南方医科大学学报 ›› 2026, Vol. 46 ›› Issue (1): 175-182.doi: 10.12122/j.issn.1673-4254.2026.01.19
赵新丽1,2( ), 王豪杰1,2, 宋银春1,2, 袁帅1,2, 张振1,2, 周星琦1,2, 李姗姗1,2, 李娴1,2(
), 王豪杰1,2, 宋银春1,2, 袁帅1,2, 张振1,2, 周星琦1,2, 李姗姗1,2, 李娴1,2( ), 李锋1,2(
), 李锋1,2( )
)
收稿日期:2025-06-22
出版日期:2026-01-20
发布日期:2026-01-16
通讯作者:
李娴,李锋
E-mail:1850466372@qq.com;Lixian0813@126.com;1583635955@qq.com
作者简介:赵新丽,在读硕士研究生,E-mail: 1850466372@qq.com
基金资助:
Xinli ZHAO1,2( ), Haojie WANG1,2, Yinchun SONG1,2, Shuai YUAN1,2, Zhen ZHANG1,2, Xingqi ZHOU1,2, Shanshan LI1,2, Xian LI1,2(
), Haojie WANG1,2, Yinchun SONG1,2, Shuai YUAN1,2, Zhen ZHANG1,2, Xingqi ZHOU1,2, Shanshan LI1,2, Xian LI1,2( ), Feng LI1,2(
), Feng LI1,2( )
)
Received:2025-06-22
Online:2026-01-20
Published:2026-01-16
Contact:
Xian LI, Feng LI
E-mail:1850466372@qq.com;Lixian0813@126.com;1583635955@qq.com
摘要:
目的 明确核糖核酸外切酶3(ERI3)在肝癌组织中的表达情况,并分析其对患者远期预后的评估价值以及对肿瘤细胞转移的影响和可能的机制。 方法 通过TCGA-LIHC数据集(377例肝癌与50例癌旁组织),采用DESeq2进行ERI3差异表达分析,结合HPA数据库免疫组化验证。通过STRING与GeneMANIA构建蛋白互作网络;利用Cox回归、KM生存分析评估预后价值;ROC曲线分析诊断效能;ssGSEA算法进行免疫浸润相关性研究;多因素Cox回归构建列线图预后模型。实验验证采用人肝癌细胞系SMMC-7721,通过敲低基因ERI3后,使用克隆形成、划痕及Transwell实验检测增殖、迁移与侵袭能力。 结果 ERI3在肝癌组织中显著高表达(P<0.001),且表达水平随TNM分期升高而递增(T1-T4:P<0.001)。高表达ERI3患者总生存期(OS,HR=2.86,95% CI:1.68-4.88,P<0.001)、疾病特异性生存期(DSS,HR=2.27,P=0.013)及无进展生存期(PFI,HR=1.83,P=0.012)均显著缩短。诊断效能分析显示ERI3的AUC达0.955(95% CI:0.931-0.978)。免疫浸润研究发现ERI3与Th2细胞呈正相关(r=0.340,P<0.001),与Th17细胞呈负相关(r=-0.284,P<0.001)。多因素Cox回归证实ERI3是独立预后因子(HR=1.987, P=0.003),据此构建的列线图预测效能良好(C-index=0.668)。体外实验表明,敲低ERI3可显著抑制SMMC-7721细胞的增殖、迁移及侵袭能力(P<0.05)。 结论 ERI3高表达可显著促进肝癌细胞增殖、迁移和侵袭,且与患者预后不良有关。
赵新丽, 王豪杰, 宋银春, 袁帅, 张振, 周星琦, 李姗姗, 李娴, 李锋. ERI3在肝癌中高表达并与患者不良预后相关[J]. 南方医科大学学报, 2026, 46(1): 175-182.
Xinli ZHAO, Haojie WANG, Yinchun SONG, Shuai YUAN, Zhen ZHANG, Xingqi ZHOU, Shanshan LI, Xian LI, Feng LI. ERI3 expression is elevated in hepatocellular carcinoma and correlates with poor patient prognosis[J]. Journal of Southern Medical University, 2026, 46(1): 175-182.
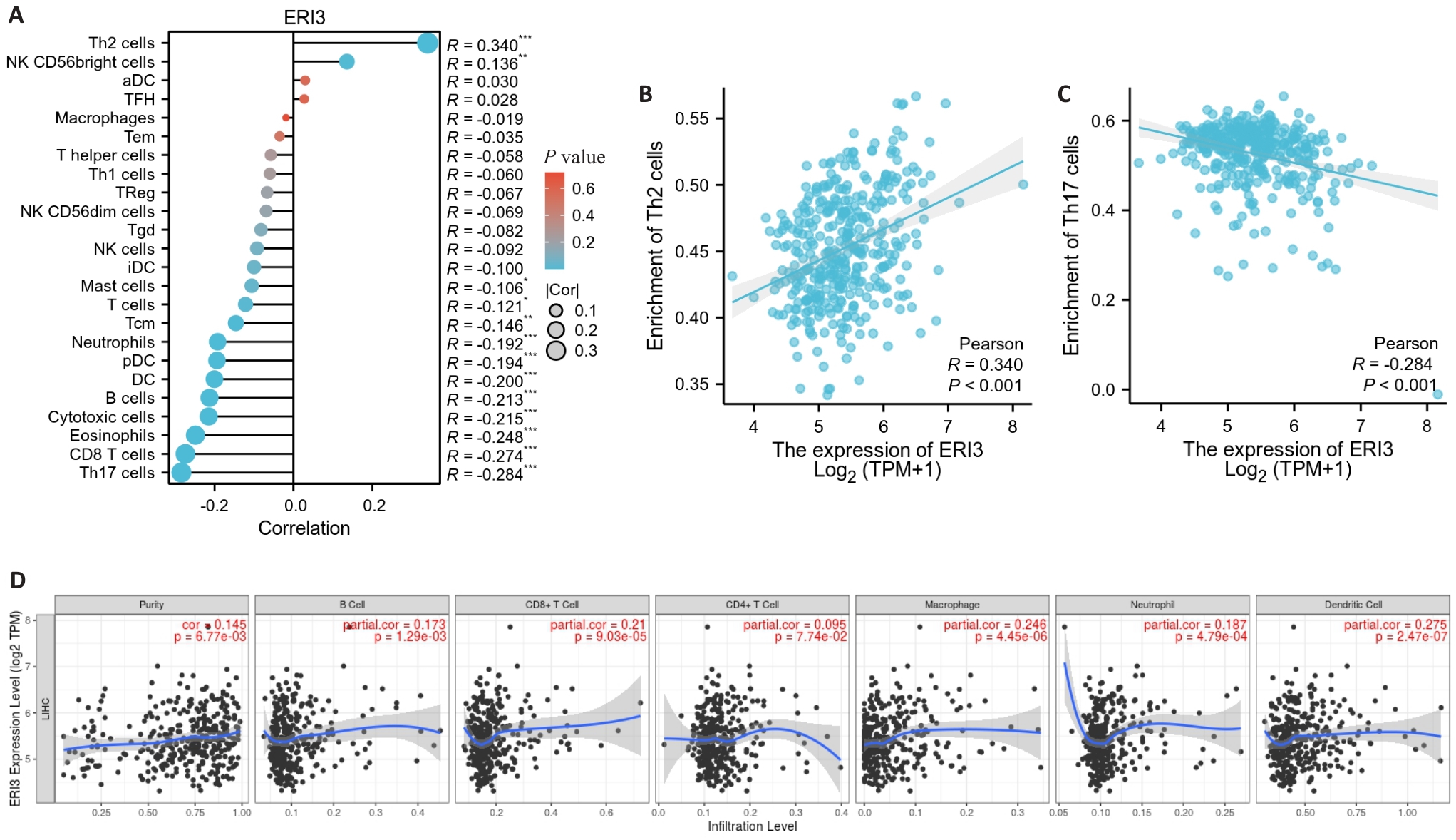
图5 免疫浸润分析
Fig.5 Tumor immune infiltration analysis. A: Lollipop chart showing the correlation between ERI3 expression and infiltration levels of 24 immune cell types. B, C: Scatter plots showing the correlation between ERI3 expression and abundance of Th2 and Th17 cells. D: Scatter plot of the correlation between ERI3 expression and B cells in HCC. n=377, *P<0.05, **P<0.01, ***P<0.001.
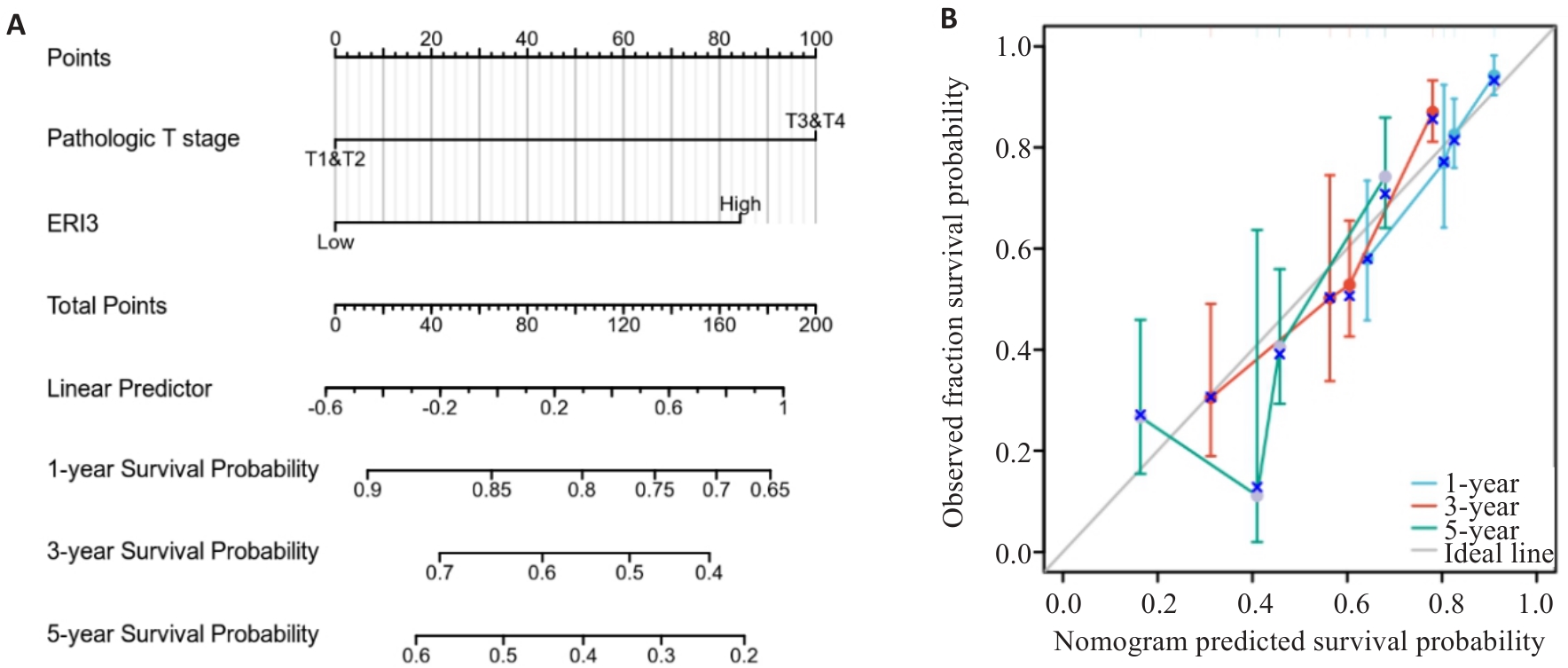
图6 临床列线图和校准曲线
Fig.6 Nomogram and calibration curve. A: Prognostic nomogram integrating ERI3 expression and clinicopathological parameters. B: Calibration curves for 1-, 3-, and 5-year survival probability of HCC patients (n=373).
| Variable | n | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Pathologic T stage | 370 | ||||
| T1&T2/T3&T4 | 277/93 | 2.598 (1.826-3.697) | <0.001 | 2.467 (1.587-3.836) | <0.001 |
| Pathologic N stage | 258 | ||||
| N0/N1 | 254/4 | 2.029 (0.497-8.281) | 0.324 | ||
| Pathologic M stage | 272 | ||||
| M0/M1 | 268/4 | 4.077 (1.281-12.973) | 0.017 | 1.739 (0.531-5.697) | 0.361 |
| Gender | 373 | ||||
| Female/Male | 121/252 | 0.793 (0.557-1.130) | 0.200 | ||
| Age | 373 | ||||
| ≤60/>60 | 177/196 | 1.205 (0.850-1.708) | 0.295 | ||
| Weight | 345 | ||||
| ≤70/>70 | 184/161 | 0.941 (0.657-1.346) | 0.738 | ||
| AFP (ng/mL) | 279 | ||||
| ≤400/>400 | 215/64 | 1.075 (0.658-1.759) | 0.772 | ||
| Fibrosis ishak score | 214 | ||||
| 0 | 75 | ||||
| 1, 2&3, 4 | 59 | 0.823 (0.436-1.554) | 0.548 | ||
| 5&6 | 80 | 0.737 (0.410-1.324) | 0.307 | ||
| Vascular invasion | 317 | ||||
| No/Yes | 208/109 | 1.344 (0.887-2.035) | 0.163 | ||
| ERI3 | 373 | ||||
| Low/High | 186/187 | 2.307 (1.608-3.310) | <0.001 | 1.987 (1.256-3.142) | 0.003 |
表1 影响肝细胞癌患者总生存期的单因素及多因素Cox比例风险回归分析
Tab.1 Univariate and multivariate Cox proportional hazards regression analysis of overall survival in HCC patients
| Variable | n | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Pathologic T stage | 370 | ||||
| T1&T2/T3&T4 | 277/93 | 2.598 (1.826-3.697) | <0.001 | 2.467 (1.587-3.836) | <0.001 |
| Pathologic N stage | 258 | ||||
| N0/N1 | 254/4 | 2.029 (0.497-8.281) | 0.324 | ||
| Pathologic M stage | 272 | ||||
| M0/M1 | 268/4 | 4.077 (1.281-12.973) | 0.017 | 1.739 (0.531-5.697) | 0.361 |
| Gender | 373 | ||||
| Female/Male | 121/252 | 0.793 (0.557-1.130) | 0.200 | ||
| Age | 373 | ||||
| ≤60/>60 | 177/196 | 1.205 (0.850-1.708) | 0.295 | ||
| Weight | 345 | ||||
| ≤70/>70 | 184/161 | 0.941 (0.657-1.346) | 0.738 | ||
| AFP (ng/mL) | 279 | ||||
| ≤400/>400 | 215/64 | 1.075 (0.658-1.759) | 0.772 | ||
| Fibrosis ishak score | 214 | ||||
| 0 | 75 | ||||
| 1, 2&3, 4 | 59 | 0.823 (0.436-1.554) | 0.548 | ||
| 5&6 | 80 | 0.737 (0.410-1.324) | 0.307 | ||
| Vascular invasion | 317 | ||||
| No/Yes | 208/109 | 1.344 (0.887-2.035) | 0.163 | ||
| ERI3 | 373 | ||||
| Low/High | 186/187 | 2.307 (1.608-3.310) | <0.001 | 1.987 (1.256-3.142) | 0.003 |
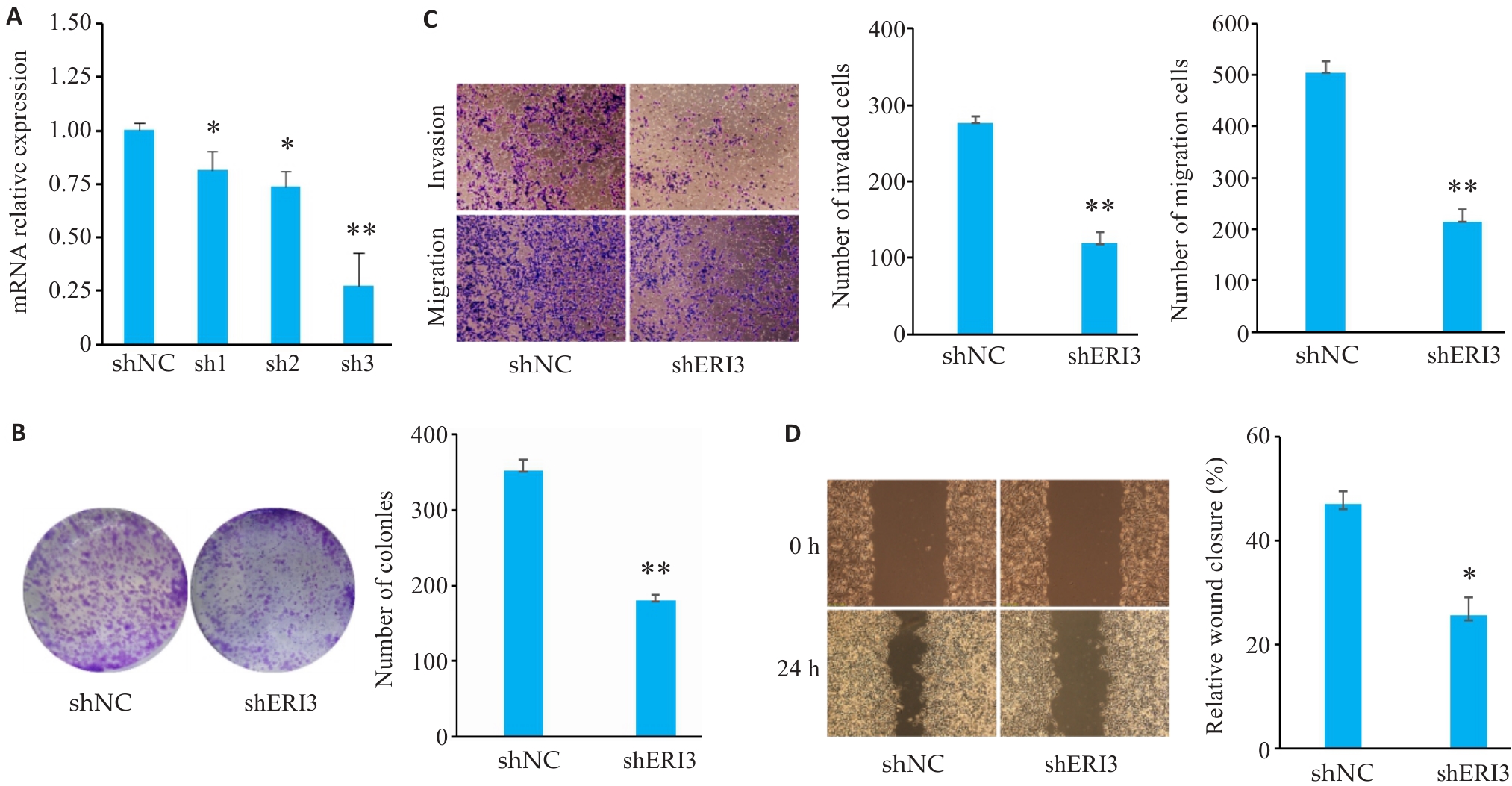
图7 敲低ERI3抑制肝癌细胞的增殖、侵袭和迁移
Fig7 ERI3 knockdown inhibits proliferation, invasion, and migration of HCC cells in vitro. A: PCR for assessing the efficiency of ERI3 knockdown. B: Colony formation assay for assessing cell proliferation. C: Transwell assay for assessing cell migration and invasion (Original magnification: ×10). D: ERI3 knockdown of ERI3 impairs wound healing in HCC Cells (×200). n=3, *P<0.05, **P<0.01 vs shNC group.
| [1] | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020[J]. CA A Cancer J Clinicians, 2020, 70(1): 7-30. doi:10.3322/caac.21590 |
| [2] | Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-49. doi:10.3322/caac.21660 |
| [3] | Sartorius K, Sartorius B, Aldous C, et al. Global and country underestimation of hepatocellular carcinoma (HCC) in 2012 and its implications[J]. Cancer Epidemiol, 2015, 39(3): 284-90. doi:10.1016/j.canep.2015.04.006 |
| [4] | Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015[J]. CA A Cancer J Clinicians, 2016, 66(2): 115-32. doi:10.3322/caac.21338 |
| [5] | de Lope CR, Tremosini S, Forner A, et al. Management of HCC[J]. J Hepatol, 2012, 56: S75-87. doi:10.1016/s0168-8278(12)60009-9 |
| [6] | Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2016, 2: 16018. doi:10.1038/nrdp.2016.18 |
| [7] | Aravalli RN, Steer CJ. Immune-mediated therapies for liver cancer[J]. Genes: Basel, 2017, 8(2): E76. doi:10.3390/genes8020076 |
| [8] | Yang H, Li G, Qiu G. Bioinformatics analysis using ATAC-seq and RNA-seq for the identification of 15 gene signatures associated with the prediction of prognosis in hepatocellular carcinoma[J]. Front Oncol, 2021, 11: 726551. doi:10.3389/fonc.2021.726551 |
| [9] | Liu FY, Feng XM, Ji XL, et al. Cluster classification and clinical prognostic modeling based on m6A RNA methylation regulators in liver cancer[J]. Zhonghua Gan Zang Bing Za Zhi, 2022, 30(9): 962-9. |
| [10] | Liu J, Qu J, Xu L, et al. Prediction of liver cancer prognosis based on immune cell marker genes[J]. Front Immunol, 2023, 14: 1147797. doi:10.3389/fimmu.2023.1147797 |
| [11] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. doi:10.1186/s13059-014-0550-8 |
| [12] | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data[J]. Bioinformatics, 2010, 26(1): 139-40. doi:10.1093/bioinformatics/btp616 |
| [13] | Liu JF, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics[J]. Cell, 2018, 173(2): 400-16. |
| [14] | Hänzelmann S, Castelo R, Guinney J. GSVA gene set variation analysis for microarray and RNA-seq data[J]. BMC Bioinformatics, 2013, 14: 7. doi:10.1186/1471-2105-14-7 |
| [15] | Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer[J]. Immunity, 2013, 39(4): 782-95. doi:10.1016/j.immuni.2013.10.003 |
| [16] | COHEN J. Statistical power analysis for the behavioral sciences [M]. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates, 1988. |
| [17] | Mukaka MM. Statistics corner: a guide to appropriate use of correlation coefficient in medical research[J]. Malawi Med J, 2012, 24(3): 69-71. |
| [18] | Meyer T. Treatment of advanced hepatocellular carcinoma: beyond sorafenib[J]. Lancet Gastroenterol Hepatol, 2018, 3(4): 218-20. doi:10.1016/s2468-1253(17)30255-8 |
| [19] | Currie E, Schulze A, Zechner R, et al. Cellular fatty acid metabolism and cancer[J]. Cell Metab, 2013, 18(2): 153-61. doi:10.1016/j.cmet.2013.05.017 |
| [20] | Bechmann LP, Hannivoort RA, Gerken G, et al. The interaction of hepatic lipid and glucose metabolism in liver diseases[J]. J Hepatol, 2012, 56(4): 952-64. doi:10.1016/j.jhep.2011.08.025 |
| [21] | Li X, Sun R, Liu RP. Natural products in licorice for the therapy of liver diseases: Progress and future opportunities[J]. Pharmacol Res, 2019, 144: 210-26. doi:10.1016/j.phrs.2019.04.025 |
| [22] | Thor Snaebjornsson M, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer[J]. Cell Metab, 2020, 31(1): 62-76. doi:10.1016/j.cmet.2019.11.010 |
| [23] | Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond[J]. Curr Opin Cell Biol, 2007, 19(2): 215-22. doi:10.1016/j.ceb.2007.02.004 |
| [24] | Li C, Sheng LN, Sun G, et al. The application of ultraviolet-induced photo-crosslinking in edible film preparation and its implication in food safety[J]. LWT, 2020, 131: 109791. doi:10.1016/j.lwt.2020.109791 |
| [25] | Wang H, Xu M, Zhang T, et al. PYCR1 promotes liver cancer cell growth and metastasis by regulating IRS1 expression through lactylation modification[J]. Clin Transl Med, 2024, 14(10): e70045. doi:10.1002/ctm2.70045 |
| [26] | Ho V, Pelland-St-Pierre L, Gravel S, et al. Endocrine disruptors: Challenges and future directions in epidemiologic research[J]. Environ Res, 2022, 204(pt a): 111969. doi:10.1016/j.envres.2021.111969 |
| [27] | Wang X, Zhang XY, Liao NQ, et al. Identification of ribosome biogenesis genes and subgroups in ischaemic stroke[J]. Front Immunol, 2024, 15: 1449158. doi:10.3389/fimmu.2024.1449158 |
| [28] | Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer[J]. Nat Rev Cancer, 2021, 21(6): 345-59. doi:10.1038/s41568-021-00347-z |
| [29] | Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: nomenclature and experimental guidelines[J]. Immunity, 2014, 41(1): 14-20. doi:10.1016/j.immuni.2014.06.008 |
| [30] | Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated macrophages as treatment targets in oncology[J]. Nat Rev Clin Oncol, 2017, 14(7): 399-416. doi:10.1038/nrclinonc.2016.217 |
| [31] | Sunakawa Y, Stintzing S, Cao S, et al. Variations in genes regulating tumor-associated macrophages (TAMs) to predict outcomes of bevacizumab-based treatment in patients with metastatic colorectal cancer: results from TRIBE and FIRE3 trials[J]. Ann Oncol, 2015, 26(12): 2450-6. doi:10.1093/annonc/mdv474 |
| [32] | Xiang X, Wang J, Lu D, et al. Targeting tumor-associated macrophages to synergize tumor immunotherapy[J]. Signal Transduct Target Ther, 2021, 6(1): 75. doi:10.1038/s41392-021-00484-9 |
| [33] | Guo MJ, Sheng WY, Yuan X, et al. Neutrophils as promising therapeutic targets in pancreatic cancer liver metastasis[J]. Int Immunopharmacol, 2024, 140: 112888. doi:10.1016/j.intimp.2024.112888 |
| [1] | 沙桐, 王文研, 宣佳斌, 吴洁, 石能贤, 何劲, 胡鸿彬, 张耀元. 基于Th1/Th2细胞因子检测的脓毒症免疫状态分型及预后分析:一项回顾性研究[J]. 南方医科大学学报, 2026, 46(1): 6-22. |
| [2] | 冯志惠, 李文月, 张铭修, 王培培, 帅阳阳, 张宏. lncRNA HClnc1通过靶向RBBP5/KAT2B复合物增强ODC1转录促进肝癌细胞生长和转移[J]. 南方医科大学学报, 2025, 45(9): 1919-1926. |
| [3] | 王莹, 李静, 王伊迪, 华明钰, 胡玮彬, 张晓智. 原发性肝癌患者的临床结局与治疗反应预测模型:基于失巢凋亡和免疫基因[J]. 南方医科大学学报, 2025, 45(9): 1967-1979. |
| [4] | 张瑜, 李海涛, 潘玉卿, 曹杰贤, 翟丽, 张曦. MZB1基因在泛癌中的表达及其与免疫浸润及预后的关系[J]. 南方医科大学学报, 2025, 45(9): 2006-2018. |
| [5] | 王子良, 陈孝华, 杨晶晶, 严晨, 张志郅, 黄炳轶, 赵萌, 刘嵩, 葛思堂, 左芦根, 陈德利. 高表达SURF4通过抑制紧密连接蛋白表达促进胃癌细胞的恶性生物学行为[J]. 南方医科大学学报, 2025, 45(8): 1732-1742. |
| [6] | 黄亚婷, 王振友. 基于耦合扩散的肿瘤转移与骨髓来源的抑制细胞相互作用模型[J]. 南方医科大学学报, 2025, 45(8): 1768-1776. |
| [7] | 于滢, 涂丽, 刘洋, 宋雪翼, 邵倩倩, 唐小龙. TGF-β通过miR-23a-3p/IRF1轴下调主要组织相容性复合体I类表达促进肝癌免疫逃逸[J]. 南方医科大学学报, 2025, 45(7): 1397-1408. |
| [8] | 庞金龙, 赵新丽, 张振, 王豪杰, 周星琦, 杨玉梅, 李姗姗, 常小强, 李锋, 李娴. 皮肤黑色素瘤中MMRN2高表达促进肿瘤细胞的侵袭和迁移并与不良预后相关[J]. 南方医科大学学报, 2025, 45(7): 1479-1489. |
| [9] | 吴璇, 方家敏, 韩玮玮, 陈琳, 孙菁, 金齐力. 高表达PRELID1促进胃癌细胞上皮间质转化并与不良预后相关[J]. 南方医科大学学报, 2025, 45(7): 1535-1542. |
| [10] | 王康, 李海宾, 余靖, 孟源, 张虹丽. ELFN1高表达是结肠癌的预后生物标志物并促进结肠癌细胞的增殖转移[J]. 南方医科大学学报, 2025, 45(7): 1543-1553. |
| [11] | 翁诺舟, 谭彬, 曾文涛, 古家宇, 翁炼基, 郑克鸿. 过表达RGL1通过激活CDC42/RAC1复合体上调运动型黏着斑组装促进结直肠癌转移[J]. 南方医科大学学报, 2025, 45(5): 1031-1038. |
| [12] | 宋添力, 王一民, 孙童, 刘绪, 黄胜, 冉云. 正肝方对二乙基亚硝胺诱导的肝癌大鼠的抗癌作用及机制:基于激活Hippo/YAP通路[J]. 南方医科大学学报, 2025, 45(4): 799-809. |
| [13] | 岳雅清, 牟召霞, 王希波, 刘艳. Aurora-A过表达通过激活NF-κBp65/ARPC4信号轴促进宫颈癌细胞的侵袭和转移[J]. 南方医科大学学报, 2025, 45(4): 837-843. |
| [14] | 张毅, 沈昱, 万志强, 陶嵩, 柳亚魁, 王栓虎. CDKN3高表达促进胃癌细胞的迁移和侵袭:基于调控p53/NF-κB信号通路和抑制胃癌细胞凋亡[J]. 南方医科大学学报, 2025, 45(4): 853-861. |
| [15] | 黄晴晴, 张文静, 张小凤, 王炼, 宋雪, 耿志军, 左芦根, 王月月, 李静, 胡建国. 高表达MYO1B促进胃癌细胞增殖、迁移和侵袭并与患者的不良预后有关[J]. 南方医科大学学报, 2025, 45(3): 622-631. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||