Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (9): 1809-1817.doi: 10.12122/j.issn.1673-4254.2025.09.01
Liying ZHANG( ), Tongzhen ZHANG, Xin ZHAO
), Tongzhen ZHANG, Xin ZHAO
Received:2025-04-09
Accepted:2025-05-17
Online:2025-09-20
Published:2025-09-28
Contact:
Liying ZHANG
E-mail:540185181@qq.com
Supported by:Liying ZHANG, Tongzhen ZHANG, Xin ZHAO. Diagnostic value of morphological features of breast lesions on DWI and T2WI assessed using Breast Imaging Reporting and Data System lexicon descriptors[J]. Journal of Southern Medical University, 2025, 45(9): 1809-1817.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.09.01
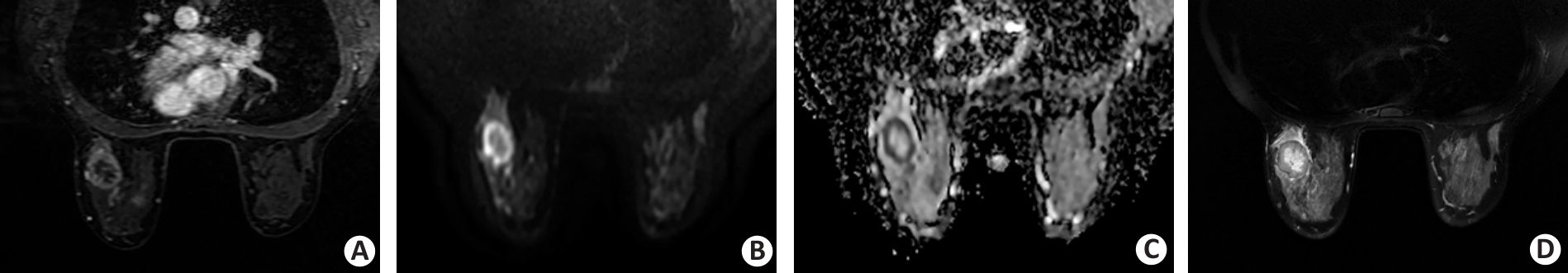
Fig.2 A 38-year-old female patient with invasive ductal breast carcinoma in the left breast. A: Dynamic contrast-enhanced image shows a round uncircumscribed mass with rim enhancement in the left breast. B, C: Diffusion-weighted image (DWI) acquired by using a b value of 800 s/mm2 shows a round, circumscribed, and high-intensity mass. The internal pattern of combined assessment of DWI and apparent diffusion coefficient is considered a rim-like pattern. D: T2-weighted image reveals the corresponding lesion as a high-intensity area surrounded by a low-intensity rim.
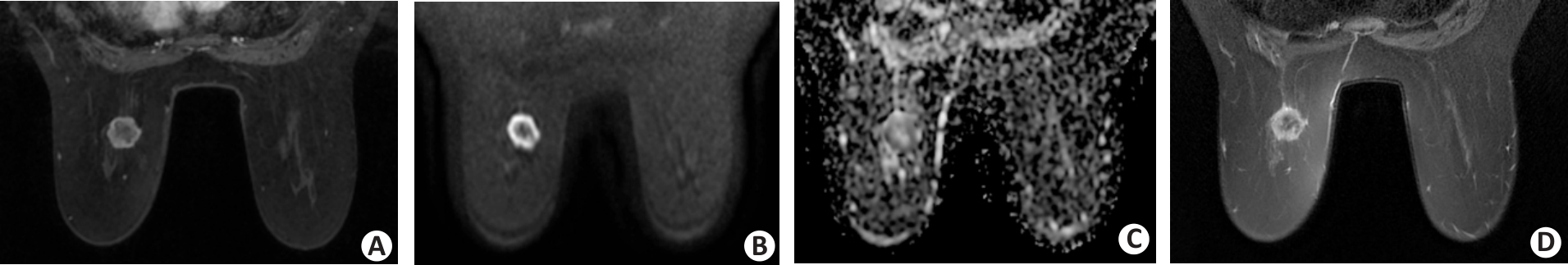
Fig.3 A 58-year-old female patient with invasive ductal breast carcinoma in the left breast. A: The dynamic contrast-enhanced image shows a round circumscribed mass with rim enhancement in the left breast. B, C: DWI acquired by using a b value of 800 s/mm2 shows a round, circumscribed, and high-intensity mass. The internal pattern of combined assessment of DWI and apparent diffusion coefficient is considered a rim-like pattern. D: T2-weighted imaging reveals the corresponding lesion as a low-intensity area surrounded by a high-intensity rim.
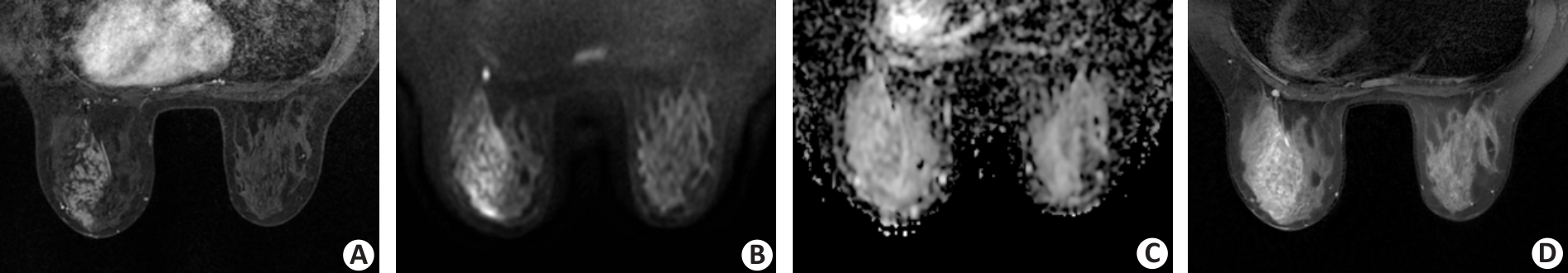
Fig.4 A 34-year-old female patient with ductal carcinoma in situ in the left breast. A: The dynamic contrast-enhanced image shows a segmental non-mass lesion with clustered ring pattern enhancement in the left breast. B, C: DWI acquired by using a b value of 800 s/mm2 shows a segmental high-intensity non-mass lesion extending from the nipple to the chest wall region anteriorly. The internal pattern of combined assessment of DWI and apparent diffusion coefficient is considered a clustered ring pattern. D: T2-weighted imaging reveals the corresponding lesion as a segmental, heterogeneous, and slight high-intensity non-mass lesion.
| Variable | Malignant (n=299) | Benign (n=95) | |||
|---|---|---|---|---|---|
| Mass (n=239) | Non-mass (n=60) | Mass (n=82) | Non-mass (n=13) | ||
| Age (year, Mean±SD) | 48.8±9.8 | 50.5±10.6 | 37.9±11.5 | 35.2±9.0 | |
| Lesion size (mm, Mean±SD) | 25.8±13.1 | 53.6±20.8 | 23.7±11.8 | 49.6±24.4 | |
| Histological type | |||||
| Invasive ductal carcinoma | 210 | 36 | |||
| Invasive lobular carcinoma | 5 | 3 | |||
| Mucinous carcinoma | 7 | 0 | |||
| Papillary carcinoma | 7 | 2 | |||
| Apocrine carcinoma | 0 | 1 | |||
| Metaplastic carcinomas | 2 | 0 | |||
| Ductal carcinoma in situ | 8 | 18 | |||
| Fibroadenoma | 54 | 0 | |||
| Fibrocystic changes | 7 | 0 | |||
| Papilloma | 6 | 2 | |||
| Sclerosing adenosis | 4 | 1 | |||
| Phyllodes tumors | 6 | 0 | |||
| Inflammatory changes | 5 | 10 | |||
Tab.1 Clinicopathological characteristics of the patients
| Variable | Malignant (n=299) | Benign (n=95) | |||
|---|---|---|---|---|---|
| Mass (n=239) | Non-mass (n=60) | Mass (n=82) | Non-mass (n=13) | ||
| Age (year, Mean±SD) | 48.8±9.8 | 50.5±10.6 | 37.9±11.5 | 35.2±9.0 | |
| Lesion size (mm, Mean±SD) | 25.8±13.1 | 53.6±20.8 | 23.7±11.8 | 49.6±24.4 | |
| Histological type | |||||
| Invasive ductal carcinoma | 210 | 36 | |||
| Invasive lobular carcinoma | 5 | 3 | |||
| Mucinous carcinoma | 7 | 0 | |||
| Papillary carcinoma | 7 | 2 | |||
| Apocrine carcinoma | 0 | 1 | |||
| Metaplastic carcinomas | 2 | 0 | |||
| Ductal carcinoma in situ | 8 | 18 | |||
| Fibroadenoma | 54 | 0 | |||
| Fibrocystic changes | 7 | 0 | |||
| Papilloma | 6 | 2 | |||
| Sclerosing adenosis | 4 | 1 | |||
| Phyllodes tumors | 6 | 0 | |||
| Inflammatory changes | 5 | 10 | |||
| Parameter | DCE | DWI | T2WI | |||||
|---|---|---|---|---|---|---|---|---|
| ICC (95% CI) | Level of concordance | ICC (95% CI) | Level of concordance | ICC (95% CI) | Level of concordance | |||
| Mass-like lesions | ||||||||
| Shape | 0.908 (0.887-0.926) | Excellent | 0.910 (0.888-0.928) | Excellent | 0.941 (0.926-0.953) | Excellent | ||
| Margin | 0.890 (0.865-0.911) | Good | 0.837 (0.786-0.875) | Good | 0.759 (0.708-0.801) | Good | ||
| Internal patterns | 0.871 (0.841-0.895) | Good | 0.907 (0.886-0.925) | Excellent | 0.833 (0.796-0.864) | Good | ||
| Signal intensity | - | - | 0.897 (0.872-0.917) | Good | 0.934 (0.918-0.948) | Excellent | ||
| Non-mass-like lesions | ||||||||
| Distribution | 0.991 (0.985-0.994) | Excellent | 0.824 (0.734-0.886) | Good | 0.791 (0.687-0.863) | Good | ||
| Internal patterns | 0.789 (0.684-0.862) | Good | 0.844 (0.763-0.899) | Good | 0.802 (0.703-0.871) | Good | ||
| Signal intensity | - | - | 0.834 (0.748-0.892) | Good | 0.846 (0.765-0.900) | Good | ||
Tab.2 Interclass correlation coefficient (ICC) for the qualitative parameters analyzed
| Parameter | DCE | DWI | T2WI | |||||
|---|---|---|---|---|---|---|---|---|
| ICC (95% CI) | Level of concordance | ICC (95% CI) | Level of concordance | ICC (95% CI) | Level of concordance | |||
| Mass-like lesions | ||||||||
| Shape | 0.908 (0.887-0.926) | Excellent | 0.910 (0.888-0.928) | Excellent | 0.941 (0.926-0.953) | Excellent | ||
| Margin | 0.890 (0.865-0.911) | Good | 0.837 (0.786-0.875) | Good | 0.759 (0.708-0.801) | Good | ||
| Internal patterns | 0.871 (0.841-0.895) | Good | 0.907 (0.886-0.925) | Excellent | 0.833 (0.796-0.864) | Good | ||
| Signal intensity | - | - | 0.897 (0.872-0.917) | Good | 0.934 (0.918-0.948) | Excellent | ||
| Non-mass-like lesions | ||||||||
| Distribution | 0.991 (0.985-0.994) | Excellent | 0.824 (0.734-0.886) | Good | 0.791 (0.687-0.863) | Good | ||
| Internal patterns | 0.789 (0.684-0.862) | Good | 0.844 (0.763-0.899) | Good | 0.802 (0.703-0.871) | Good | ||
| Signal intensity | - | - | 0.834 (0.748-0.892) | Good | 0.846 (0.765-0.900) | Good | ||
| Lesion type | DCE | P | DWI | P | T2WI | P | |||
|---|---|---|---|---|---|---|---|---|---|
| Benign | Malignant | Benign | Malignant | Benign | Malignant | ||||
| Mass-like lesions (n=321) | |||||||||
| Shape | |||||||||
| Oval | 33 (40.2) | 17 (7.1) | <0.001 | 34 (41.5) | 27 (11.3) | <0.001 | 33 (40.2) | 19 (7.9) | <0.001 |
| Round | 28 (34.1) | 115 (48.1) | 29 (35.4) | 111 (46.4) | 30 (36.6) | 120 (50.2) | |||
| Irregular | 21 (25.6) | 107 (44.8) | 19 (23.2) | 101 (42.3) | 19 (23.2) | 100 (41.8) | |||
| Margin | |||||||||
| Circumscribed | 48 (58.5) | 59 (24.7) | <0.001 | 50 (61.0) | 100 (41.8) | 0.003 | 49 (59.8) | 63 (26.4) | <0.001 |
| Not circumscribed | 34 (41.5) | 180 (75.3) | 32 (39.0) | 139 (58.2) | 33 (40.2) | 176 (73.6) | |||
| Internal patterns | |||||||||
| Homogeneous | 11 (13.4) | 25 (10.5) | <0.001 | 17 (20.7) | 43 (18.0) | <0.001 | 23 (28.0) | 69 (28.9) | <0.001 |
| Heterogeneous | 62 (75.6) | 131 (54.8) | 60 (73.2) | 127 (53.1) | 57 (69.5) | 119 (49.8) | |||
| Rim | 9 (11.0) | 83 (34.7) | 5 (6.1) | 69 (28.9) | 2 (2.4) | 51 (21.3) | |||
| Signal intensity | |||||||||
| Low-Iso | - | - | - | 6 (7.3) | 0 (0) | <0.001 | 19 (23.2) | 95 (39.7) | 0.002 |
| Slightly high | - | - | 10 (12.2) | 9 (3.8) | 9 (11.0) | 41 (17.2) | |||
| High | - | - | 66 (80.5) | 230 (96.2) | 54 (65.9) | 103 (43.1) | |||
| Non-mass-like lesions (n=73) | |||||||||
| Distribution | |||||||||
| Focal | 3 (23.1) | 6 (10.0) | 0.153 | 3 (23.1) | 6 (10.0) | 0.085 | 3 (23.1) | 8 (13.3) | 0.134 |
| Segmental | 5 (38.5) | 32 (53.3) | 4 (30.8) | 31 (51.7) | 4 (30.8) | 29 (48.3) | |||
| Regional | 5 (38.5) | 13 (21.7) | 6 (46.2) | 14 (23.3) | 6 (46.2) | 14 (23.3) | |||
| Diffuse | 0 (0) | 9 (15.0) | 0 (0) | 9 (15.0) | 0 (0) | 9 (15.0) | |||
| Internal patterns | |||||||||
| Homogeneous | 0 (0) | 1 (1.7) | 0.171 | 0 (0) | 0 (0) | 0.289 | 0 (0) | 1 (1.7) | 1.0 |
| Heterogeneous | 8 (61.5) | 50 (83.3) | 11 (84.6) | 56 (93.3) | 13 (100.0) | 58 (96.7) | |||
| Clumped/clustered ring | 5 (38.5) | 9 (15.0) | 2 (15.4) | 4 (6.7) | 0 (0) | 1 (1.7) | |||
| Signal intensity | |||||||||
| Low-Iso | - | - | - | 2 (15.4) | 2 (3.3) | 0.036 | 6 (46.2) | 27 (45.0) | 0.608 |
| Slightly high | - | - | 5 (38.5) | 11 (18.3) | 6 (46.2) | 21 (35.0) | |||
| High | - | - | 6 (46.2) | 47 (78.3) | 1 (7.7) | 12 (20.0) | |||
Tab.3 Comparison between benign and malignant lesions for qualitative imaging findings with DCE, DWI and T2WI sequences ([n (%)])
| Lesion type | DCE | P | DWI | P | T2WI | P | |||
|---|---|---|---|---|---|---|---|---|---|
| Benign | Malignant | Benign | Malignant | Benign | Malignant | ||||
| Mass-like lesions (n=321) | |||||||||
| Shape | |||||||||
| Oval | 33 (40.2) | 17 (7.1) | <0.001 | 34 (41.5) | 27 (11.3) | <0.001 | 33 (40.2) | 19 (7.9) | <0.001 |
| Round | 28 (34.1) | 115 (48.1) | 29 (35.4) | 111 (46.4) | 30 (36.6) | 120 (50.2) | |||
| Irregular | 21 (25.6) | 107 (44.8) | 19 (23.2) | 101 (42.3) | 19 (23.2) | 100 (41.8) | |||
| Margin | |||||||||
| Circumscribed | 48 (58.5) | 59 (24.7) | <0.001 | 50 (61.0) | 100 (41.8) | 0.003 | 49 (59.8) | 63 (26.4) | <0.001 |
| Not circumscribed | 34 (41.5) | 180 (75.3) | 32 (39.0) | 139 (58.2) | 33 (40.2) | 176 (73.6) | |||
| Internal patterns | |||||||||
| Homogeneous | 11 (13.4) | 25 (10.5) | <0.001 | 17 (20.7) | 43 (18.0) | <0.001 | 23 (28.0) | 69 (28.9) | <0.001 |
| Heterogeneous | 62 (75.6) | 131 (54.8) | 60 (73.2) | 127 (53.1) | 57 (69.5) | 119 (49.8) | |||
| Rim | 9 (11.0) | 83 (34.7) | 5 (6.1) | 69 (28.9) | 2 (2.4) | 51 (21.3) | |||
| Signal intensity | |||||||||
| Low-Iso | - | - | - | 6 (7.3) | 0 (0) | <0.001 | 19 (23.2) | 95 (39.7) | 0.002 |
| Slightly high | - | - | 10 (12.2) | 9 (3.8) | 9 (11.0) | 41 (17.2) | |||
| High | - | - | 66 (80.5) | 230 (96.2) | 54 (65.9) | 103 (43.1) | |||
| Non-mass-like lesions (n=73) | |||||||||
| Distribution | |||||||||
| Focal | 3 (23.1) | 6 (10.0) | 0.153 | 3 (23.1) | 6 (10.0) | 0.085 | 3 (23.1) | 8 (13.3) | 0.134 |
| Segmental | 5 (38.5) | 32 (53.3) | 4 (30.8) | 31 (51.7) | 4 (30.8) | 29 (48.3) | |||
| Regional | 5 (38.5) | 13 (21.7) | 6 (46.2) | 14 (23.3) | 6 (46.2) | 14 (23.3) | |||
| Diffuse | 0 (0) | 9 (15.0) | 0 (0) | 9 (15.0) | 0 (0) | 9 (15.0) | |||
| Internal patterns | |||||||||
| Homogeneous | 0 (0) | 1 (1.7) | 0.171 | 0 (0) | 0 (0) | 0.289 | 0 (0) | 1 (1.7) | 1.0 |
| Heterogeneous | 8 (61.5) | 50 (83.3) | 11 (84.6) | 56 (93.3) | 13 (100.0) | 58 (96.7) | |||
| Clumped/clustered ring | 5 (38.5) | 9 (15.0) | 2 (15.4) | 4 (6.7) | 0 (0) | 1 (1.7) | |||
| Signal intensity | |||||||||
| Low-Iso | - | - | - | 2 (15.4) | 2 (3.3) | 0.036 | 6 (46.2) | 27 (45.0) | 0.608 |
| Slightly high | - | - | 5 (38.5) | 11 (18.3) | 6 (46.2) | 21 (35.0) | |||
| High | - | - | 6 (46.2) | 47 (78.3) | 1 (7.7) | 12 (20.0) | |||
| Evaluation metrics | Mass-like lesions | Non-mass-like lesions | |||
|---|---|---|---|---|---|
| DCE | DWI | T2WI | DCE+DWI+T2WI | DWI | |
| Sensitivity (%) | 81.2 | 74.1 | 73.6 | 74.9 | 78.3 |
| Specificity (%) | 67.1 | 73.2 | 72.0 | 85.4 | 53.8 |
| PPV (%) | 87.8 | 88.9 | 88.4 | 93.7 | 88.7 |
| NPV (%) | 55.0 | 49.2 | 48.4 | 53.8 | 35.0 |
| Accuracy (%) | 77.6 | 73.8 | 73.2 | 77.6 | 74.0 |
| AUC (95% CI) | 0.786 (0.728-0.844) | 0.793 (0.735-0.850) | 0.809 (0.755-0.864) | 0.865 (0.816-0.914) | 0.669 (0.514-0.823) |
Tab.4 Diagnostic performance of the different prediction models for evaluating benign and malignant mass-like and non-mass-like lesions
| Evaluation metrics | Mass-like lesions | Non-mass-like lesions | |||
|---|---|---|---|---|---|
| DCE | DWI | T2WI | DCE+DWI+T2WI | DWI | |
| Sensitivity (%) | 81.2 | 74.1 | 73.6 | 74.9 | 78.3 |
| Specificity (%) | 67.1 | 73.2 | 72.0 | 85.4 | 53.8 |
| PPV (%) | 87.8 | 88.9 | 88.4 | 93.7 | 88.7 |
| NPV (%) | 55.0 | 49.2 | 48.4 | 53.8 | 35.0 |
| Accuracy (%) | 77.6 | 73.8 | 73.2 | 77.6 | 74.0 |
| AUC (95% CI) | 0.786 (0.728-0.844) | 0.793 (0.735-0.850) | 0.809 (0.755-0.864) | 0.865 (0.816-0.914) | 0.669 (0.514-0.823) |
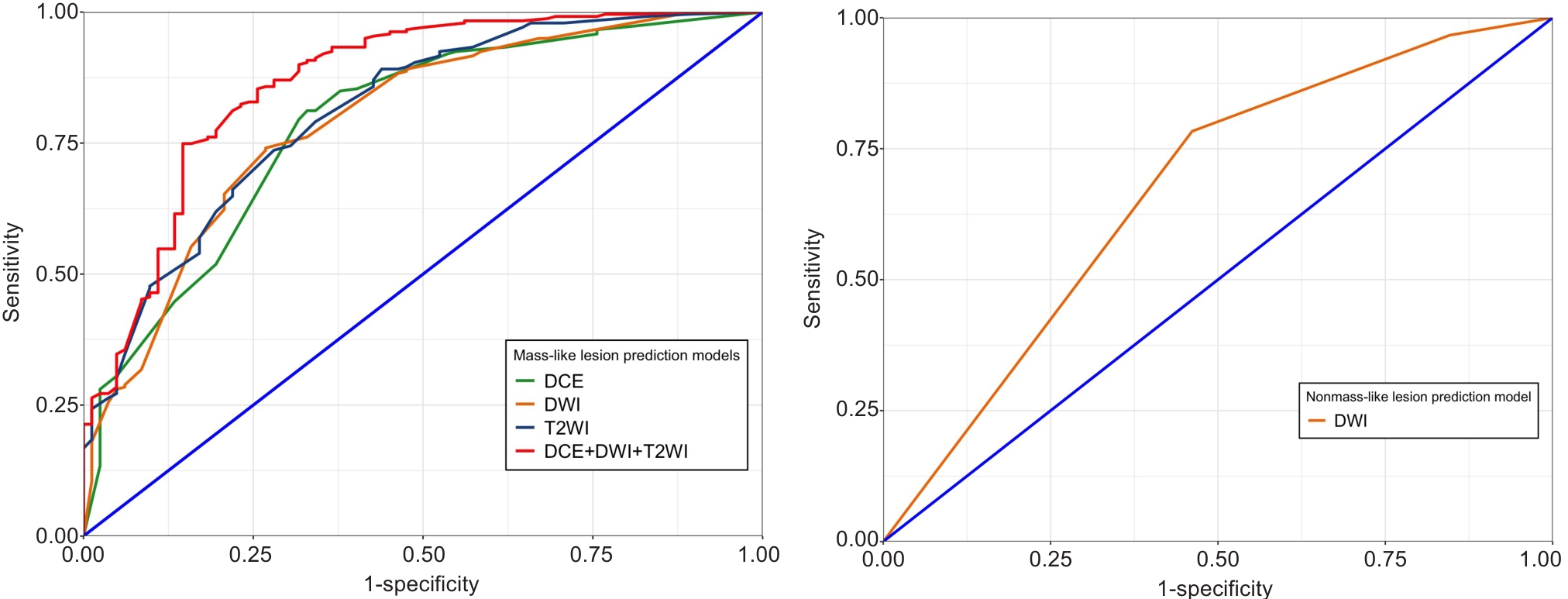
Fig.5 Receiver operating characteristic (ROC) curves of the different prediction models for evaluating benign and malignant mass-like and non-mass-like lesions.
| Model | DCE | DWI | T2WI | DCE+DWI+T2WI |
|---|---|---|---|---|
| DCE | N/A | 0.837 | 0.293 | 0.001 |
| DWI | - | N/A | 0.522 | <0.001 |
| T2WI | - | - | N/A | 0.001 |
Tab.5 P-values for comparisons of areas under the curve between different mass-like lesion prediction models
| Model | DCE | DWI | T2WI | DCE+DWI+T2WI |
|---|---|---|---|---|
| DCE | N/A | 0.837 | 0.293 | 0.001 |
| DWI | - | N/A | 0.522 | <0.001 |
| T2WI | - | - | N/A | 0.001 |
| [1] | Barkhausen J, Bischof A, Haverstock D, et al. Diagnostic efficacy of contrast-enhanced breast MRI versus X-ray mammography in women with different degrees of breast density [J]. Acta Radiol, 2021, 62(5): 586-93. doi:10.1177/0284185120936271 |
| [2] | Kim YE, Cha JH, Kim HH, et al. Analysis of false-negative findings of breast cancer on previous magnetic resonance imaging [J]. Acta Radiol, 2021, 62(6): 722-34. doi:10.1177/0284185120941830 |
| [3] | Tsarouchi MI, Vlachopoulos GF, Karahaliou AN, et al. Multi-parametric MRI lesion heterogeneity biomarkers for breast cancer diagnosis [J]. Phys Med, 2020, 80: 101-10. doi:10.1016/j.ejmp.2020.10.007 |
| [4] | Yamaguchi K, Nakazono T, Egashira R, et al. Maximum slope of ultrafast dynamic contrast-enhanced MRI of the breast: comparisons with prognostic factors of breast cancer [J]. Jpn J Radiol, 2021, 39(3): 246-53. doi:10.1007/s11604-020-01049-6 |
| [5] | Sippo DA, Rutledge GM, Mercaldo SF, et al. Impact of background parenchymal enhancement on diagnostic performance in screening breast MRI [J]. Acad Radiol, 2020, 27(5): 663-71. doi:10.1016/j.acra.2019.06.020 |
| [6] | Rella R, Bufi E, Belli P, et al. Background parenchymal enhancement in breast magnetic resonance imaging: A review of current evidences and future trends [J]. Diagn Interv Imaging, 2018, 99(12): 815-26. doi:10.1016/j.diii.2018.08.011 |
| [7] | Kanao S, Kataoka M, Iima M, et al. Differentiating benign and malignant inflammatory breast lesions: value of T2 weighted and diffusion weighted MR images [J]. Magn Reson Imaging, 2018, 50: 38-44. doi:10.1016/j.mri.2018.03.012 |
| [8] | Nita Amornsiripanitch M. Diffusion-weighted MRI for unenhanced breast cancer screening [J]. Radiology, 2019, 293(3): 504-20. doi:10.1148/radiol.2019182789 |
| [9] | Ha SM, Chang JM, Lee SH, et al. Detection of contralateral breast cancer using diffusion-weighted magnetic resonance imaging in women with newly diagnosed breast cancer: comparison with combined mammography and whole-breast ultrasound [J]. Korean J Radiol, 2021, 22(6): 867-79. doi:10.3348/kjr.2020.1183 |
| [10] | Kul S, Metin Y, Bekircavusoglu S, et al. Qualitative characterization of breast tumors with diffusion-weighted imaging has comparable accuracy to quantitative analysis [J]. Clin Imaging, 2021, 77: 17-24. doi:10.1016/j.clinimag.2021.02.025 |
| [11] | Kim JY, Kim JJ, Hwangbo L, et al. Diffusion-weighted imaging of invasive breast cancer: relationship to distant metastasis-free survival [J]. Radiology, 2019, 291(2): 300-07. doi:10.1148/radiol.2019181706 |
| [12] | Arponen O, Sudah M, Masarwah A, et al. Diffusion-weighted imaging in 3.0 Tesla breast MRI: diagnostic performance and tumor characterization using small subregions vs. whole tumor regions of interest [J]. PLoS One, 2015, 10(10): e0138702. doi:10.1371/journal.pone.0138702 |
| [13] | Baltzer P, Mann RM, Iima M, et al. Diffusion-weighted imaging of the breast-a consensus and mission statement from the EUSOBI International Breast Diffusion-Weighted Imaging working group [J]. Eur Radiol, 2020, 30(3): 1436-50. doi:10.1007/s00330-019-06510-3 |
| [14] | Ota R, Kataoka M, Iima M, et al. Evaluation of breast lesions based on modified BI-RADS using high-resolution readout-segmented diffusion-weighted echo-planar imaging and T2/T1-weighted image [J]. Magn Reson Imaging, 2023, 98: 132-9. doi:10.1016/j.mri.2022.12.024 |
| [15] | Naranjo ID, Sogani J, Saccarelli C, et al. MRI Screening of BRCA mutation carriers: comparison of standard protocol and abbreviated protocols with and without T2-weighted images [J]. AJR Am J Roentgenol, 2022, 218(5): 810-20. doi:10.2214/ajr.21.27022 |
| [16] | Mann RM, Cho N, Moy L. Breast MRI: State of the Art [J]. Radiology, 2019, 292(3): 520-36. doi:10.1148/radiol.2019182947 |
| [17] | Mann RM, Kuhl CK, Kinkel K, et al. Breast MRI: guidelines from the European Society of Breast Imaging [J]. Eur Radiol, 2008, 18(7): 1307-18. doi:10.1007/s00330-008-0863-7 |
| [18] | Wu LM, Chen J, Hu J, et al. Diffusion-weighted magnetic resonance imaging combined with T2-weighted images in the detection of small breast cancer: a single-center multi-observer study [J]. Acta Radiol, 2014, 55(1): 24-31. doi:10.1177/0284185113492458 |
| [19] | Kawashima H, Kobayashi-Yoshida M, Matsui O, et al. Peripheral hyperintense pattern on T2-weighted magnetic resonance imaging (MRI) in breast carcinoma: correlation with early peripheral enhancement on dynamic MRI and histopathologic findings [J]. J Magn Reson Imaging, 2010, 32(5): 1117-23. doi:10.1002/jmri.22279 |
| [20] | Yamaguchi R, Furusawa H, Nakahara H, et al. Clinicopathological study of invasive ductal carcinoma with large central acellular zone: special reference to magnetic resonance imaging findings [J]. Pathol Int, 2008, 58(1): 26-30. doi:10.1111/j.1440-1827.2007.02184.x |
| [21] | Niko Radovic M. Evaluation of breast cancer morphology using diffusion-weighted and dynamic contrast-enhanced MRI: intermethod and interobserver agreement [J]. J Magn Reson Imaging, 2019, 49(5): 1381-90. doi:10.1002/jmri.26332 |
| [22] | Kishimoto AO, Kataoka M, Iima M, et al. The comparison of high-resolution diffusion weighted imaging (DWI) with high-resolution contrast-enhanced MRI in the evaluation of breast cancers [J]. Magn Reson Imaging, 2020, 71: 161-9. doi:10.1016/j.mri.2020.03.007 |
| [23] | Christner SA, Grunz JP, Schlaiss T, et al. Breast lesion morphology assessment with high and standard b values in diffusion-weighted imaging at 3 Tesla [J]. Magn Reson Imaging, 2024, 107: 100-10. doi:10.1016/j.mri.2024.01.005 |
| [24] | Yabuuchi H, Matsuo Y, Sunami S, et al. Detection of non-palpable breast cancer in asymptomatic women by using unenhanced diffusion-weighted and T2-weighted MR imaging: comparison with mammography and dynamic contrast-enhanced MR imaging [J]. Eur Radiol, 2011, 21(1): 11-7. doi:10.1007/s00330-010-1890-8 |
| [25] | An YY. Differentiation of malignant and benign breast lesions: added value of the qualitative analysis of breast lesions on diffusion-weighted imaging (DWI) using readout-segmented echo-planar imaging at 3.0 T [J]. PLOS ONE, 2017, 12(3): e0174681. doi:10.1371/journal.pone.0174681 |
| [26] | Kul S, Eyuboglu I, Cansu A, et al. Diagnostic efficacy of the diffusion weighted imaging in the characterization of different types of breast lesions [J]. J Magn Reson Imaging, 2014, 40(5): 1158-64. doi:10.1002/jmri.24491 |
| [27] | Kuroki-suzuli S. Detecting breast cancer with non-contrast MR imaging: combining diffusion-weighted and STIR imaging [J]. Magn Reson Med Sci, 2007, 6(1): 21-7. doi:10.2463/mrms.6.21 |
| [28] | Heacock L, Melsaether AN, Heller SL, et al. Evaluation of a known breast cancer using an abbreviated breast MRI protocol: correlation of imaging characteristics and pathology with lesion detection and conspicuity [J]. Eur J Radiol, 2016, 85(4): 815-23. doi:10.1016/j.ejrad.2016.01.005 |
| [29] | Ao F, Yan Y, Zhang ZL, et al. The value of dynamic contrast-enhanced magnetic resonance imaging combined with apparent diffusion coefficient in the differentiation of benign and malignant diseases of the breast [J]. Acta Radiol, 2022, 63(7): 891-900. doi:10.1177/02841851211024002 |
| [30] | Westra C, Dialani V, Mehta TS, et al. Using T2-weighted sequences to more accurately characterize breast masses seen on MRI [J]. AJR Am J Roentgenol, 2014, 202(3): W183-190. doi:10.2214/ajr.13.11266 |
| [31] | Yuen S, Monzawa S, Yanai S, et al. The association between MRI findings and breast cancer subtypes: focused on the combination patterns on diffusion-weighted and T2-weighted images [J]. Breast Cancer, 2020, 27(5): 1029-37. doi:10.1007/s12282-020-01105-z |
| [32] | Santamaría G. Radiologic and pathologic findings in breast tumors with high signal intensity on T2-weighted MR images [J]. Radio Graphics, 2010, 30(2): 533-48. doi:10.1148/rg.302095044 |
| [33] | Baltzer PA, Benndorf M, Dietzel M, et al. Sensitivity and specificity of unenhanced MR mammography (DWI combined with T2-weighted TSE imaging, ueMRM) for the differentiation of mass lesions [J]. Eur Radiol, 2010, 20(5): 1101-10. doi:10.1007/s00330-009-1654-5 |
| [1] | Siyuan MA, Bochao ZHANG, Chun PU. Circ_0000437 promotes proliferation, invasion, migration and epithelial-mesenchymal transition of breast cancer cells by targeting the let-7b-5p/CTPS1 axis [J]. Journal of Southern Medical University, 2025, 45(8): 1682-1696. |
| [2] | Zhaojun ZHANG, Qiong WU, Miaomiao XIE, Ruyin YE, Chenchen GENG, Jiwen SHI, Qingling YANG, Wenrui WANG, Yurong SHI. Layered double hydroxide-loaded si-NEAT1 regulates paclitaxel resistance and tumor-associated macrophage polarization in breast cancer by targeting miR-133b/PD-L1 [J]. Journal of Southern Medical University, 2025, 45(8): 1718-1731. |
| [3] | Jiahao LI, Ruiting XIAN, Rong LI. Down-regulation of ACADM-mediated lipotoxicity inhibits invasion and metastasis of estrogen receptor-positive breast cancer cells [J]. Journal of Southern Medical University, 2025, 45(6): 1163-1173. |
| [4] | Zhenxiang DONG, Yihao GUO, Qiang LIU, Yizhe ZHANG, Qianyi QIU, Xiaodong ZHANG, Yanqiu FENG. A single repetition time quantitative magnetic susceptibility imaging method for the lumbar spine using bipolar readout gradient [J]. Journal of Southern Medical University, 2025, 45(6): 1336-1342. |
| [5] | Di CHEN, Ying LÜ, Yixin GUO, Yirong ZHANG, Ruixuan WANG, Xiaoruo ZHOU, Yuxin CHEN, Xiaohui WU. Dihydroartemisinin enhances doxorubicin-induced apoptosis of triple negative breast cancer cells by negatively regulating the STAT3/HIF-1α pathway [J]. Journal of Southern Medical University, 2025, 45(2): 254-260. |
| [6] | Qiao CHU, Xiaona WANG, Jiaying XU, Huilin PENG, Yulin ZHAO, Jing ZHANG, Guoyu LU, Kai WANG. Pulsatilla saponin D inhibits invasion and metastasis of triple-negative breast cancer cells through multiple targets and pathways [J]. Journal of Southern Medical University, 2025, 45(1): 150-161. |
| [7] | Weiyang FANG, Hui XIAO, Shuang WANG, Xiaoming LIN, Chaomin CHEN. A deep learning model based on magnetic resonance imaging and clinical feature fusion for predicting preoperative cytokeratin 19 status in hepatocellular carcinoma [J]. Journal of Southern Medical University, 2024, 44(9): 1738-1751. |
| [8] | Mingzi OUYANG, Jiaqi CUI, Hui WANG, Zheng LIANG, Dajin PI, Liguo CHEN, Qianjun CHEN, Yingchao WU. Kaixinsan alleviates adriamycin-induced depression-like behaviors in mice by reducing ferroptosis in the prefrontal cortex [J]. Journal of Southern Medical University, 2024, 44(8): 1441-1449. |
| [9] | Jincun FANG, Liwei LIU, Junhao LIN, Fengsheng CHEN. Overexpression of CDHR2 inhibits proliferation of breast cancer cells by inhibiting the PI3K/Akt pathway [J]. Journal of Southern Medical University, 2024, 44(6): 1117-1125. |
| [10] | Zhi CUI, Cuijiao MA, Qianru WANG, Jinhao CHEN, Ziyang YAN, Jianlin YANG, Yafeng LÜ, Chunyu CAO. A recombinant adeno-associated virus expressing secretory TGF‑β type II receptor inhibits triple-negative murine breast cancer 4T1 cell proliferation and lung metastasis in mice [J]. Journal of Southern Medical University, 2024, 44(5): 818-826. |
| [11] | Youqin ZENG, Siyu CHEN, Yan LIU, Yitong LIU, Ling ZHANG, Jiao XIA, Xinyu WU, Changyou WEI, Ping LENG. AKBA combined with doxorubicin inhibits proliferation and metastasis of triple-negative breast cancer MDA-MB-231 cells and xenograft growth in nude mice [J]. Journal of Southern Medical University, 2024, 44(12): 2449-2460. |
| [12] | Xiaoyin HUANG, Fenglian CHEN, Yu ZHANG, Shujun LIANG. A predictive model for survival outcomes of glioma patients based on multi-parametric, multi-regional MRI radiomics features and clinical features [J]. Journal of Southern Medical University, 2024, 44(10): 2004-2014. |
| [13] | HE Huishan, GUO Erjia, MENG Wenyi, WANG Yu, WANG Wen, HE Wenle, WU Yuankui, YANG Wei. Predicting cerebral glioma enhancement pattern using a machine learning-based magnetic resonance imaging radiomics model [J]. Journal of Southern Medical University, 2024, 44(1): 194-200. |
| [14] | XU Mengqi, SHI Yutong, LIU Junping, WU Minmin, ZHANG Fengmei, HE Zhiqiang, TANG Min. JAG1 affects monocytes-macrophages to reshape the pre-metastatic niche of triple-negative breast cancer through LncRNA MALAT1 in exosomes [J]. Journal of Southern Medical University, 2023, 43(9): 1525-1535. |
| [15] | XU Jiaming, LIN Long, CHEN Qionghui, LI Lan. Hsa-miR-148a-3p promotes malignant behavior of breast cancer cells by downregulating DUSP1 [J]. Journal of Southern Medical University, 2023, 43(9): 1515-1524. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||