Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (8): 1718-1731.doi: 10.12122/j.issn.1673-4254.2025.08.16
Zhaojun ZHANG1( ), Qiong WU1, Miaomiao XIE1, Ruyin YE1, Chenchen GENG1, Jiwen SHI1, Qingling YANG1, Wenrui WANG1, Yurong SHI2(
), Qiong WU1, Miaomiao XIE1, Ruyin YE1, Chenchen GENG1, Jiwen SHI1, Qingling YANG1, Wenrui WANG1, Yurong SHI2( )
)
Received:2024-12-24
Online:2025-08-20
Published:2025-09-05
Contact:
Yurong SHI
E-mail:zzj17318530948@163.com;shiyr@126.com
Zhaojun ZHANG, Qiong WU, Miaomiao XIE, Ruyin YE, Chenchen GENG, Jiwen SHI, Qingling YANG, Wenrui WANG, Yurong SHI. Layered double hydroxide-loaded si-NEAT1 regulates paclitaxel resistance and tumor-associated macrophage polarization in breast cancer by targeting miR-133b/PD-L1[J]. Journal of Southern Medical University, 2025, 45(8): 1718-1731.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.08.16
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
| NEAT1 | CTTCCTCCCTTTAACTTATCCATTCAC | CTCTTCCTCCACCATTACCAACAATAC |
| miR-133b | TCCCCTTCAACCAGCTAA | Universal primer |
| PD-L1 | GTAAGACCACCACCACCAATTC | AGTTGTTGTGTTGATTCTCAGTGTG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Arg-1 | GGACCTGCCCTTTGCTGACATC | TCTTCTTGACTTCTGCCACCTTGC |
| CD163 | ACAATGAAGATGCTGGCGTGAC | ACAATGAAGATGCTGGCGTGAC |
| IL-10 | ACCAAGACCCAGACATCAAGGC | AGGCATTCTTCACCTGCTCCAC |
| CD68 | CCCACCTGCTTCTCTCATTCCC | TTGTACTCCACCGCCATGTAGC |
Tab.1 PCR primer sequences
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | CAGCCTCAAGATCATCAGCA | TGTGGTCATGAGTCCTTCCA |
| NEAT1 | CTTCCTCCCTTTAACTTATCCATTCAC | CTCTTCCTCCACCATTACCAACAATAC |
| miR-133b | TCCCCTTCAACCAGCTAA | Universal primer |
| PD-L1 | GTAAGACCACCACCACCAATTC | AGTTGTTGTGTTGATTCTCAGTGTG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Arg-1 | GGACCTGCCCTTTGCTGACATC | TCTTCTTGACTTCTGCCACCTTGC |
| CD163 | ACAATGAAGATGCTGGCGTGAC | ACAATGAAGATGCTGGCGTGAC |
| IL-10 | ACCAAGACCCAGACATCAAGGC | AGGCATTCTTCACCTGCTCCAC |
| CD68 | CCCACCTGCTTCTCTCATTCCC | TTGTACTCCACCGCCATGTAGC |

Fig.1 LncRNA NEAT1 regulates paclitaxel resistance of breast cancer cells. A, B: Western blotting and immunofluorescence detection of expression of drug resistance-related proteins in resistant breast cancer cells (scale bar=50 μm). C: qRT-PCR technology for detecting the expression of NEAT1. D: Western blotting for detecting drug resistance of the cells. E: Flow cytometry for analyzing apoptosis rate of the resistant breast cancer cells. F, G: Scratch and Transwell assays for assessing the migration and invasion capabilities of resistant cells (scale bar=200 μm). Scratch and Transwell assays for assessing migration and invasion capabilities of resistant breast cancer cells. *P<0.05, **P<0.01, ***P<0.001.
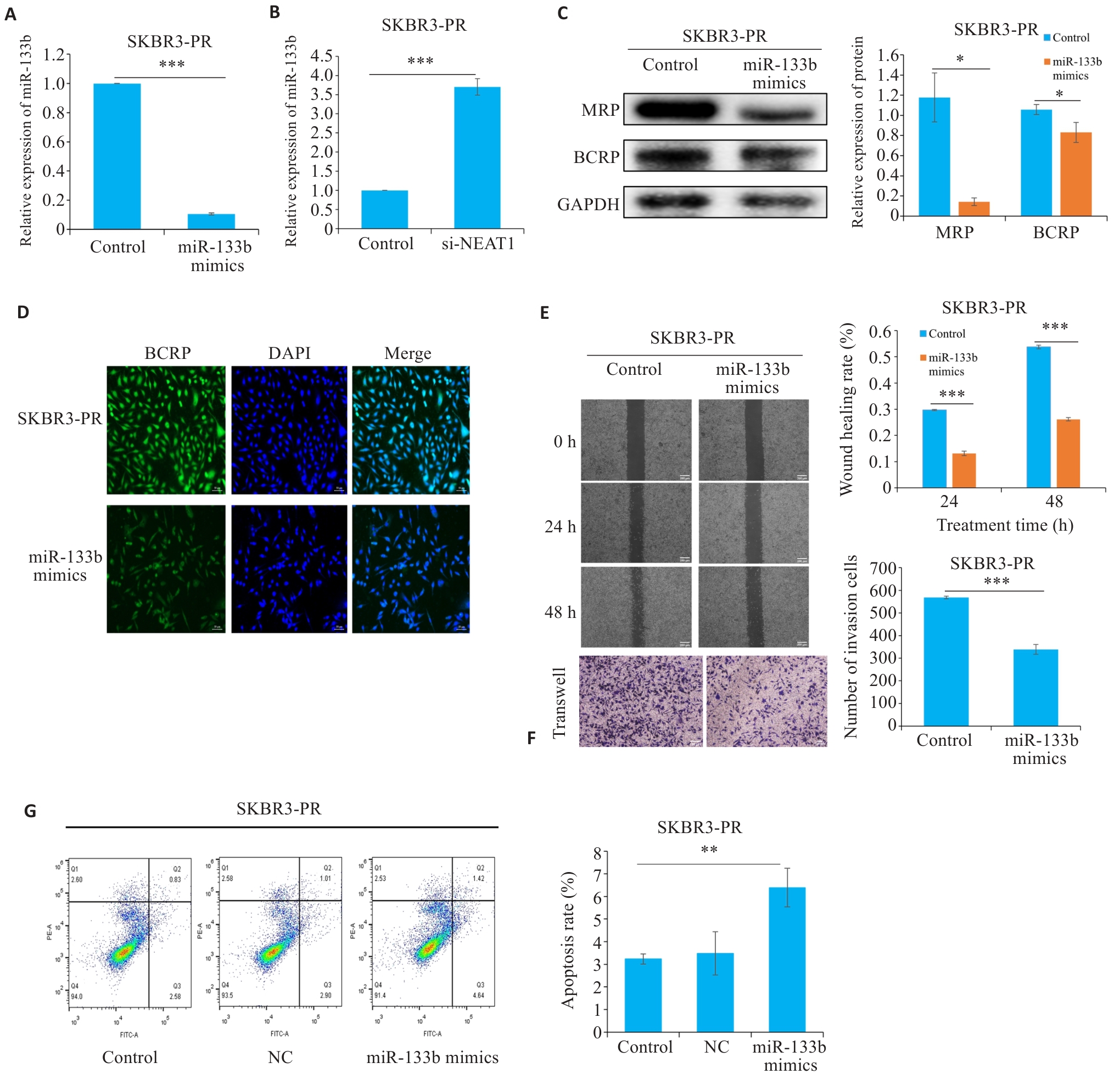
Fig.2 LncRNA NEAT1 affects drug resistance in breast cancer cells by regulating miR-133b. A, B: qRT-PCR for detecting the expression of miR-133b. C: Western blotting for detecting expressions of drug resistance proteins. D: Immunofluorescence for detecting the expression of resistance proteins. E, F: Scratch and Transwell assays for assessing the migration and invasiveness of resistant cells. G: Flow cytometry for detecting apoptosis rate of resistant cells. *P<0.05, **P<0.01, ***P<0.001.
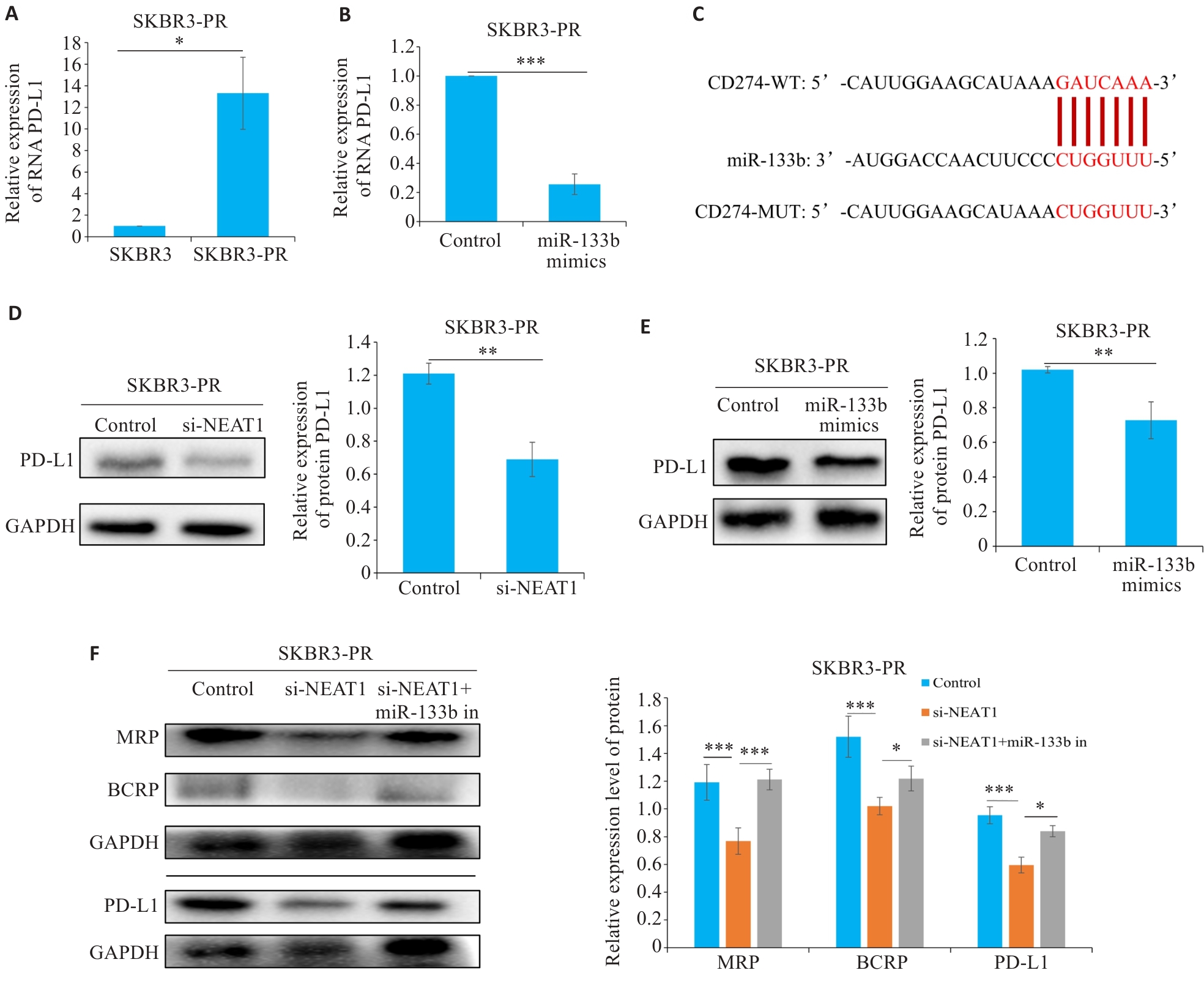
Fig.3 LncRNA NEAT1 regulates miR-133b to target PD-L1 and affect paclitaxel resistance in breast cancer cells. A, B: qRT-PCR for detecting the expression of PD-L1. C: Bioinformatics analysis reveals a regulatory relationship between miR-133b and PD-L1. D, E: Western blotting for detecting the expression of PD-L1. F: Western blotting for detecting the expressions of MRP, BCRP, and PD-L1 in drug-resistant breast cancer cells. *P<0.05, **P<0.01, ***P<0.001.
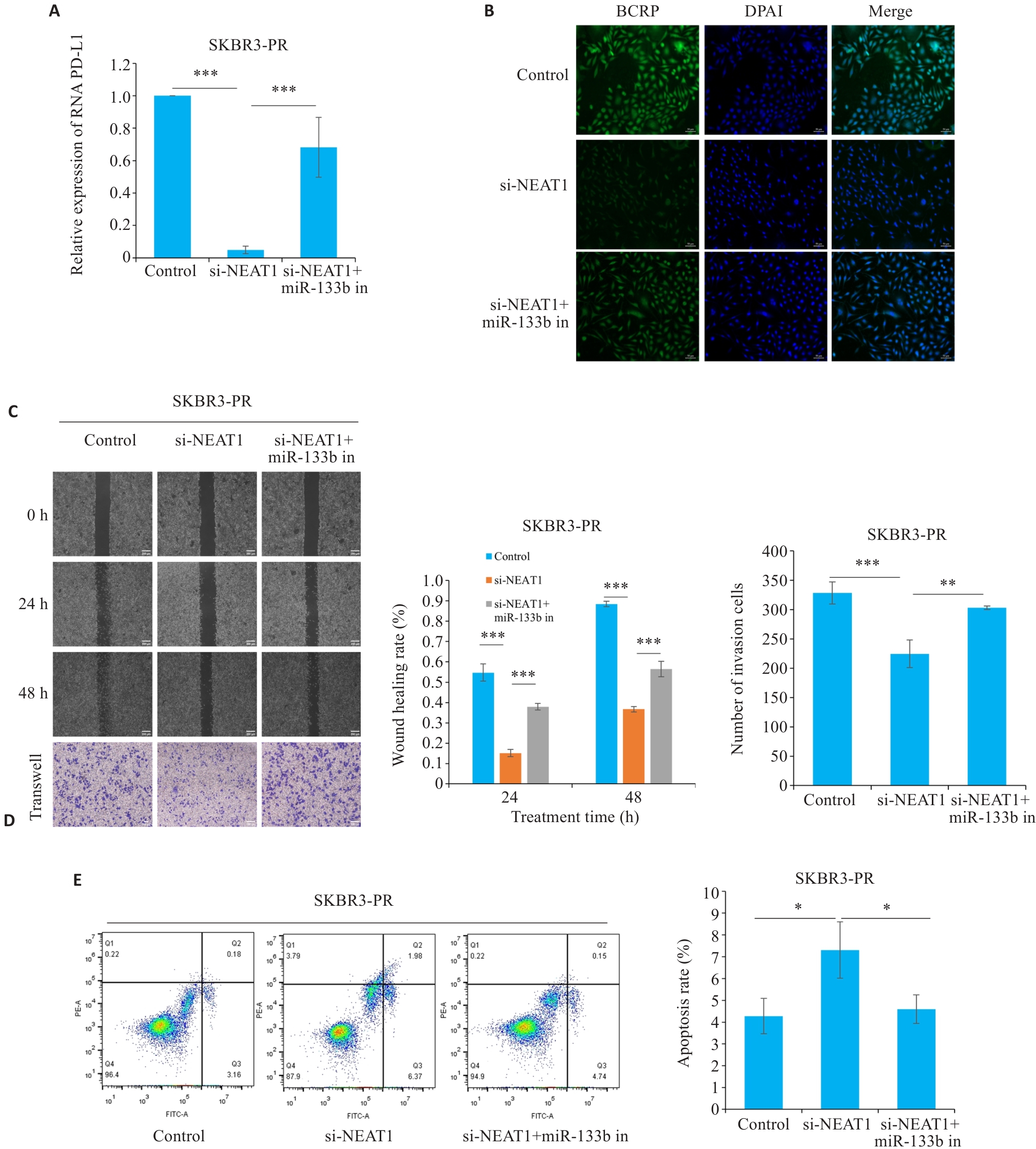
Fig.4 LncRNA NEAT1 regulates miR-133b to target PD-L1 and affect paclitaxel resistance in breast cancer cells. A: qRT-PCR for detecting the expression of PD-L1. B: Immunofluorescence for detecting the expression of BCRP in resistant cells (scale bar=50 μm). C, D: Scratch and Transwell assays for assessing migration and invasion capabilities of resistant cells (scale bar=200 μm). E: Flow cytometry for detecting apoptosis rate of the resistant cells. *P<0.05, **P<0.01, ***P<0.001.
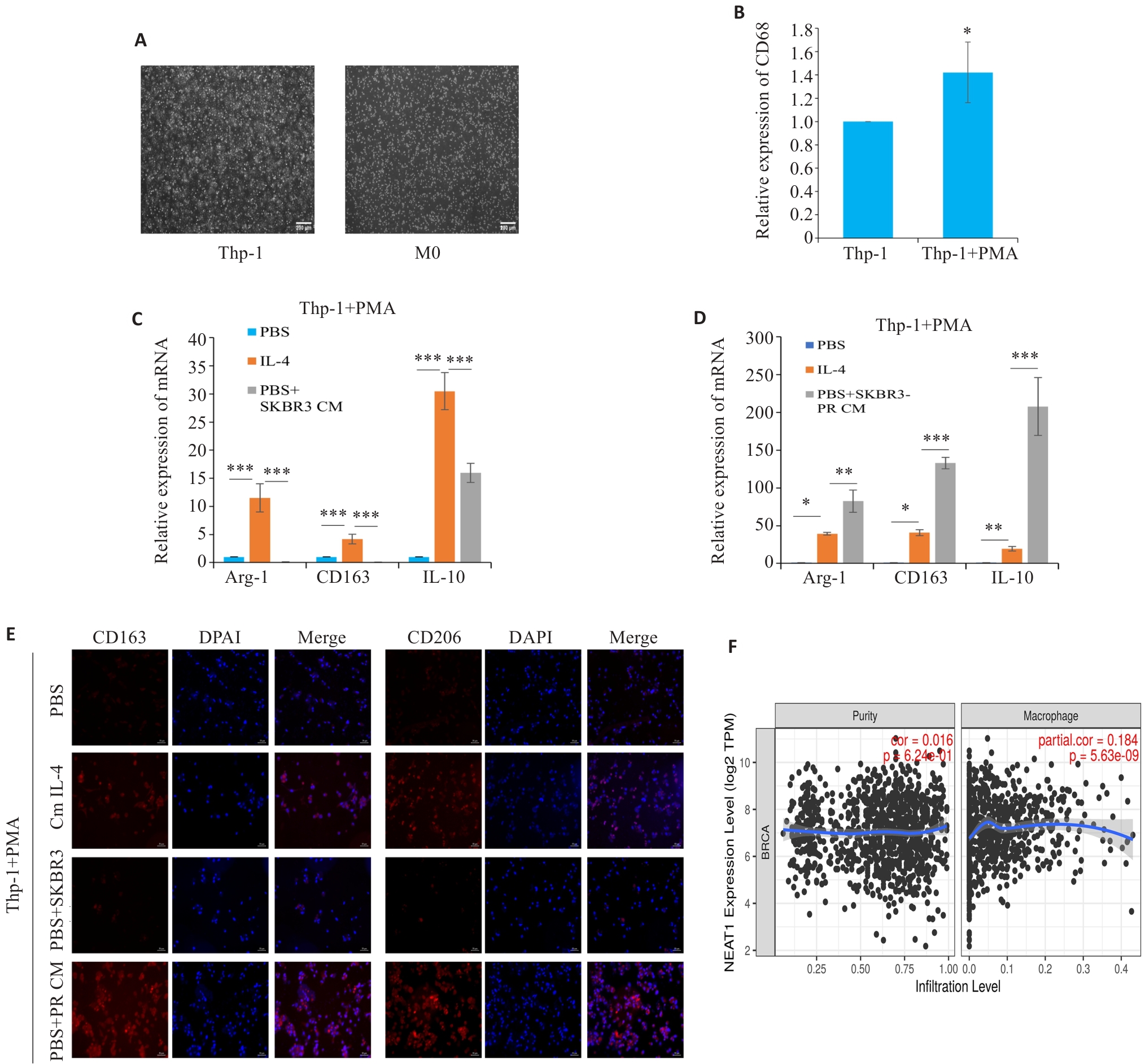
Fig.5 Resistance cell supernatant can regulate the polarization of tumor-associated macrophages. A: Morphology of Thp-1 and M0-type macrophages (scale bar=200 μm ). B: qRT-PCR for detecting the expression of CD68. C, D: qRT-PCR for detecting the expression of M2 macrophage polarization markers Arg-1, CD163, and IL-10. E: Immunofluorescence staining for detecting M2 polarization markers CD163 and CD206 in the macrophages (scale bar=50 μm). F: Timer2.0 website predicts that the expression of NEAT1 is positively correlated with macrophage infiltration. *P<0.05, **P<0.01, ***P<0.001.
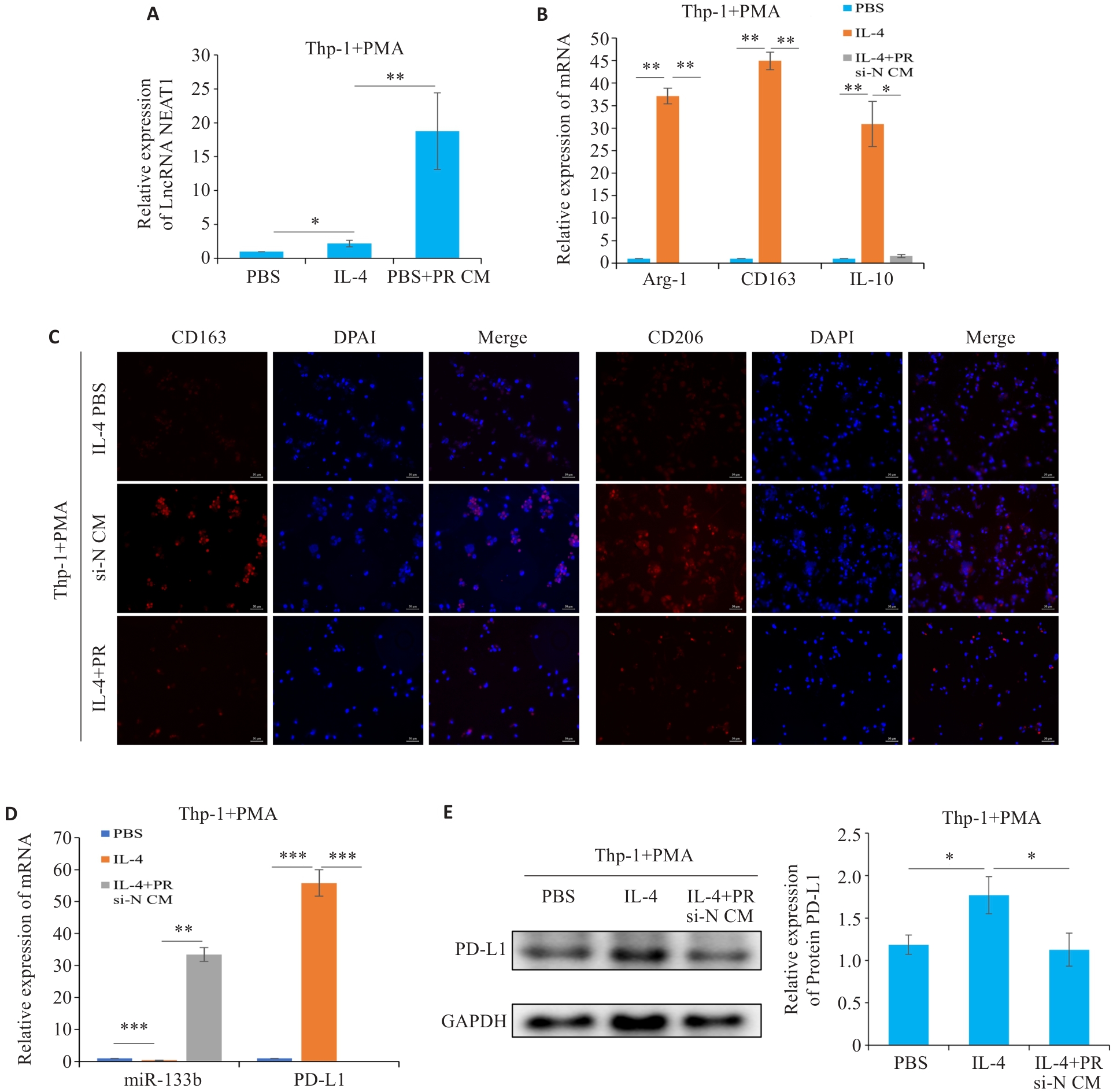
Fig.6 LncRNA NEAT1 regulates polarization of tumor-associated macrophages in breast cancer. A, D: qRT-PCR for detecting the expressions of NEAT1, miR-133b, and PD-L1, respectively. B, C: qRT-PCR and Immunofluorescence for detecting M2 macrophage polarization markers (scale bar=50 μm). E: Western blotting for detecting the expression of PD-L1 in the macrophages. *P<0.05, **P<0.01, ***P<0.001.
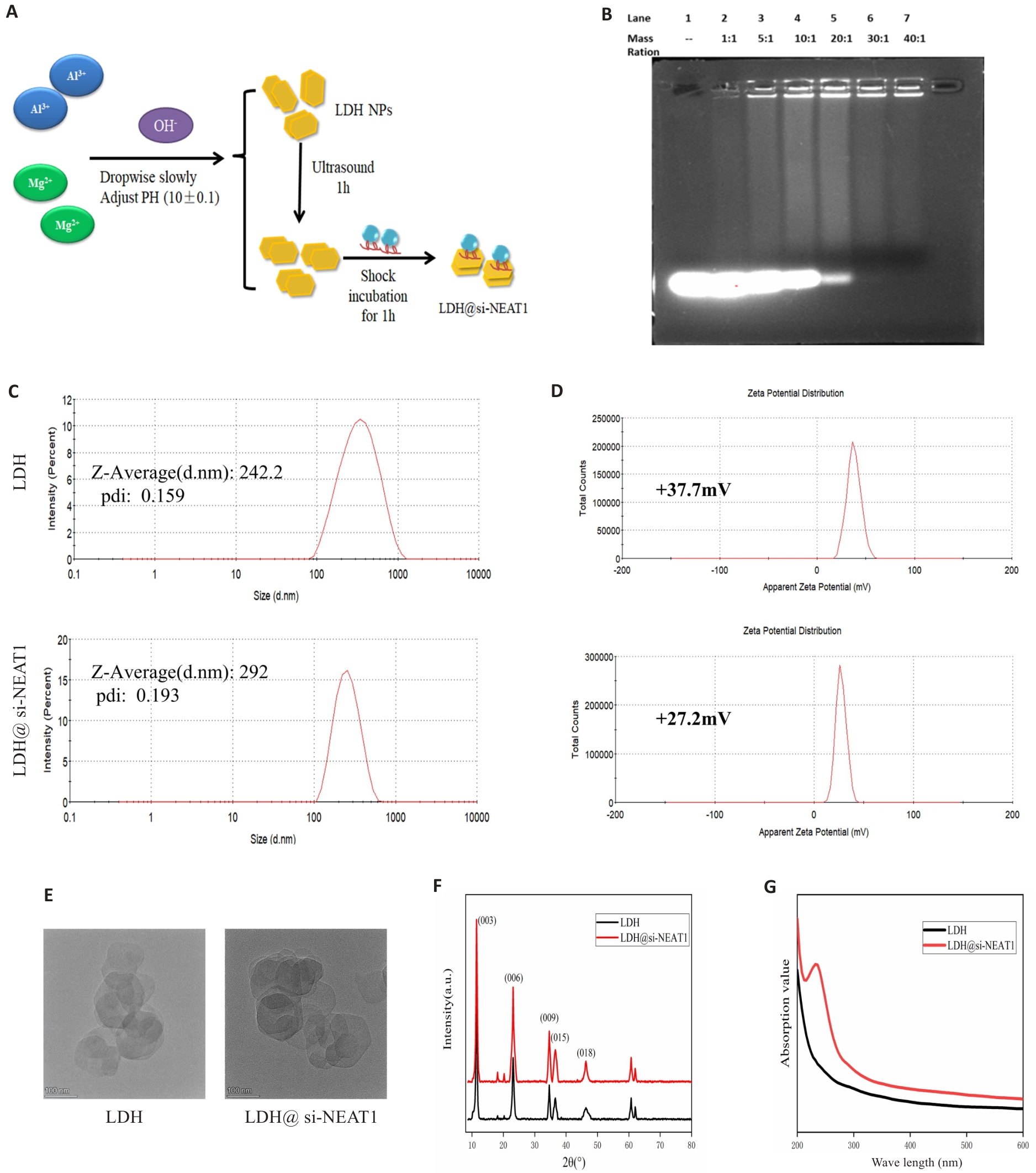
Fig.7 Construction of LDH Nanoparticles Loaded with si-NEAT1. A: Synthesis and loading of LDH. B: Agarose gel electrophoresis to determine the optimal loading ratio of LDH with si-NEAT1. C, D: Malvern particle size analysis for assessing particle size distribution and zeta potential of LDH and LDH@si-NEAT1. E: Transmission electron microscopy images of LDH and LDH@si-NEAT1. F, G: X-ray diffraction and UV-Vis spectra of LDH and LDH@si-NEAT1.
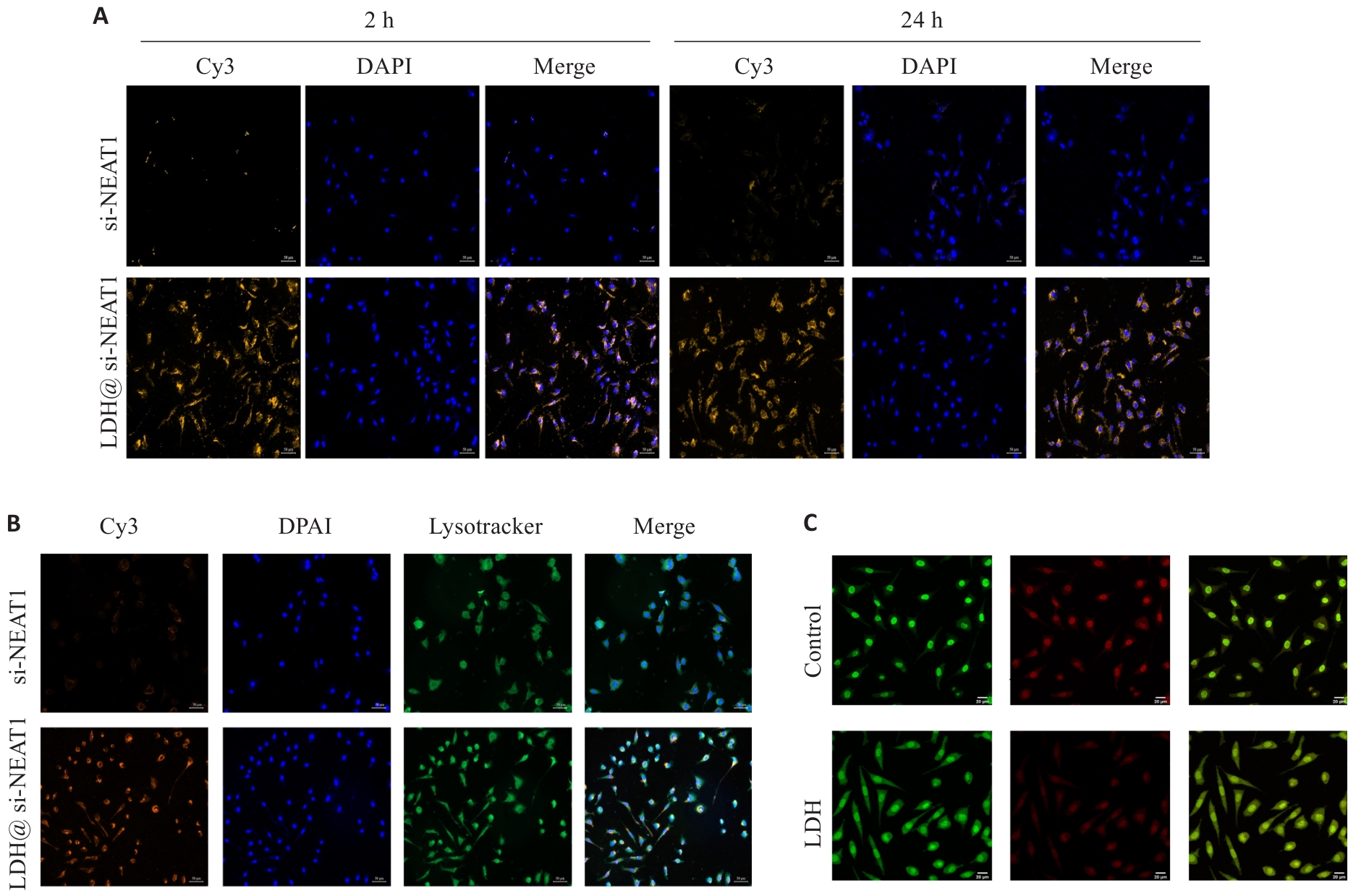
Fig.8 LDH@si-NEAT1 characterization experiments. A: Uptake experiment of LDH@si-NEAT1 (Original magnification: ×200). B: Confocal microscopy to detect lysosomal escape of LDH@si-NEAT1 (×200). C: Acridine orange staining to test lysosomal membrane permeability (×600).
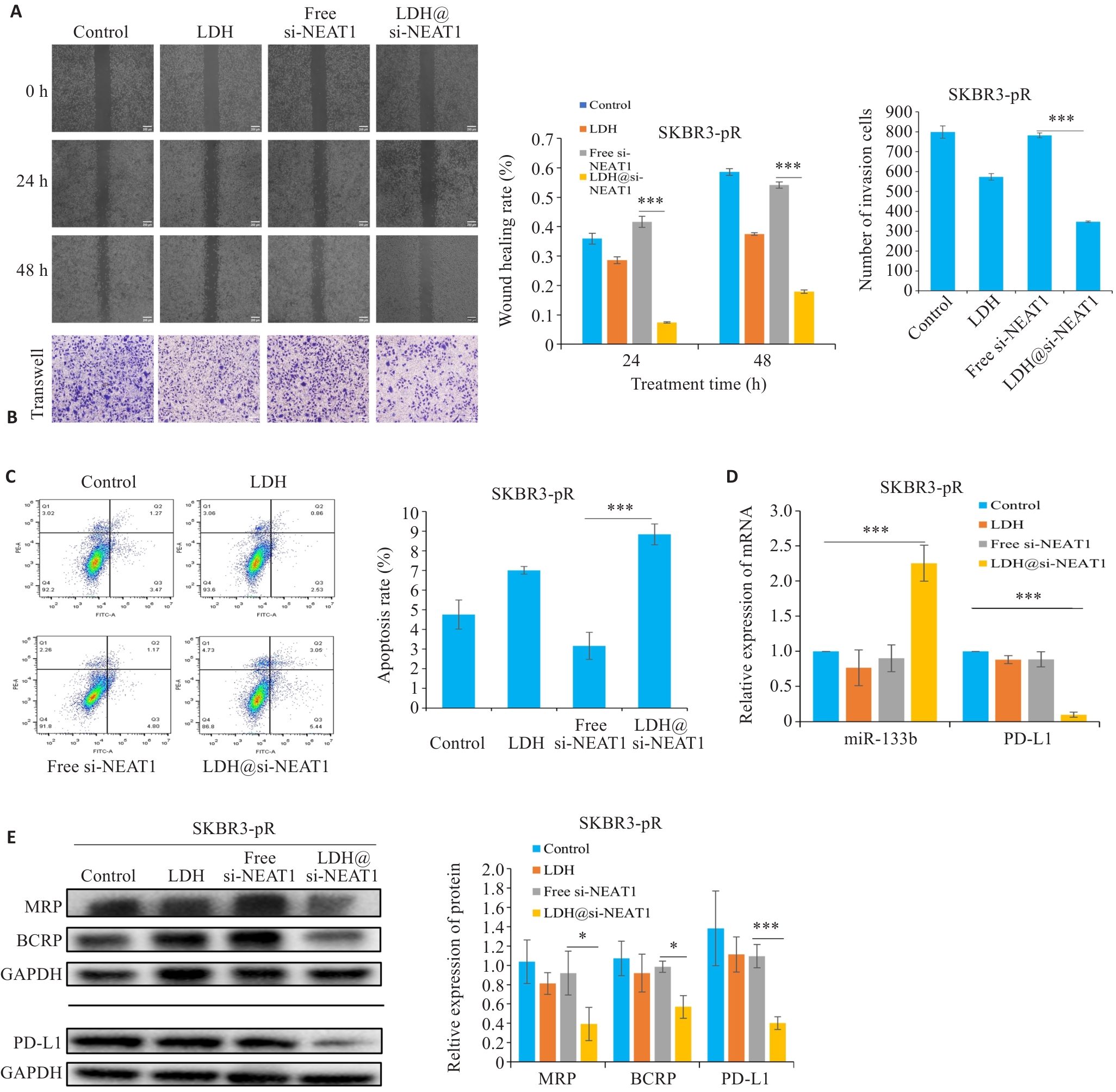
Fig.9 LDH@si-NEAT1 regulate paclitaxel resistance in breast cancer cells. A, B: Scratch and Transwell assays to assess the migration and invasion capabilities of drug-resistant cells. C: Flow cytometry for detecting apoptosis rate of drug-resistant cells. D: qRT-PCR for detecting expressions of miR-133b and PD-L1, respectively. E: Western blotting for detecting expressions of drug resistance proteins and PD-L1. *P<0.05, ***P<0.001.
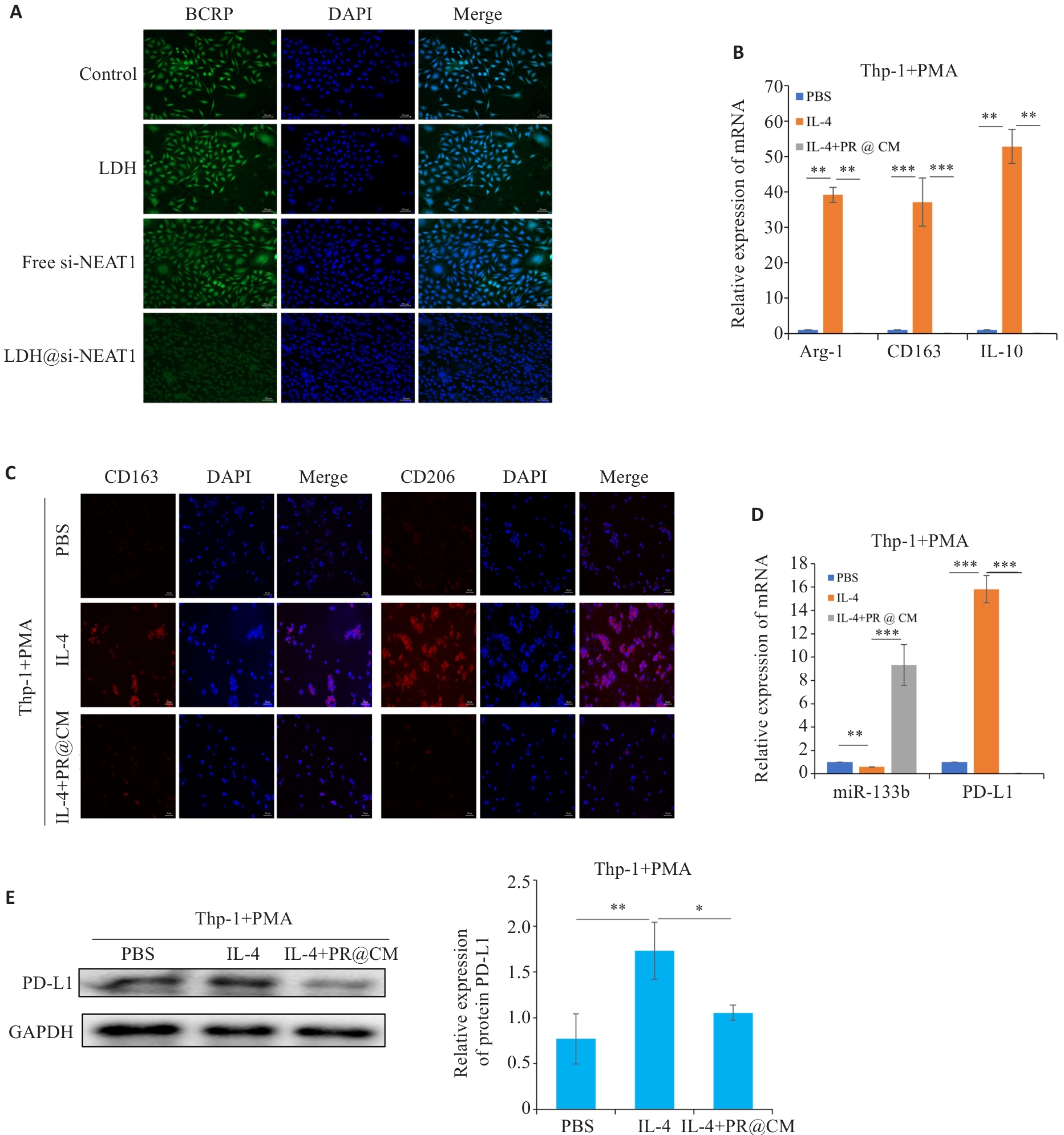
Fig.10 LDH@si-NEAT1 regulate M2 polarization state. A: Immunofluorescence staining for detecting expressions of drug resistance proteins (scale bar=50 μm). B, C: qRT-PCR and immunofluorescence staining for detecting M2 macrophage polarization markers (scale bar=50 μm). D: qRT-PCR of the expressions of miR-133b and PD-L1. E: Western blotting of the expression of PD-L1 in macrophages. *P<0.05, **P<0.01, ***P<0.001.
| [1] | Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-63. doi:10.3322/caac.21834 |
| [2] | 石英香, 王 婧, 石玉荣, 等. 乳腺癌紫杉醇耐药机制的研究进展[J]. 齐齐哈尔医学院学报, 2022, 43(6): 557-61. |
| [3] | Abu Samaan TM, Samec M, Liskova A, et al. Paclitaxel's mechanistic and clinical effects on breast cancer[J]. Biomolecules, 2019, 9(12): 789. doi:10.3390/biom9120789 |
| [4] | Ghafouri-Fard S, Taheri M. Nuclear enriched abundant transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis[J]. Biomed Pharmacother, 2019, 111: 51-9. doi:10.1016/j.biopha.2018.12.070 |
| [5] | Wang C, Duan YJ, Duan G, et al. Stress induces dynamic, cytotoxicity-antagonizing TDP-43 nuclear bodies via paraspeckle LncRNA NEAT1-mediated liquid-liquid phase separation[J]. Mol Cell, 2020, 79(3): 443-58.e7. doi:10.1016/j.molcel.2020.06.019 |
| [6] | Wei XB, Jiang WQ, Zeng JH, et al. Exosome-derived lncRNA NEAT1 exacerbates sepsis-associated encephalopathy by promoting ferroptosis through regulating miR-9-5p/TFRC and GOT1 axis[J]. Mol Neurobiol, 2022, 59(3): 1954-69. doi:10.1007/s12035-022-02738-1 |
| [7] | Luo X, Wei QL, Jiang XY, et al. CSTF3 contributes to platinum resistance in ovarian cancer through alternative polyadenylation of lncRNA NEAT1 and generating the short isoform NEAT1_1[J]. Cell Death Dis, 2024, 15(6): 432. doi:10.1038/s41419-024-06816-1 |
| [8] | Puccio N, Manzotti G, Mereu E, et al. Combinatorial strategies targeting NEAT1 and AURKA as new potential therapeutic options for multiple myeloma[J]. Haematologica, 2024, 109(12): 4040-55. |
| [9] | Li MY, Yang YH, Xiong LT, et al. Metabolism, metabolites, and macrophages in cancer[J]. J Hematol Oncol, 2023, 16(1): 80. doi:10.1186/s13045-023-01478-6 |
| [10] | Borisenko GG, Matsura T, Liu SX, et al. Macrophage recognition of externalized phosphatidylserine and phagocytosis of apoptotic Jurkat cells: existence of a threshold[J]. Arch Biochem Biophys, 2003, 413(1): 41-52. doi:10.1016/s0003-9861(03)00083-3 |
| [11] | Liu DL, Wei YH, Liu YD, et al. The long non-coding RNA NEAT1/miR-224-5p/IL-33 axis modulates macrophage M2a polarization and A1 astrocyte activation[J]. Mol Neurobiol, 2021, 58(9): 4506-19. doi:10.1007/s12035-021-02405-x |
| [12] | 彭 威, 徐 旸, 王文锐. 层状双氢氧化物作为基因药物递送载体的研究进展[J]. 中国医药工业杂志, 2020, 51(10): 1243-53. doi:10.16522/j.cnki.cjph.2020.10.003 |
| [13] | Li L, Soyhan I, Warszawik E, et al. Layered double hydroxides: recent progress and promising perspectives toward biomedical applications[J]. Adv Sci (Weinh), 2024, 11(20): e2306035. doi:10.1002/advs.202306035 |
| [14] | 徐惠敏, 史竞彤, 孙轶华. 肿瘤相关巨噬细胞的极化方式及其对乳腺癌进展的影响[J]. 现代肿瘤医学, 2021, 29(22): 4049-54. doi:10.3969/j.issn.1672-4992.2021.22.037 |
| [15] | Mou SJ, Yang PF, Liu YP, et al. BCLAF1 promotes cell proliferation, invasion and drug-resistance though targeting lncRNA NEAT1 in hepatocellular carcinoma[J]. Life Sci, 2020, 242: 117177. doi:10.1016/j.lfs.2019.117177 |
| [16] | Wei XY, Tao S, Mao HL, et al. Exosomal lncRNA NEAT1 induces paclitaxel resistance in breast cancer cells and promotes cell migration by targeting miR-133b[J]. Gene, 2023, 860: 147230. doi:10.1016/j.gene.2023.147230 |
| [17] | Long F, Li X, Pan JY, et al. The role of lncRNA NEAT1 in human cancer chemoresistance[J]. Cancer Cell Int, 2024, 24(1): 236. doi:10.1186/s12935-024-03426-x |
| [18] | Wang JY, Zhang JH, Liu H, et al. N6-methyladenosine reader hnRNPA2B1 recognizes and stabilizes NEAT1 to confer chemoresistance in gastric cancer[J]. Cancer Commun (Lond), 2024, 44(4): 469-90. doi:10.1002/cac2.12534 |
| [19] | Zhang SJ, Kim EJ, Huang JF, et al. NEAT1 repression by MED12 creates chemosensitivity in p53 wild-type breast cancer cells[J]. FEBS J, 2024, 291(9): 1909-24. doi:10.1111/febs.17097 |
| [20] | Zhen SM, Jia YL, Zhao Y, et al. NEAT1_1 confers gefitinib resistance in lung adenocarcinoma through promoting AKR1C1-mediated ferroptosis defence[J]. Cell Death Discov, 2024, 10(1): 131. doi:10.1038/s41420-024-01892-w |
| [21] | Wang XW, Xu Y, Zhu YC, et al. LncRNA NEAT1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting miR-27b-3p and ZEB1 in diabetic nephropathy[J]. J Cell Physiol, 2019, 234(8): 12926-33. doi:10.1002/jcp.27959 |
| [22] | Lin SP, Wen ZK, Li SX, et al. LncRNA Neat1 promotes the macrophage inflammatory response and acts as a therapeutic target in titanium particle-induced osteolysis[J]. Acta Biomater, 2022, 142: 345-60. doi:10.1016/j.actbio.2022.02.007 |
| [23] | Wei FF, Yan ZY, Zhang XM, et al. LncRNA-NEAT1 inhibits the occurrence and development of pancreatic cancer through spongy miR-146b-5p/traf6[J]. Biotechnol Genet Eng Rev, 2024, 40(2): 1094-112. doi:10.1080/02648725.2023.2192059 |
| [24] | Geng F, Jia WC, Li T, et al. Knockdown of lncRNA NEAT1 suppresses proliferation and migration, and induces apoptosis of cervical cancer cells by regulating the miR-377/FGFR1 axis[J]. Mol Med Rep, 2022, 25(1): 10. doi:10.3892/mmr.2021.12526 |
| [25] | Zhang K, Zhou H, Yan B, et al. TUG1/miR-133b/CXCR4 axis regulates cisplatin resistance in human tongue squamous cell carcinoma[J]. Cancer Cell Int, 2020, 20: 148. doi:10.1186/s12935-020-01224-9 |
| [26] | Li PF, Sun Q, Bai SP, et al. Combination of the cuproptosis inducer disulfiram and anti-PD-L1 abolishes NSCLC resistance by ATP7B to regulate the HIF-1 signaling pathway[J]. Int J Mol Med, 2024, 53(2): 19. doi:10.3892/ijmm.2023.5343 |
| [27] | Rabiee N, Bagherzadeh M, Ghadiri AM, et al. ZnAl nano layered double hydroxides for dual functional CRISPR/Cas9 delivery and enhanced green fluorescence protein biosensor[J]. Sci Rep, 2020, 10(1): 20672. doi:10.1038/s41598-020-77809-1 |
| [28] | Lee J, Seo HS, Park W, et al. Biofunctional layered double hydroxide nanohybrids for cancer therapy[J]. Materials (Basel), 2022, 15(22): 7977. doi:10.3390/ma15227977 |
| [29] | Zhang SW, Pang SY, Pei WH, et al. Layered double hydroxide-loaded miR-30a for the treatment of breast cancer in vitro and in vivo [J]. ACS Omega, 2023, 8(21): 18435-48. doi:10.1021/acsomega.2c07866 |
| [30] | Li L, Qian YJ, Sun LY, et al. Albumin-stabilized layered double hydroxide nanoparticles synergized combination chemotherapy for colorectal cancer treatment[J]. Nanomedicine, 2021, 34: 102369. doi:10.1016/j.nano.2021.102369 |
| [31] | Chang MY, Wang M, Liu B, et al. A cancer nanovaccine based on an FeAl-layered double hydroxide framework for reactive oxygen species-augmented metalloimmunotherapy[J]. ACS Nano, 2024, 18(11): 8143-56. doi:10.1021/acsnano.3c11960 |
| [1] | Siyuan MA, Bochao ZHANG, Chun PU. Circ_0000437 promotes proliferation, invasion, migration and epithelial-mesenchymal transition of breast cancer cells by targeting the let-7b-5p/CTPS1 axis [J]. Journal of Southern Medical University, 2025, 45(8): 1682-1696. |
| [2] | Jiahao LI, Ruiting XIAN, Rong LI. Down-regulation of ACADM-mediated lipotoxicity inhibits invasion and metastasis of estrogen receptor-positive breast cancer cells [J]. Journal of Southern Medical University, 2025, 45(6): 1163-1173. |
| [3] | Di CHEN, Ying LÜ, Yixin GUO, Yirong ZHANG, Ruixuan WANG, Xiaoruo ZHOU, Yuxin CHEN, Xiaohui WU. Dihydroartemisinin enhances doxorubicin-induced apoptosis of triple negative breast cancer cells by negatively regulating the STAT3/HIF-1α pathway [J]. Journal of Southern Medical University, 2025, 45(2): 254-260. |
| [4] | Qiao CHU, Xiaona WANG, Jiaying XU, Huilin PENG, Yulin ZHAO, Jing ZHANG, Guoyu LU, Kai WANG. Pulsatilla saponin D inhibits invasion and metastasis of triple-negative breast cancer cells through multiple targets and pathways [J]. Journal of Southern Medical University, 2025, 45(1): 150-161. |
| [5] | Mingzi OUYANG, Jiaqi CUI, Hui WANG, Zheng LIANG, Dajin PI, Liguo CHEN, Qianjun CHEN, Yingchao WU. Kaixinsan alleviates adriamycin-induced depression-like behaviors in mice by reducing ferroptosis in the prefrontal cortex [J]. Journal of Southern Medical University, 2024, 44(8): 1441-1449. |
| [6] | Jincun FANG, Liwei LIU, Junhao LIN, Fengsheng CHEN. Overexpression of CDHR2 inhibits proliferation of breast cancer cells by inhibiting the PI3K/Akt pathway [J]. Journal of Southern Medical University, 2024, 44(6): 1117-1125. |
| [7] | Zhi CUI, Cuijiao MA, Qianru WANG, Jinhao CHEN, Ziyang YAN, Jianlin YANG, Yafeng LÜ, Chunyu CAO. A recombinant adeno-associated virus expressing secretory TGF‑β type II receptor inhibits triple-negative murine breast cancer 4T1 cell proliferation and lung metastasis in mice [J]. Journal of Southern Medical University, 2024, 44(5): 818-826. |
| [8] | CHEN Shoufeng, ZHANG Shuchao, FAN Weilin, SUN Wei, LIU Beibei, LIU Jianmin, GUO Yuanyuan. Efficacy of combined treatment with pirfenidone and PD-L1 inhibitor in mice bearing ectopic bladder cancer xenograft [J]. Journal of Southern Medical University, 2024, 44(2): 210-216. |
| [9] | Youqin ZENG, Siyu CHEN, Yan LIU, Yitong LIU, Ling ZHANG, Jiao XIA, Xinyu WU, Changyou WEI, Ping LENG. AKBA combined with doxorubicin inhibits proliferation and metastasis of triple-negative breast cancer MDA-MB-231 cells and xenograft growth in nude mice [J]. Journal of Southern Medical University, 2024, 44(12): 2449-2460. |
| [10] | Jinhong ZHANG, Xin LIU, Jian LIU. PHPS1 enhances PD-L1 serine phosphorylation by regulating ROS/SHP-2/AMPK activity to promote apoptosis of oral squamous cell carcinoma cells [J]. Journal of Southern Medical University, 2024, 44(12): 2469-2476. |
| [11] | XU Mengqi, SHI Yutong, LIU Junping, WU Minmin, ZHANG Fengmei, HE Zhiqiang, TANG Min. JAG1 affects monocytes-macrophages to reshape the pre-metastatic niche of triple-negative breast cancer through LncRNA MALAT1 in exosomes [J]. Journal of Southern Medical University, 2023, 43(9): 1525-1535. |
| [12] | XU Jiaming, LIN Long, CHEN Qionghui, LI Lan. Hsa-miR-148a-3p promotes malignant behavior of breast cancer cells by downregulating DUSP1 [J]. Journal of Southern Medical University, 2023, 43(9): 1515-1524. |
| [13] | WANG Li, YAN Zhirui, XIA Yaoxiong. Silencing RAB27a inhibits proliferation, invasion and adhesion of triple-negative breast cancer cells [J]. Journal of Southern Medical University, 2023, 43(4): 560-567. |
| [14] | Guangdong Medical Industry Association(GMIA). GMIA-Breast Oncoplastic and Reconstruction Society consensus on operative standards of breast cancer surgery [J]. Journal of Southern Medical University, 2023, 43(10): 1827-1832. |
| [15] | GUO Li, MA Yinling, LI Ting, LI Jinping. CD40LG is a novel immune- and stroma-related prognostic biomarker in the tumor microenvironment of invasive breast cancer [J]. Journal of Southern Medical University, 2022, 42(9): 1267-1278. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||