南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (9): 1967-1979.doi: 10.12122/j.issn.1673-4254.2025.09.16
• • 上一篇
王莹1( ), 李静1, 王伊迪2, 华明钰1, 胡玮彬1, 张晓智1(
), 李静1, 王伊迪2, 华明钰1, 胡玮彬1, 张晓智1( )
)
收稿日期:2025-02-19
出版日期:2025-09-20
发布日期:2025-09-28
通讯作者:
张晓智
E-mail:wangying123456@stu.xjtu.edu.cn;zhangxiaozhi@xjtu.edu.cn
作者简介:王 莹,在读硕士研究生,E-mail: wangying123456@stu.xjtu.edu.cn
基金资助:
Ying WANG1( ), Jing LI1, Yidi WANG2, Mingyu HUA1, Weibin HU1, Xiaozhi ZHANG1(
), Jing LI1, Yidi WANG2, Mingyu HUA1, Weibin HU1, Xiaozhi ZHANG1( )
)
Received:2025-02-19
Online:2025-09-20
Published:2025-09-28
Contact:
Xiaozhi ZHANG
E-mail:wangying123456@stu.xjtu.edu.cn;zhangxiaozhi@xjtu.edu.cn
摘要:
目的 利用生物信息学方法构建原发性肝癌(PLC)的预后模型。 方法 在The Cancer Genome Atlas (TCGA)数据库中纳入了404名PLC患者。使用单变量 Cox 回归及LASSO-Cox方法构建失巢凋亡和免疫相关基因(DAIs)的预后模型。Kaplan-Meier法和受试者工作特征曲线用于评估模型的预测能力,建立列线图以方便临床应用。进行基因集富集分析(GSEA)揭示相关通路,并使用CIBERSORT和TIDE方法进一步探讨DAIs与肿瘤免疫微环境之间的关系。使用“pRRophetic”R包来计算PLC药物的半数最大抑制浓度(IC50)。检测了PLC患者组织中SEMA7A的表达。 结果 构建并验证了基于7个DAIs(NR4A3、SEMA7A、IL11、AR、BIRC5、EGF和SPP1)的预后模型,在TCGA训练队列和GEO验证队列(GSE14520)中均发现一致的结果,即低风险组患者具有更好的临床预后和良好的免疫状态。将该预后模型与临床信息相结合,生成复合列线图以促进临床实践。体细胞突变分析显示TTN、TP53和CTNNB1突变占总突变比例最大,低风险-低TMB组生存率更高。药物敏感性分析显示高风险与低风险组之间以及TP53突变与非突变之间对化疗药物的敏感性存在差异。免疫组化结果显示SEMA7A在PLC中的表达高于癌旁正常肝组织(P<0.05)。 结论 本研究建立了一个基于DAIs的新的预测模型,用于预测PLC患者的临床结局和治疗反应,为个体化治疗提供新思路。
王莹, 李静, 王伊迪, 华明钰, 胡玮彬, 张晓智. 原发性肝癌患者的临床结局与治疗反应预测模型:基于失巢凋亡和免疫基因[J]. 南方医科大学学报, 2025, 45(9): 1967-1979.
Ying WANG, Jing LI, Yidi WANG, Mingyu HUA, Weibin HU, Xiaozhi ZHANG. Construction and verification of a prognostic model combining anoikis and immune prognostic signatures for primary liver cancer[J]. Journal of Southern Medical University, 2025, 45(9): 1967-1979.
| Clinical | Total (n=576) | TCGA (n=351) | GEO (n=225) |
|---|---|---|---|
| Gender [n (%)] | |||
| Female | 149 (25.87) | 119 (33.90) | 30 (13.33) |
| Male | 427 (74.13) | 232 (66.10) | 195 (86.67) |
| Age (mean year) | 55.78 | 59.06 | 50.66 |
| Age (median year) | 56 (16-82) | 61 (16-82) | 50 (21-77) |
| Stage [n (%)] | |||
| I | 273 (47.40) | 177 (50.43) | 96 (42.67) |
| II | 163 (28.30) | 85 (24.22) | 78 (34.67) |
| III | 7 (1.22) | 4 (1.14) | 3 (1.33) |
| IIIA | 89 (15.45) | 60 (17.09) | 29 (12.89) |
| IIIB | 23 (3.99) | 8 (2.28) | 15 (6.67) |
| IIIC | 13 (2.26) | 9 (2.56) | 4 (1.78) |
| IV | 2 (0.35) | 2 (0.57) | 0 (0.00) |
| IVA | 2 (0.35) | 2 (0.57) | 0 (0.00) |
| IVB | 4 (0.69) | 4 (1.14) | 0 (0.00) |
表1 TCGA和GEO数据库中肝癌患者的临床资料
Tab.1 Clinical data of liver cancer patients in TCGA and GEO databases
| Clinical | Total (n=576) | TCGA (n=351) | GEO (n=225) |
|---|---|---|---|
| Gender [n (%)] | |||
| Female | 149 (25.87) | 119 (33.90) | 30 (13.33) |
| Male | 427 (74.13) | 232 (66.10) | 195 (86.67) |
| Age (mean year) | 55.78 | 59.06 | 50.66 |
| Age (median year) | 56 (16-82) | 61 (16-82) | 50 (21-77) |
| Stage [n (%)] | |||
| I | 273 (47.40) | 177 (50.43) | 96 (42.67) |
| II | 163 (28.30) | 85 (24.22) | 78 (34.67) |
| III | 7 (1.22) | 4 (1.14) | 3 (1.33) |
| IIIA | 89 (15.45) | 60 (17.09) | 29 (12.89) |
| IIIB | 23 (3.99) | 8 (2.28) | 15 (6.67) |
| IIIC | 13 (2.26) | 9 (2.56) | 4 (1.78) |
| IV | 2 (0.35) | 2 (0.57) | 0 (0.00) |
| IVA | 2 (0.35) | 2 (0.57) | 0 (0.00) |
| IVB | 4 (0.69) | 4 (1.14) | 0 (0.00) |
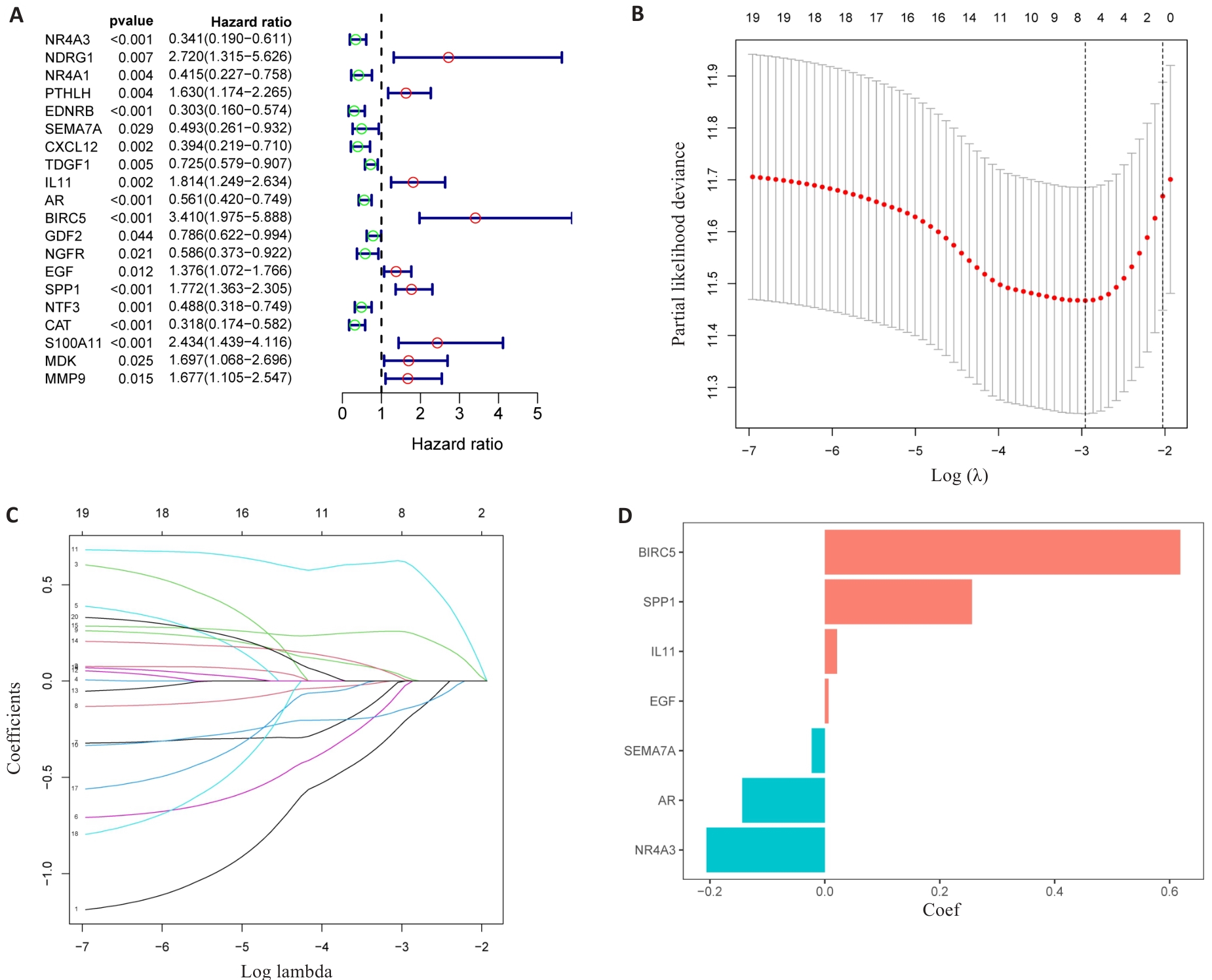
图3 DAIs的识别
Fig.3 Identification of the DAIs. A: Forest maps of the predictive power of 20 characteristic genes. B: LASSO regression analysis based on DAIs. C: LASSO coefficient of DAIs gene in PLC. D: LASSO gene coefficient histogram.
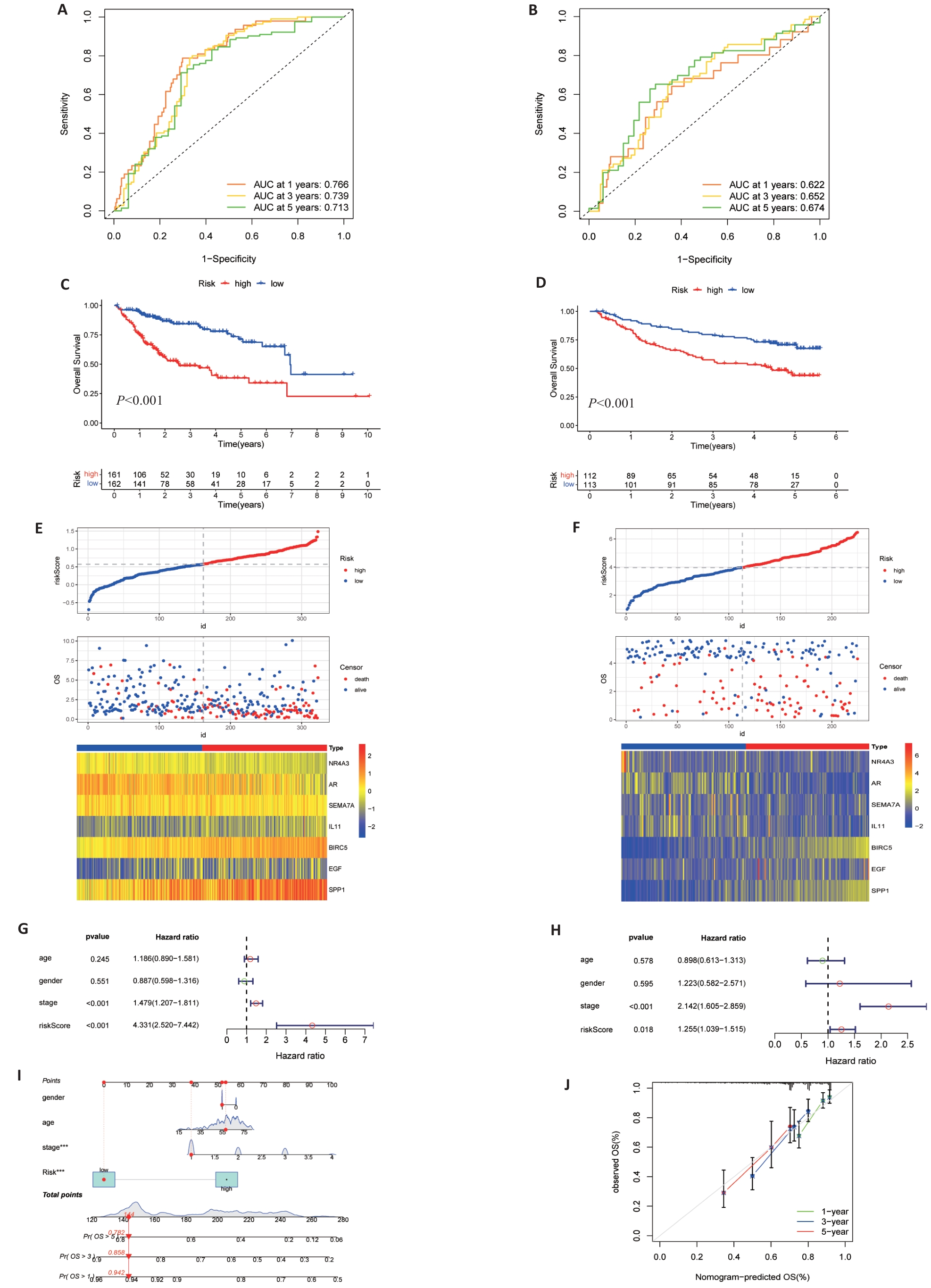
图4 风险模型的验证
Fig.4 Validation of risk signature. A, B: Prediction of time-dependent ROC for 1, 3, and 5-year OS in TCGA cohort and GEO cohort. C, D: Kaplan-Meier survival curves showed that there were differences in OS between high and low TCGA and GEO groups. E, F: Risk scores, survival status and heat maps of 7 DAIs between high and low groups of TCGA (E) and GSE14520 (F). G, H: Multivariate cox regression analysis of TCGA (G) and GEO (H) risk scores and clinical data. I: Nomogram of clinical data and risk groups of TCGA. J: Standard curves showing good nomogram accuracy.
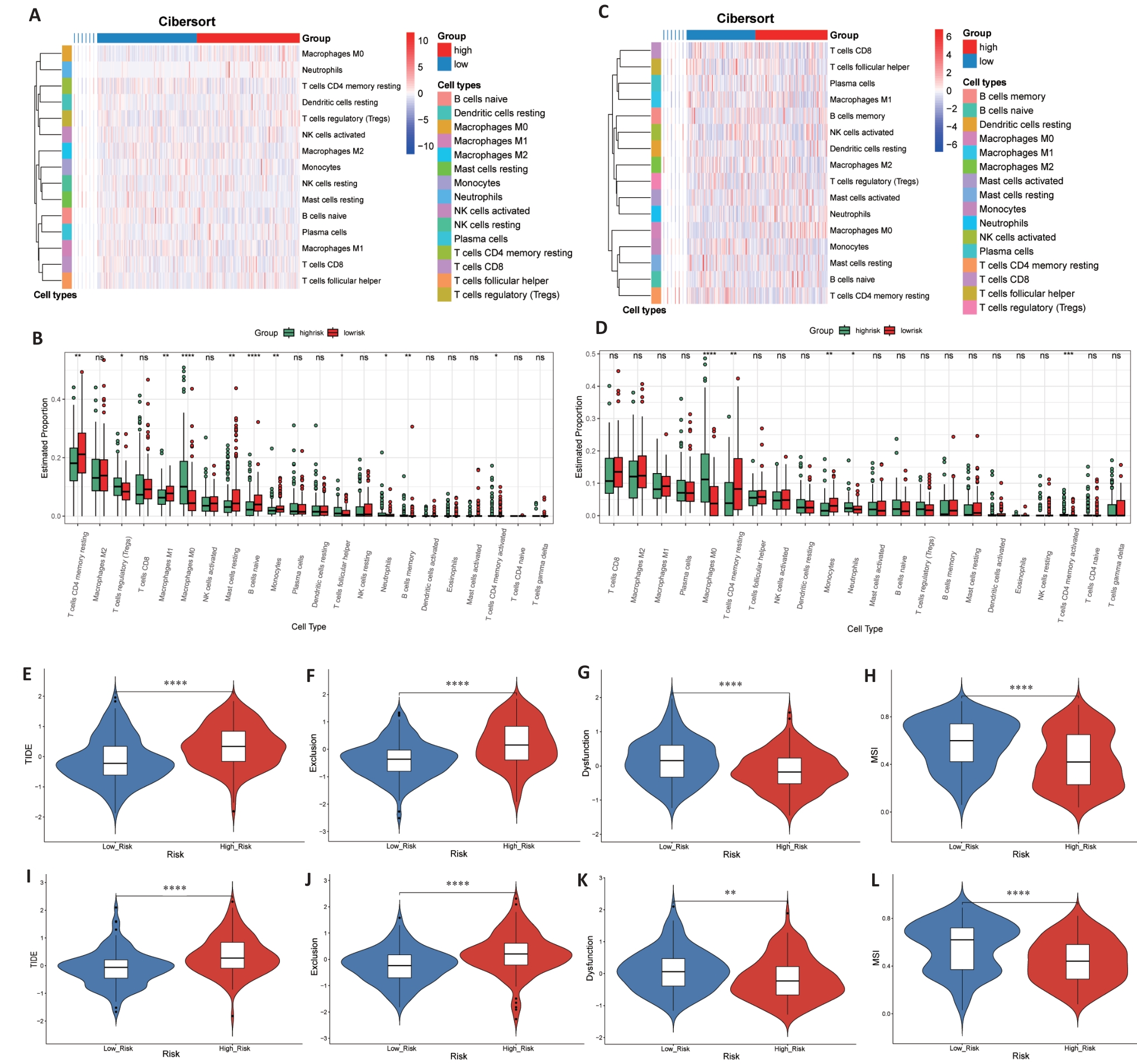
图6 免疫状态和免疫治疗反应预测
Fig. 6 Immune status and prediction of response to immunotherapy. A-D: Heatmaps and enrichment scores of 22 immune cells in TCGA (A, B) and GEO (C, D) high-risk and low-risk groups. E-L: TIDE score, rejection score, dysfunction score and MSI score between TCGA (E-H) and GEO (I-L) groups. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
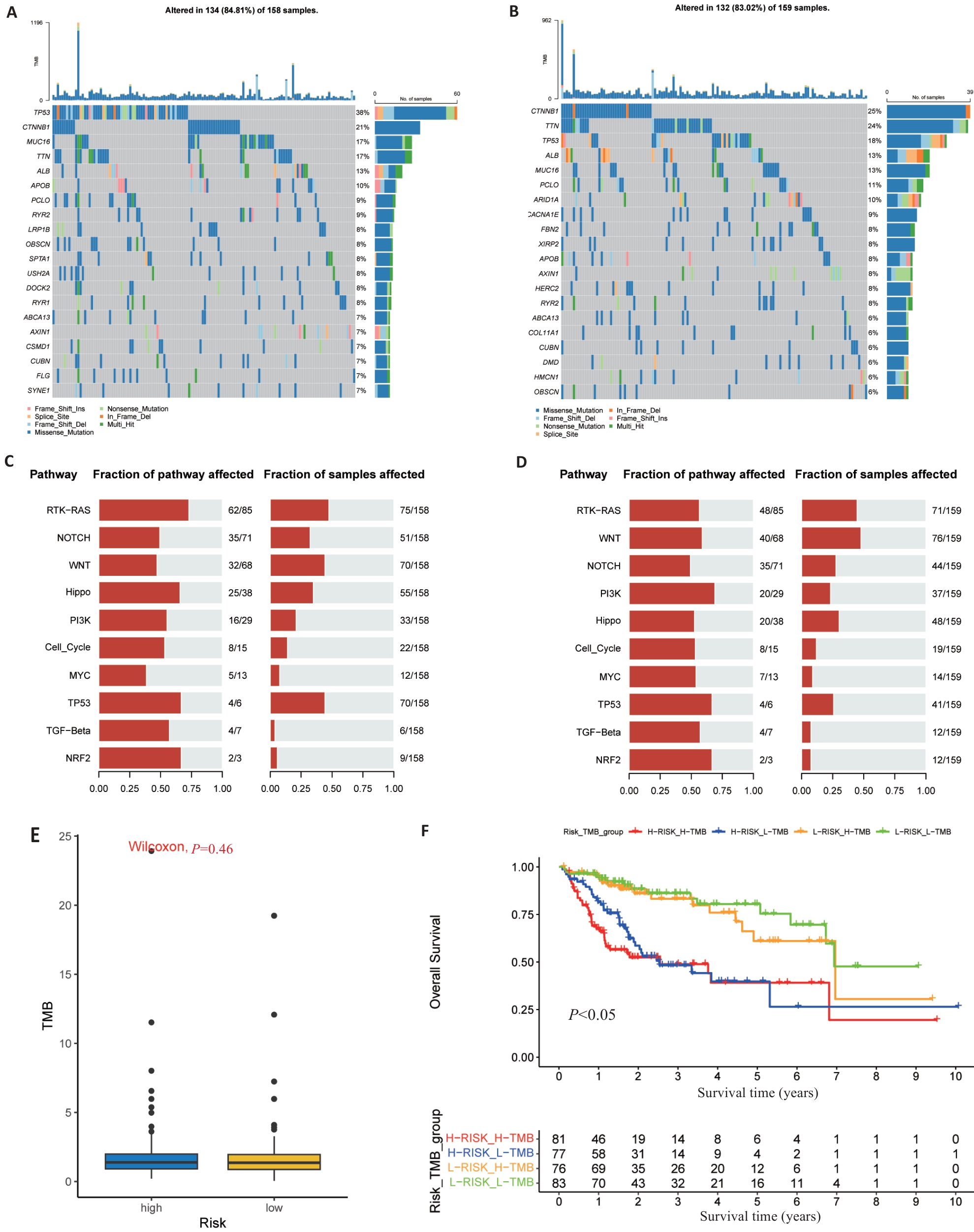
图7 基于风险模型的体细胞突变和TMB分析
Fig.7 Somatic mutations and TMB in risk signature. A, B: The waterfall map shows the top 20 genes with the highest mutation frequency in the high-risk group (A) and low-risk group (B) of TCGA. C, D: The bar chart shows the top 10 high mutation pathways in the high-risk (C) and low-risk (D) TCGA groups. E: Box chart showing TMB score comparison between high- and low-risk groups. F: Kaplan-Meier survival curves show OS differences between groups classified according to TMB and TCGA risk.
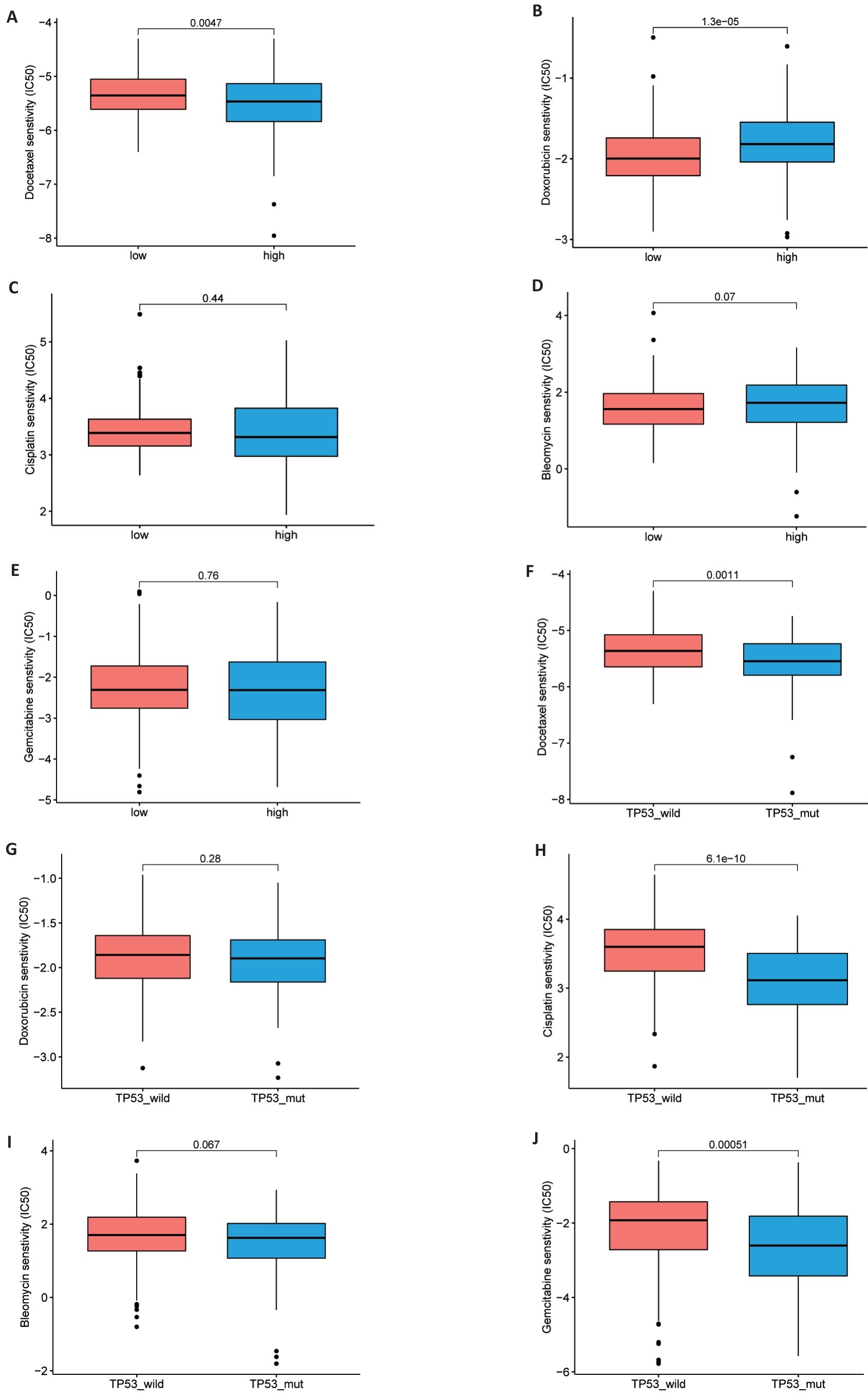
图8 药物敏感性分析
Fig. 8 Drug sensitivity analysis. A-J: Comparison of half maximum inhibitory concentrations (IC50) of docetaxel, adriamycin, cisplatin and bleomycin in high and low risk TCGA groups (A-E) and TP53 mutant and non-mutant groups (F-J).
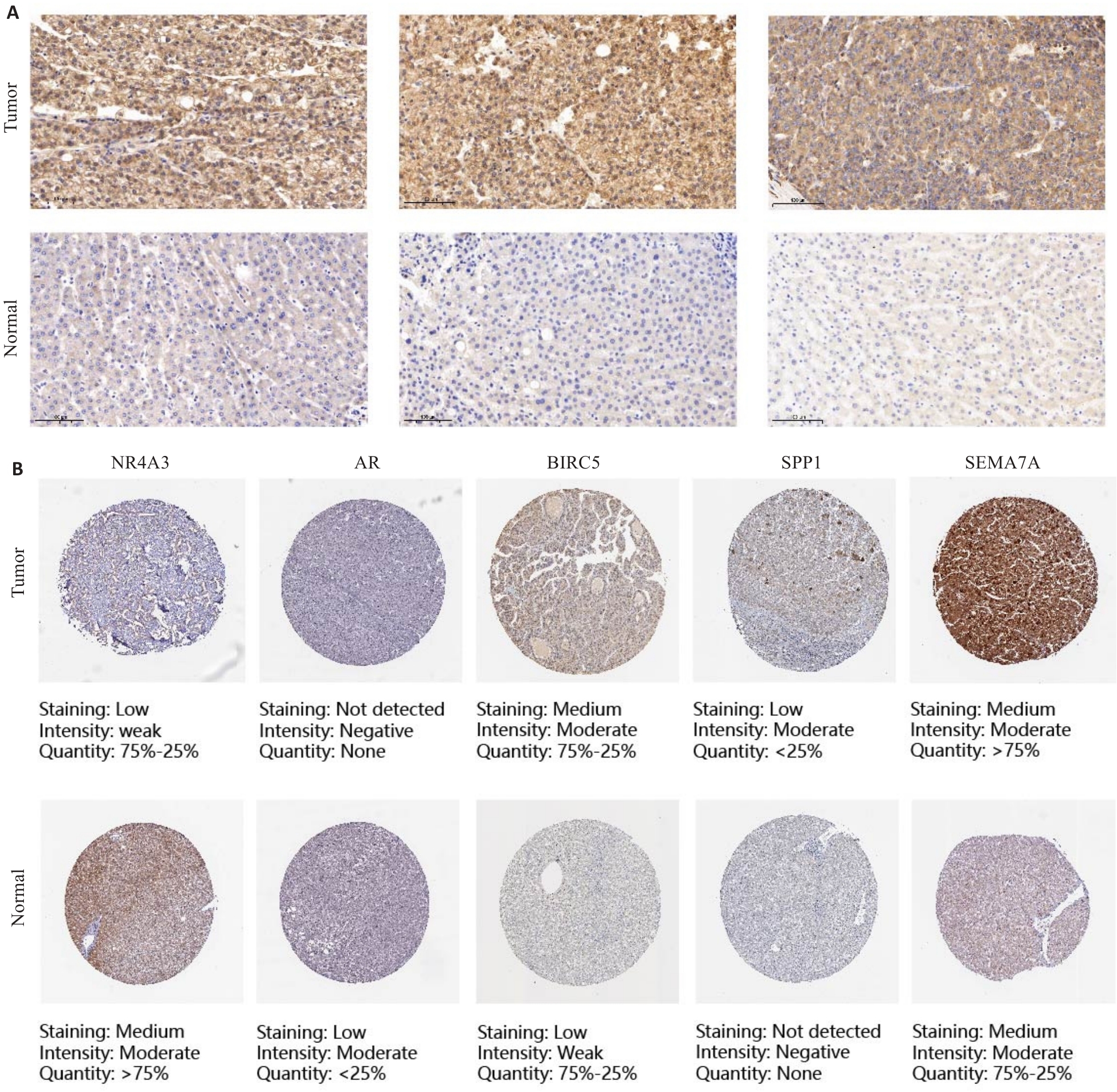
图9 IHC和HPA网站验证了DAIs在PLC及邻近组织中的表达
Fig.9 Immunohistochemistry and HPA website for verifying the expression of DAIs in primary liver cancer and adjacent tissues (Original magnification: ×100). A: Representative immunohistochemical images of SEMA7A in PLC tissues and adjacent tissues. B: Representative immunohistochemical images of NR4A3, AR, BIRC5, SPP1 and SEMA7A in PLC tissues and normal tissues on the HPA website.
| Gene | Expression level | Tissue | χ2 | P | |
|---|---|---|---|---|---|
| Cancer | Normal | ||||
| SEMA7A | High expression | 28 | 7 | 7.72 | <0.01 |
| Low expression | 6 | 9 | |||
表2 SEMA7A在肝癌组织及邻近正常肝组织中的表达情况
Tab.2 Expression of SEMA7A in liver cancer tissues and adjacent normal liver tissues (n)
| Gene | Expression level | Tissue | χ2 | P | |
|---|---|---|---|---|---|
| Cancer | Normal | ||||
| SEMA7A | High expression | 28 | 7 | 7.72 | <0.01 |
| Low expression | 6 | 9 | |||
| [1] | Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-63. doi:10.3322/caac.21834 |
| [2] | Gilmore AP. Anoikis[J]. Cell Death Differ, 2005, 12(S2): 1473-7. doi:10.1038/sj.cdd.4401723 |
| [3] | Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: Beyond the migration of single cells[J]. J Biol Chem, 2020, 295(8): 2495-505. doi:10.1074/jbc.rev119.007759 |
| [4] | Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression[J]. Biochim Biophys Acta, 2013, 1833(12): 3481-98. doi:10.1016/j.bbamcr.2013.06.026 |
| [5] | Kakavandi E, Shahbahrami R, Goudarzi H, et al. Anoikis resistance and oncoviruses[J]. J Cell Biochem, 2018, 119(3): 2484-91. doi:10.1002/jcb.26363 |
| [6] | Adeshakin FO, Adeshakin AO, Afolabi LO, et al. Mechanisms for modulating anoikis resistance in cancer and the relevance of metabolic reprogramming[J]. Front Oncol, 2021, 11: 626577. doi:10.3389/fonc.2021.626577 |
| [7] | Guadamillas MC, Cerezo A, Del Pozo MA. Overcoming anoikis: pathways to anchorage-independent growth in cancer[J]. J Cell Sci, 2011, 124(Pt 19): 3189-97. doi:10.1242/jcs.072165 |
| [8] | Angell H, Galon J. From the immune contexture to the immunoscore: the role of prognostic and predictive immune markers in cancer[J]. Curr Opin Immunol, 2013, 25(2): 261-7. doi:10.1016/j.coi.2013.03.004 |
| [9] | Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point[J]. Nature, 2017, 541(7637): 321-30. doi:10.1038/nature21349 |
| [10] | El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial[J]. Lancet, 2017, 389(10088): 2492-502. doi:10.1016/s0140-6736(17)31046-2 |
| [11] | Zhao ZH, Li C, Peng Y, et al. Construction of an original anoikis-related prognostic model closely related to immune infiltration in gastric cancer[J]. Front Genet, 2023, 13: 1087201. doi:10.3389/fgene.2022.1087201 |
| [12] | Chen Z, Liu X, Zhu ZJ, et al. A novel anoikis-related prognostic signature associated with prognosis and immune infiltration landscape in clear cell renal cell carcinoma[J]. Front Genet, 2022, 13: 1039465. doi:10.3389/fgene.2022.1039465 |
| [13] | Chen S, Gu JM, Zhang QF, et al. Development of biomarker signatures associated with anoikis to predict prognosis in endometrial carcinoma patients[J]. J Oncol, 2021, 2021: 3375297. doi:10.1155/2021/3375297 |
| [14] | Sun ZZ, Zhao YQ, Wei Y, et al. Identification and validation of an anoikis-associated gene signature to predict clinical character, stemness, IDH mutation, and immune filtration in glioblastoma[J]. Front Immunol, 2022, 13: 939523. doi:10.3389/fimmu.2022.939523 |
| [15] | Moujalled D, Strasser A, Liddell JR. Molecular mechanisms of cell death in neurological diseases[J]. Cell Death Differ, 2021, 28(7): 2029-44. doi:10.1038/s41418-021-00814-y |
| [16] | Chen YT, Huang WR, Ouyang J, et al. Identification of anoikis-related subgroups and prognosis model in liver hepatocellular carcinoma[J]. Int J Mol Sci, 2023, 24(3): 2862. doi:10.3390/ijms24032862 |
| [17] | Chi H, Jiang PY, Xu K, et al. A novel anoikis-related gene signature predicts prognosis in patients with head and neck squamous cell carcinoma and reveals immune infiltration[J]. Front Genet, 2022, 13: 984273. doi:10.3389/fgene.2022.984273 |
| [18] | Pan Q, Luo G, Qu JQ, et al. A homozygous R148W mutation in Semaphorin 7A causes progressive familial intrahepatic cholestasis[J]. EMBO Mol Med, 2021, 13(11): e14563. doi:10.15252/emmm.202114563 |
| [19] | Li X, Xie WL, Pan Q, et al. Semaphorin 7A interacts with nuclear factor NF-kappa-B p105 via integrin β1 and mediates inflammation[J]. Cell Commun Signal, 2023, 21(1): 24. doi:10.1186/s12964-022-01024-w |
| [20] | Yeh CM, Chang LY, Lin SH, et al. Epigenetic silencing of the NR4A3 tumor suppressor, by aberrant JAK/STAT signaling, predicts prognosis in gastric cancer[J]. Sci Rep, 2016, 6: 31690. doi:10.1038/srep31690 |
| [21] | Fedorova O, Petukhov A, Daks A, et al. Orphan receptor NR4A3 is a novel target of p53 that contributes to apoptosis[J]. Oncogene, 2019, 38(12): 2108-22. doi:10.1038/s41388-018-0566-8 |
| [22] | Wang HH, Guo QN, Nampoukime KB, et al. Long non-coding RNA LINC00467 drives hepatocellular carcinoma progression via inhibiting NR4A3[J]. J Cell Mol Med, 2020, 24(7): 3822-36. doi:10.1111/jcmm.14942 |
| [23] | Shi DM, Dong SS, Zhou HX, et al. Genomic and transcriptomic profiling reveals key molecules in metastatic potentials and organ-tropisms of hepatocellular carcinoma[J]. Cell Signal, 2023, 104: 110565. doi:10.1016/j.cellsig.2022.110565 |
| [24] | Wang DY, Zheng XH, Fu BQ, et al. Hepatectomy promotes recurrence of liver cancer by enhancing IL-11-STAT3 signaling[J]. EBioMedicine, 2019, 46: 119-32. doi:10.1016/j.ebiom.2019.07.058 |
| [25] | Yu LD, Wang S, Lin XJ, et al. microRNA-124a inhibits cell proliferation and migration in liver cancer by regulating interleukin-11[J]. Mol Med Rep, 2018, 17(3): 3972-8. |
| [26] | Sun RF, Zhao CY, Chen S, et al. Androgen receptor stimulates hexokinase 2 and induces glycolysis by PKA/CREB signaling in hepatocellular carcinoma[J]. Dig Dis Sci, 2021, 66(3): 802-13. doi:10.1007/s10620-020-06229-y |
| [27] | Xu RZ, Lin LB, Zhang B, et al. Identification of prognostic markers for hepatocellular carcinoma based on the epithelial-mesenchymal transition-related gene BIRC5 [J]. BMC Cancer, 2021, 21(1): 687. doi:10.1186/s12885-021-08390-7 |
| [28] | Zhang LY, Yuan LY, Li DH, et al. Identification of potential prognostic biomarkers for hepatocellular carcinoma[J]. J Gastrointest Oncol, 2022, 13(2): 812-21. doi:10.21037/jgo-22-303 |
| [29] | Wang X, Liang C, Yao X, et al. PKM2-induced the phosphorylation of histone H3 contributes to EGF-mediated PD-L1 transcription in HCC[J]. Front Pharmacol, 2020, 11: 577108. doi:10.3389/fphar.2020.577108 |
| [30] | Zhao HL, Chen Q, Alam A, et al. The role of osteopontin in the progression of solid organ tumour[J]. Cell Death Dis, 2018, 9: 356. doi:10.1038/s41419-018-0391-6 |
| [31] | Wang JQ, Hao FJ, Fei XC, et al. SPP1 functions as an enhancer of cell growth in hepatocellular carcinoma targeted by miR-181c[J]. Am J Transl Res, 2019, 11(11): 6924-37. |
| [32] | Eun JW, Yoon JH, Ahn HR, et al. Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib[J]. Cancer Commun (Lond), 2023, 43(4): 455-79. doi:10.1002/cac2.12414 |
| [33] | Fu JX, Li KR, Zhang WB, et al. Large-scale public data reuse to model immunotherapy response and resistance[J]. Genome Med, 2020, 12(1): 21. doi:10.1186/s13073-020-0721-z |
| [34] | Hou ZQ, Liu J, Jin ZX, et al. Use of chemotherapy to treat hepatocellular carcinoma[J]. Biosci Trends, 2022, 16(1): 31-45. doi:10.5582/bst.2022.01044 |
| [1] | 冯志惠, 李文月, 张铭修, 王培培, 帅阳阳, 张宏. lncRNA HClnc1通过靶向RBBP5/KAT2B复合物增强ODC1转录促进肝癌细胞生长和转移[J]. 南方医科大学学报, 2025, 45(9): 1919-1926. |
| [2] | 陈鑫源, 吴成挺, 李瑞迪, 潘雪芹, 张耀丹, 陶俊宇, 林才志. 双术汤通过P53/SLC7A11/GPX4通路诱导胃癌细胞铁死亡[J]. 南方医科大学学报, 2025, 45(7): 1363-1371. |
| [3] | 于滢, 涂丽, 刘洋, 宋雪翼, 邵倩倩, 唐小龙. TGF-β通过miR-23a-3p/IRF1轴下调主要组织相容性复合体I类表达促进肝癌免疫逃逸[J]. 南方医科大学学报, 2025, 45(7): 1397-1408. |
| [4] | 庞金龙, 赵新丽, 张振, 王豪杰, 周星琦, 杨玉梅, 李姗姗, 常小强, 李锋, 李娴. 皮肤黑色素瘤中MMRN2高表达促进肿瘤细胞的侵袭和迁移并与不良预后相关[J]. 南方医科大学学报, 2025, 45(7): 1479-1489. |
| [5] | 王康, 李海宾, 余靖, 孟源, 张虹丽. ELFN1高表达是结肠癌的预后生物标志物并促进结肠癌细胞的增殖转移[J]. 南方医科大学学报, 2025, 45(7): 1543-1553. |
| [6] | 宋添力, 王一民, 孙童, 刘绪, 黄胜, 冉云. 正肝方对二乙基亚硝胺诱导的肝癌大鼠的抗癌作用及机制:基于激活Hippo/YAP通路[J]. 南方医科大学学报, 2025, 45(4): 799-809. |
| [7] | 高志, 吴傲, 胡仲翔, 孙培养. 类风湿性关节炎中氧化应激与免疫浸润的生物信息学分析[J]. 南方医科大学学报, 2025, 45(4): 862-870. |
| [8] | 邹金华, 王惠, 张冬艳. SLC1A5通过促进M2型巨噬细胞极化促进肝癌进展[J]. 南方医科大学学报, 2025, 45(2): 269-284. |
| [9] | 许怀文, 翁丽, 薛鸿. CXCL12可作为2型糖尿病合并慢性阻塞性肺疾病的潜在治疗靶点[J]. 南方医科大学学报, 2025, 45(1): 100-109. |
| [10] | 徐朦, 陈丽娜, 吴金玉, 刘丽丽, 施美, 周灏, 张国梁. “白花蛇舌草-半枝莲”治疗原发性肝癌的机制研究:基于网络药理学、分子对接及体外实验验证[J]. 南方医科大学学报, 2025, 45(1): 80-89. |
| [11] | 叶梦楠, 武鸿美, 梅琰, 张庆玲. CREM在胃癌中高表达并与患者的不良预后相关[J]. 南方医科大学学报, 2024, 44(9): 1776-1782. |
| [12] | 张力苹, 刘喜娟, 胡潇, 王嘉丽, 余锡贺, 栗国梁, 游海敏, 张启周, 张海波. 经动脉化疗栓塞续贯肝动脉灌注化疗联合TKI和PD-1单抗在晚期肝癌一线治疗中的疗效观察[J]. 南方医科大学学报, 2024, 44(9): 1831-1838. |
| [13] | 纪凯, 于冠宇, 周乐其, 张天帅, 凌潜龙, 满文江, 朱冰, 张卫. HNRNPA1基因在结直肠癌组织中高表达及其潜在的诊断和治疗价值[J]. 南方医科大学学报, 2024, 44(9): 1685-1695. |
| [14] | 何华星, 刘璐琳, 刘颖茵, 陈纳川, 孙素霞. 丁酸钠与索拉非尼可能通过YAP诱导铁死亡协同抑制肝癌细胞增殖[J]. 南方医科大学学报, 2024, 44(7): 1425-1430. |
| [15] | 陈莉莉, 吴天宇, 张铭, 丁子夏, 张妍, 杨依清, 郑佳倩, 张小楠. 类风湿关节炎的潜在生物标志物及其免疫调控机制:基于GEO数据库[J]. 南方医科大学学报, 2024, 44(6): 1098-1108. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||