ultrasound images[J]. IEEE J Biomed Health Inform, 2024, 28(3): 1516-27. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||