Journal of Southern Medical University ›› 2026, Vol. 46 ›› Issue (1): 219-230.doi: 10.12122/j.issn.1673-4254.2026.01.24
Li ZHUO1( ), Min ZENG1, Shunqian TAN1, Tao LIANG1, Weiwei XIAO2, Xin ZHEN1(
), Min ZENG1, Shunqian TAN1, Tao LIANG1, Weiwei XIAO2, Xin ZHEN1( )
)
Received:2025-06-13
Online:2026-01-20
Published:2026-01-16
Contact:
Xin ZHEN
E-mail:zhuoli0901@163.com;xinzhen@smu.edu.cn
Supported by:Li ZHUO, Min ZENG, Shunqian TAN, Tao LIANG, Weiwei XIAO, Xin ZHEN. Diffusion cycle-consistent generative adversarial networks for pelvic active bone marrow segmentation[J]. Journal of Southern Medical University, 2026, 46(1): 219-230.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2026.01.24
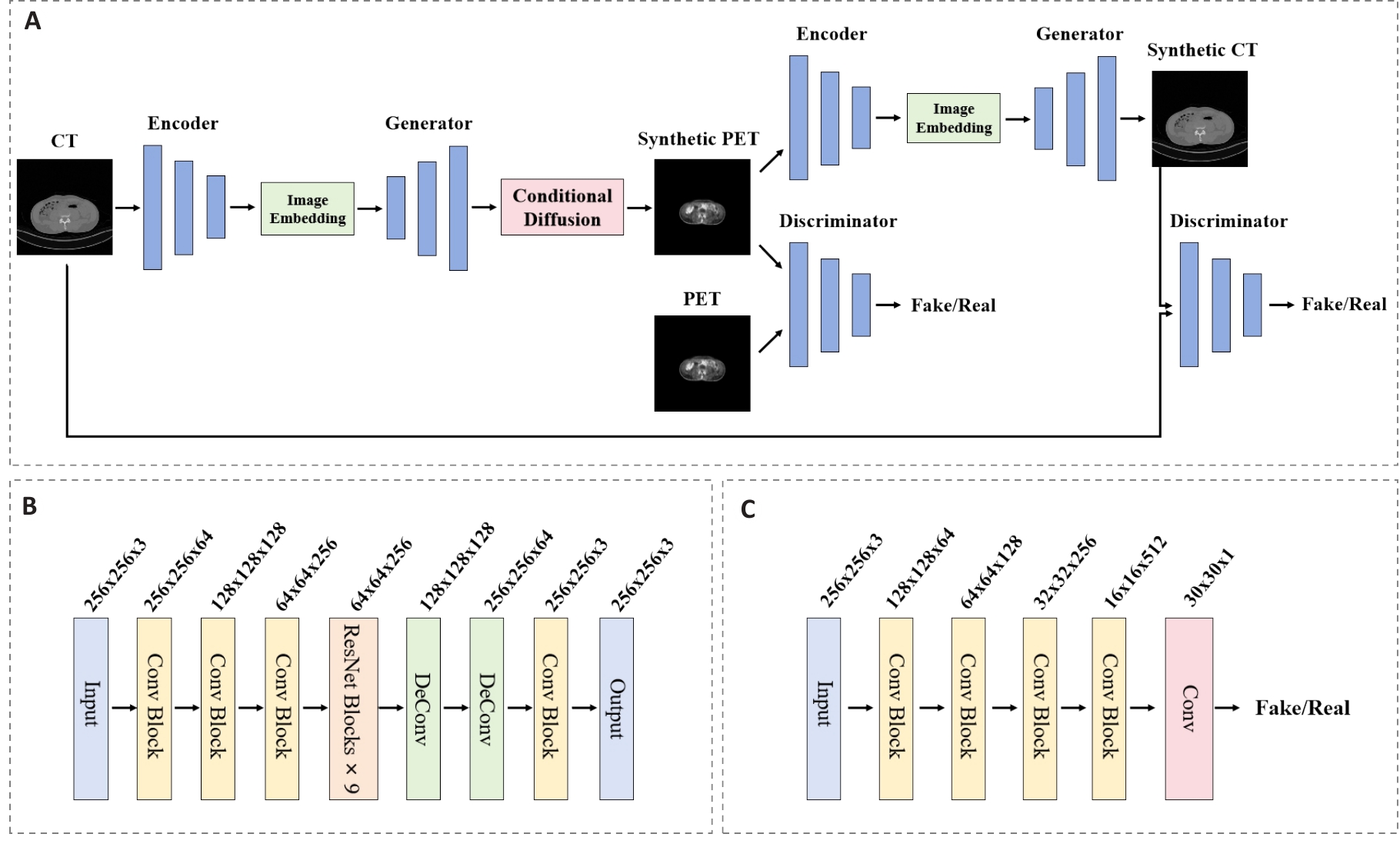
Fig.2 Architecture diagram of the cycle-consistent generative adversarial network module. A: Overall architecture of the CycleGAN establishing bidirectional mapping. B: Generator network architecture containing 9 residual blocks. C: Discriminator network architecture employing spectral normalization.
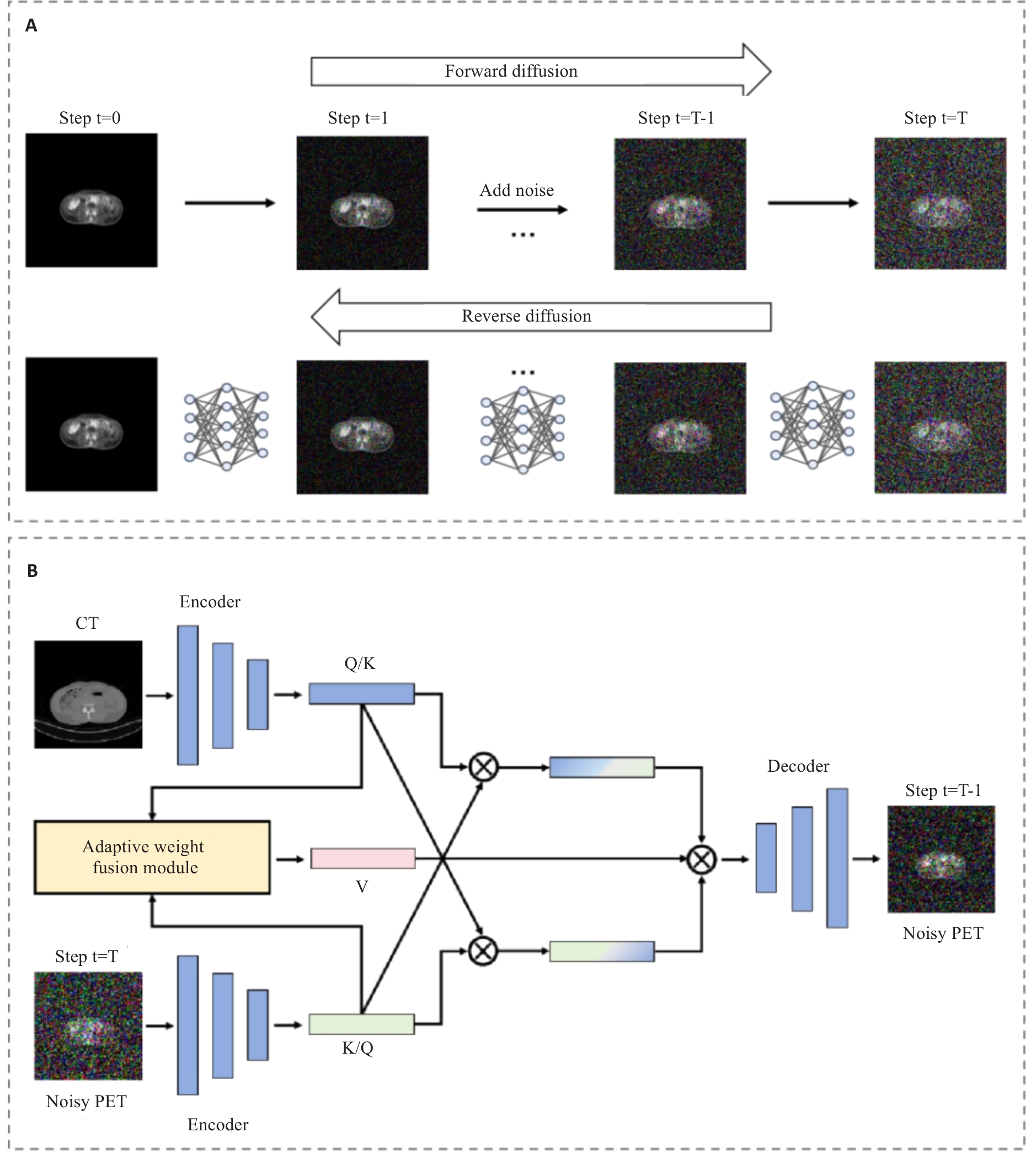
Fig.3 Cross-modal dynamic denoising method of the conditional diffusion module. A: Forward noise addition and reverse denoising reconstruction processes based on Markov chain. B: Structure of the cross-modal dynamic denoising network integrating Adaptive Weight Fusion Module and Bidirectional Cross-Attention
| Methods | PSNR | SSIM | NMSE | Average time (Epoch/s) |
|---|---|---|---|---|
| U-net[ | 21.27±1.31 | 0.787±0.031 | 0.0438±0.0062 | 57.1 |
| Pix2pix[ | 22.61±1.04 | 0.815±0.026 | 0.0359±0.0045 | 48.9 |
| CycleGAN[ | 24.33±0.91 | 0.861±0.022 | 0.0297±0.0041 | 53.4 |
| WGAN[ | 20.42±1.52 | 0.771±0.035 | 0.0481±0.0073 | 65.3 |
| DCGAN[ | 19.08±1.89 | 0.738±0.044 | 0.0541±0.0095 | 44.8 |
| DDPM[ | 23.71±0.97 | 0.849±0.024 | 0.0324±0.0042 | 197.2 |
| Ours | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 239.8 |
Tab.1 Quantitative comparison of different methods for image synthesis quality
| Methods | PSNR | SSIM | NMSE | Average time (Epoch/s) |
|---|---|---|---|---|
| U-net[ | 21.27±1.31 | 0.787±0.031 | 0.0438±0.0062 | 57.1 |
| Pix2pix[ | 22.61±1.04 | 0.815±0.026 | 0.0359±0.0045 | 48.9 |
| CycleGAN[ | 24.33±0.91 | 0.861±0.022 | 0.0297±0.0041 | 53.4 |
| WGAN[ | 20.42±1.52 | 0.771±0.035 | 0.0481±0.0073 | 65.3 |
| DCGAN[ | 19.08±1.89 | 0.738±0.044 | 0.0541±0.0095 | 44.8 |
| DDPM[ | 23.71±0.97 | 0.849±0.024 | 0.0324±0.0042 | 197.2 |
| Ours | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 239.8 |
| Methods | Dice | ASSD | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Lumbosacral spine | Ilium | Lower pelvis | Average | Lumbosacral spine | Ilium | Lower pelvis | Average | ||
| GAN+U-net[ | 0.696±0.031 | 0.652±0.039 | 0.638±0.035 | 0.662±0.035 | 4.21±0.48 | 5.08±0.63 | 5.61±0.71 | 4.97±0.61 | |
| SynSeg-Net[ | 0.718±0.027 | 0.675±0.034 | 0.661±0.032 | 0.685±0.031 | 3.89±0.44 | 4.68±0.58 | 5.15±0.64 | 4.57±0.55 | |
| Robust-Mseg[ | 0.731±0.024 | 0.693±0.030 | 0.675±0.028 | 0.700±0.027 | 3.67±0.40 | 4.35±0.52 | 4.83±0.58 | 4.28±0.50 | |
| SCM[ | 0.715±0.029 | 0.677±0.033 | 0.661±0.031 | 0.684±0.031 | 3.87±0.43 | 4.62±0.56 | 5.10±0.62 | 4.53±0.54 | |
| PEMMA[ | 0.748±0.022 | 0.709±0.026 | 0.688±0.027 | 0.715±0.025 | 3.45±0.38 | 4.12±0.49 | 4.56±0.55 | 4.04±0.47 | |
| Ours | 0.806±0.019 | 0.771±0.023 | 0.754±0.026 | 0.777±0.023 | 3.02±0.33 | 3.58±0.41 | 3.95±0.49 | 3.52±0.41 | |
Tab.2 Comparison of segmentation performance of different methods in different ABM regions.
| Methods | Dice | ASSD | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Lumbosacral spine | Ilium | Lower pelvis | Average | Lumbosacral spine | Ilium | Lower pelvis | Average | ||
| GAN+U-net[ | 0.696±0.031 | 0.652±0.039 | 0.638±0.035 | 0.662±0.035 | 4.21±0.48 | 5.08±0.63 | 5.61±0.71 | 4.97±0.61 | |
| SynSeg-Net[ | 0.718±0.027 | 0.675±0.034 | 0.661±0.032 | 0.685±0.031 | 3.89±0.44 | 4.68±0.58 | 5.15±0.64 | 4.57±0.55 | |
| Robust-Mseg[ | 0.731±0.024 | 0.693±0.030 | 0.675±0.028 | 0.700±0.027 | 3.67±0.40 | 4.35±0.52 | 4.83±0.58 | 4.28±0.50 | |
| SCM[ | 0.715±0.029 | 0.677±0.033 | 0.661±0.031 | 0.684±0.031 | 3.87±0.43 | 4.62±0.56 | 5.10±0.62 | 4.53±0.54 | |
| PEMMA[ | 0.748±0.022 | 0.709±0.026 | 0.688±0.027 | 0.715±0.025 | 3.45±0.38 | 4.12±0.49 | 4.56±0.55 | 4.04±0.47 | |
| Ours | 0.806±0.019 | 0.771±0.023 | 0.754±0.026 | 0.777±0.023 | 3.02±0.33 | 3.58±0.41 | 3.95±0.49 | 3.52±0.41 | |
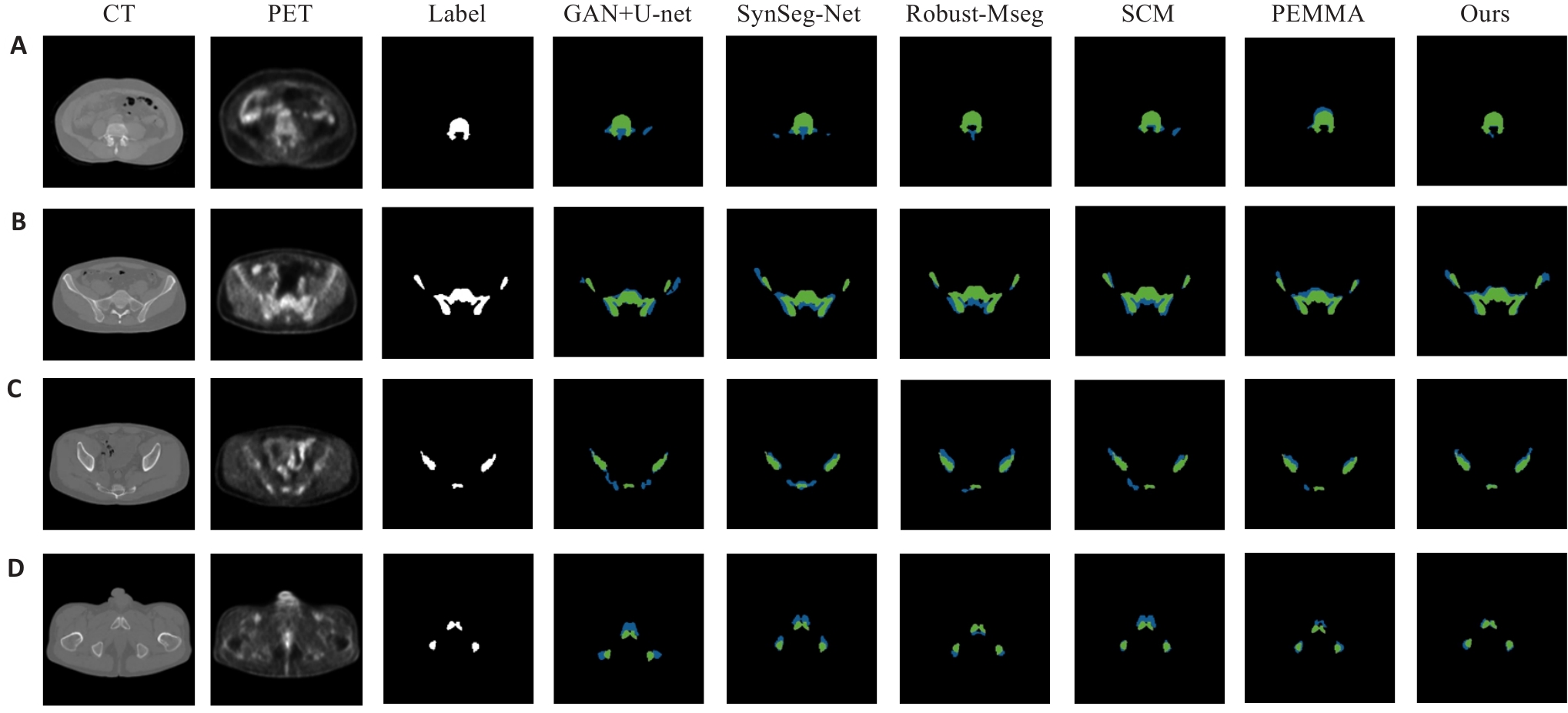
Fig.7 Visual examples of ABM segmentation results. (A) Lumbosacral spine region; (B) Ilium region; (C) Lower pelvis region; (D) Representative case with ground truth (green) and predictions (blue).
| Diffusion steps T | PSNR (dB) | SSIM | NMSE | Dice | Average time (Epoch/s) |
|---|---|---|---|---|---|
| 100 | 22.28±1.29 | 0.797±0.043 | 0.0449±0.0076 | 0.548±0.049 | 113.5 |
| 250 | 23.94±1.05 | 0.827±0.036 | 0.0383±0.0063 | 0.625±0.041 | 142.6 |
| 500 | 25.58±0.86 | 0.861±0.029 | 0.0305±0.0050 | 0.715±0.020 | 184.0 |
| 1000 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 0.777±0.023 | 239.8 |
| 1500 | 26.21±0.70 | 0.888±0.025 | 0.0238±0.0039 | 0.768±0.030 | 337.1 |
| 2000 | 26.03±0.76 | 0.884±0.028 | 0.0245±0.0043 | 0.761±0.033 | 431.0 |
Tab.3 Comparison of image generation effects and segmentation prediction under different diffusion steps
| Diffusion steps T | PSNR (dB) | SSIM | NMSE | Dice | Average time (Epoch/s) |
|---|---|---|---|---|---|
| 100 | 22.28±1.29 | 0.797±0.043 | 0.0449±0.0076 | 0.548±0.049 | 113.5 |
| 250 | 23.94±1.05 | 0.827±0.036 | 0.0383±0.0063 | 0.625±0.041 | 142.6 |
| 500 | 25.58±0.86 | 0.861±0.029 | 0.0305±0.0050 | 0.715±0.020 | 184.0 |
| 1000 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 0.777±0.023 | 239.8 |
| 1500 | 26.21±0.70 | 0.888±0.025 | 0.0238±0.0039 | 0.768±0.030 | 337.1 |
| 2000 | 26.03±0.76 | 0.884±0.028 | 0.0245±0.0043 | 0.761±0.033 | 431.0 |
| Weight strategy | Weight function | PSNR (dB) | SSIM | NMSE | Dice |
|---|---|---|---|---|---|
| Fixed weight (0.3:0.7) | 24.91±0.92 | 0.851±0.026 | 0.0339±0.0048 | 0.728±0.037 | |
| Fixed weight (0.5:0.5) | 25.07±0.84 | 0.862±0.022 | 0.0323±0.0044 | 0.745±0.033 | |
| Fixed weight (0.7:0.3) | 24.73±0.99 | 0.845±0.029 | 0.0357±0.0055 | 0.719±0.040 | |
| Linear decay | 25.58±0.75 | 0.871±0.018 | 0.0287±0.0036 | 0.763±0.026 | |
| Cosine decay | 25.84±0.70 | 0.878±0.016 | 0.0271±0.0033 | 0.769±0.024 | |
| Exponential decay | 26.08±0.66 | 0.885±0.014 | 0.0253±0.0030 | 0.772±0.023 | |
| Exponential decay+error feedback | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 0.777±0.023 |
Tab.4 Impact of different weighting strategies on model performance
| Weight strategy | Weight function | PSNR (dB) | SSIM | NMSE | Dice |
|---|---|---|---|---|---|
| Fixed weight (0.3:0.7) | 24.91±0.92 | 0.851±0.026 | 0.0339±0.0048 | 0.728±0.037 | |
| Fixed weight (0.5:0.5) | 25.07±0.84 | 0.862±0.022 | 0.0323±0.0044 | 0.745±0.033 | |
| Fixed weight (0.7:0.3) | 24.73±0.99 | 0.845±0.029 | 0.0357±0.0055 | 0.719±0.040 | |
| Linear decay | 25.58±0.75 | 0.871±0.018 | 0.0287±0.0036 | 0.763±0.026 | |
| Cosine decay | 25.84±0.70 | 0.878±0.016 | 0.0271±0.0033 | 0.769±0.024 | |
| Exponential decay | 26.08±0.66 | 0.885±0.014 | 0.0253±0.0030 | 0.772±0.023 | |
| Exponential decay+error feedback | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 | 0.777±0.023 |
| Configuration | Input modality | Dice | ASSD | PSNR (dB) | SSIM | NMSE |
|---|---|---|---|---|---|---|
| Baseline | CT only | 0.664±0.036 | 4.97±0.62 | - | - | - |
| +CycleGAN | CT+pseudo-PET | 0.702±0.028 | 4.28±0.51 | 24.35±0.81 | 0.861±0.017 | 0.0281±0.0033 |
| +Conditional diffusion | CT+enhanced-PET | 0.726±0.031 | 4.11±0.55 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
| +Multi-scale fusion | CT+enhanced-PET+advanced fusion | 0.758±0.025 | 3.76±0.44 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
| Complete method | Full pipeline with joint optimization | 0.777±0.023 | 3.52±0.41 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
Tab.5 Ablation study results
| Configuration | Input modality | Dice | ASSD | PSNR (dB) | SSIM | NMSE |
|---|---|---|---|---|---|---|
| Baseline | CT only | 0.664±0.036 | 4.97±0.62 | - | - | - |
| +CycleGAN | CT+pseudo-PET | 0.702±0.028 | 4.28±0.51 | 24.35±0.81 | 0.861±0.017 | 0.0281±0.0033 |
| +Conditional diffusion | CT+enhanced-PET | 0.726±0.031 | 4.11±0.55 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
| +Multi-scale fusion | CT+enhanced-PET+advanced fusion | 0.758±0.025 | 3.76±0.44 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
| Complete method | Full pipeline with joint optimization | 0.777±0.023 | 3.52±0.41 | 26.42±0.63 | 0.894±0.011 | 0.0235±0.0026 |
| [1] | Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020[J]. CA A Cancer J Clinicians, 2020, 70(3): 145-64. doi:10.3322/caac.21601 |
| [2] | Conroy T, Bosset JF, Etienne PL, et al. Neoadjuvant chemotherapy with FOLFIRINOX and preoperative chemoradiotherapy for patients with locally advanced rectal cancer (UNICANCER-PRODIGE 23): a multicentre, randomised, open-label, phase 3 trial[J]. Lancet Oncol, 2021, 22(5): 702-15. |
| [3] | Garcia-Aguilar J, Patil S, Gollub MJ, et al. Organ preservation in patients with rectal adenocarcinoma treated with total neoadjuvant therapy[J]. J Clin Oncol, 2022, 40(23): 2546-56. |
| [4] | Jin J, Tang Y, Hu C, et al. Multicenter, randomized, phase III trial of short-term radiotherapy plus chemotherapy versus long-term chemoradiotherapy in locally advanced rectal cancer (STELLAR)[J]. J Clin Oncol, 2022, 40(15): 1681-92. doi:10.1200/jco.2021.39.15_suppl.3510 |
| [5] | Bahadoer RR, Dijkstra EA, van Etten B, et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): a randomised, open-label, phase 3 trial[J]. Lancet Oncol, 2021, 22(1): 29-42. |
| [6] | Diefenhardt M, Ludmir EB, Hofheinz RD, et al. Association of treatment adherence with oncologic outcomes for patients with rectal cancer: a post hoc analysis of the CAO/ARO/AIO-04 phase 3 randomized clinical trial[J]. JAMA Oncol, 2020, 6(9): 1416-21. doi:10.1001/jamaoncol.2020.2394 |
| [7] | Hayman JA, Callahan JW, Herschtal A, et al. Distribution of proliferating bone marrow in adult cancer patients determined using FLT-PET imaging[J]. Int J Radiat Oncol Biol Phys, 2011, 79(3): 847-52. doi:10.1016/j.ijrobp.2009.11.040 |
| [8] | McGuire SM, Menda Y, Ponto LL, et al. A methodology for incorporating functional bone marrow sparing in IMRT planning for pelvic radiation therapy[J]. Radiother Oncol, 2011, 99(1): 49-54. doi:10.1016/j.radonc.2011.01.025 |
| [9] | Li N, Noticewala SS, Williamson CW, et al. Feasibility of atlas-based active bone marrow sparing intensity modulated radiation therapy for cervical cancer[J]. Radiother Oncol, 2017, 123(2): 325-30. doi:10.1016/j.radonc.2017.02.017 |
| [10] | Yusufaly T, Miller A, Medina-Palomo A, et al. A multi-atlas approach for active bone marrow sparing radiation therapy: implementation in the NRG-GY006 trial[J]. Int J Radiat Oncol Biol Phys, 2020, 108(5): 1240-7. doi:10.1016/j.ijrobp.2020.06.071 |
| [11] | Czernin J, Allen-Auerbach M, Nathanson D, et al. PET/CT in oncology: current status and perspectives[J]. Curr Radiol Rep, 2013, 1(3): 177-90. doi:10.1007/s40134-013-0016-x |
| [12] | Yang TJ, Oh JH, Apte A, et al. Clinical and dosimetric predictors of acute hematologic toxicity in rectal cancer patients undergoing chemoradiotherapy[J]. Radiother Oncol, 2014, 113(1): 29-34. doi:10.1016/j.radonc.2014.09.002 |
| [13] | Zhu JD, Wu H, Chen YL, et al. The correlation between the change of Hounsfield units value and Modic changes in the lumbar vertebral endplate[J]. BMC Musculoskelet Disord, 2021, 22(1): 509. doi:10.1186/s12891-021-04330-5 |
| [14] | Meyer HJ, Pönisch W, Monecke A, et al. Can diagnostic low-dose whole-body CT reflect bone marrow findings in systemic mastocytosis [J]. Anticancer Res, 2020, 40(2): 1015-22. doi:10.21873/anticanres.14036 |
| [15] | Goodsitt MM, Shenoy A, Shen JC, et al. Evaluation of dual energy quantitative CT for determining the spatial distributions of red marrow and bone for dosimetry in internal emitter radiation therapy[J]. Med Phys, 2014, 41(5): 051901. doi:10.1118/1.4870378 |
| [16] | Isola P, Zhu JY, Zhou TH, et al. Image-to-image translation with conditional adversarial networks[C]//2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). July 21-26, 2017, Honolulu, HI, USA. IEEE, 2017: 5967-76. doi:10.1109/cvpr.2017.632 |
| [17] | Salehjahromi M, Karpinets TV, Sujit SJ, et al. Synthetic PET from CT improves diagnosis and prognosis for lung cancer: Proof of concept[J]. Cell Rep Med, 2024, 5(3): 101463. doi:10.1016/j.xcrm.2024.101463 |
| [18] | Kazerouni A, Aghdam EK, Heidari M, et al. Diffusion models in medical imaging: a comprehensive survey[J]. Med Image Anal, 2023, 88: 102846. doi:10.1016/j.media.2023.102846 |
| [19] | DhariwalPrafulla, NicholAlex. Diffusion models beat GANs on image synthesis[C]. Proceedings of the 35th International Conference on Neural Information Processing Systems, 2021: 8780-94. |
| [20] | Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation[M]//Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015. Cham: Springer International Publishing, 2015: 234-41. doi:10.1007/978-3-319-24574-4_28 |
| [21] | Zhu JY, Park T, Isola P, et al. Unpaired image-to-image translation using cycle-consistent adversarial networks[C]//2017 IEEE International Conference on Computer Vision (ICCV). October 22-29, 2017, Venice, Italy. IEEE, 2017: 2242-51. doi:10.1109/iccv.2017.244 |
| [22] | Arjovsky M, Chintala S, Bottou L. Wasserstein generative adversarial networks[C]. Proceedings of the 34th International Conference on Machine Learning, 2017: 214-23. |
| [23] | Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks[J]. arXiv preprint arXiv:, 2015. |
| [24] | Huo YK, Xu ZB, Moon H, et al. SynSeg-net: synthetic segmentation without target modality ground truth[J]. IEEE Trans Med Imaging, 2018: 10.1109/TMI.2018.2876633. doi:10.1109/tmi.2018.2876633 |
| [25] | Chen C, Dou Q, Jin YM, et al. Robust multimodal brain tumor segmentation via feature disentanglement and gated fusion[M]//Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Cham: Springer International Publishing, 2019: 447-56. doi:10.1007/978-3-030-32248-9_50 |
| [26] | Diao ZS, Jiang HY, Shi TY, et al. Siamese semi-disentanglement network for robust PET-CT segmentation[J]. Expert Syst Appl, 2023, 223: 119855. doi:10.1016/j.eswa.2023.119855 |
| [27] | Saadi N, Saeed N, Yaqub M, et al. PEMMA: parameter-efficient multi-modal adaptation for medical image segmentation[M]//Medical Image Computing and Computer Assisted Intervention- MICCAI 2024. Cham: Springer Nature Switzerland, 2024: 262-71. doi:10.1007/978-3-031-72390-2_43 |
| [28] | Ponnusamy R, Zhang M, Wang Y, et al. Automatic segmentation of bone marrow lesions on MRI using a deep learning method[J]. Bioengineering (Basel), 2024, 11(4): 374. doi:10.3390/bioengineering11040374 |
| [29] | Sizikova E, Badal A, Delfino JG, et al. Synthetic data in radiological imaging: current state and future outlook[J]. Bjr|artificial Intell, 2024, 1: ubae007. doi:10.1093/bjrai/ubae007 |
| [30] | Hu C, Cao N, Li XH, et al. CBCT-to-CT synthesis using a hybrid U-Net diffusion model based on transformers and information bottleneck theory[J]. Sci Rep, 2025, 15: 10816. doi:10.1038/s41598-025-92094-6 |
| [31] | Shi YQ, Abulizi A, Wang H, et al. Diffusion models for medical image computing: a survey[J]. Tsinghua Sci Technol, 2025, 30(1): 357-83. doi:10.26599/tst.2024.9010047 |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||