南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (1): 150-161.doi: 10.12122/j.issn.1673-4254.2025.01.18
褚乔1( ), 王小娜2, 续佳颖3, 彭荟林3, 赵裕琳3, 张静3, 陆国玉1, 王恺3(
), 王小娜2, 续佳颖3, 彭荟林3, 赵裕琳3, 张静3, 陆国玉1, 王恺3( )
)
收稿日期:2024-08-29
出版日期:2025-01-20
发布日期:2025-01-20
通讯作者:
王恺
E-mail:chuqiao670906094@163.com;wangkai@swmu.edu.cn
作者简介:褚 乔,主管护师,E-mail: chuqiao670906094@163.com
基金资助:
Qiao CHU1( ), Xiaona WANG2, Jiaying XU3, Huilin PENG3, Yulin ZHAO3, Jing ZHANG3, Guoyu LU1, Kai WANG3(
), Xiaona WANG2, Jiaying XU3, Huilin PENG3, Yulin ZHAO3, Jing ZHANG3, Guoyu LU1, Kai WANG3( )
)
Received:2024-08-29
Online:2025-01-20
Published:2025-01-20
Contact:
Kai WANG
E-mail:chuqiao670906094@163.com;wangkai@swmu.edu.cn
摘要:
目的 采用网络药理学和计算机模拟探讨白头翁皂苷D(PSD)抑制三阴性乳腺癌(TNBC)侵袭转移的作用机制,并进行实验验证。 方法 运用Super-PRED、Swiss Target Prediction、PharmMapper、STITCH和BATMAN-TCM数据库收集PSD潜在靶点,利用GeneCards和OMIM数据库获得TNBC侵袭转移靶点,将两者相交获得药物-疾病交集靶点,利用Cytoscape3.10.1软件绘制“PSD-靶点-疾病”互作网络。运用Cytoscape3.10.1中的Centiscape2.2插件设定阈值并获得核心靶点,使用String数据库进行蛋白质互作(PPI)分析,通过David数据库对核心靶点进行KEGG通路和GO功能富集分析。最后将核心靶点依次与PSD进行分子对接。通过Transwell和Western blotting法对PSD的作用及机制进行验证。 结果 网络药理学结果显示,共筛选出PSD潜在靶点285个及药物与疾病核心靶点26个。GO分析获得175个条目,涉及生物大分子(蛋白质、DNA、RNA)的结合、酶活性、基因转录调控等方面。KEGG分析获得46个条目,涉及癌症途径、化学致癌-受体活化、癌症中的微小RNA、化学致癌-活性氧、癌症中PD-L1表达和PD-1检查点通路等。分子对接显示,PSD与MTOR、HDAC2、ABL1、CDK1、TLR4、TERT、PIK3R1、NFE2L2、PTPN1有较高的结合性。Transwell和Western blotting结果显示,PSD抑制TNBC细胞侵袭迁移且降低其MMP2、MMP9、N-cadherin及关键蛋白p-mTOR、ABL1、TERT、PTPN1、HDAC2、PIK3R1、CDK1、TLR4和细胞核内NFE2L2的表达(P<0.05),PSD可通过这些靶点阻碍TNBC侵袭转移。 结论 PSD通过多靶点和多途径抑制TNBC侵袭转移,为后续深入的机制研究提供前期基础,为TNBC药物研发提供新思路。
褚乔, 王小娜, 续佳颖, 彭荟林, 赵裕琳, 张静, 陆国玉, 王恺. 白头翁皂苷D通过多靶点和多途径抑制三阴性乳腺癌侵袭转移[J]. 南方医科大学学报, 2025, 45(1): 150-161.
Qiao CHU, Xiaona WANG, Jiaying XU, Huilin PENG, Yulin ZHAO, Jing ZHANG, Guoyu LU, Kai WANG. Pulsatilla saponin D inhibits invasion and metastasis of triple-negative breast cancer cells through multiple targets and pathways[J]. Journal of Southern Medical University, 2025, 45(1): 150-161.
| Compound | Target genes | Number of targets |
|---|---|---|
| PSD | BLM, PTPN1, GPR6, APEX1, KDM1A, GPR55, PTGS1, KLF5, TOP2A, TRIM24, NFKB1, SLC6A5, CLK4, ADAM10, CTSD, PDGFRA, NTRK3, NR3C2, CSNK2B, SCN2A, PTPN2, MTOR, TFPI, TLR4, SCN3A, PIK3R1, DUSP3, CYP3A4, PDE3A, GLRA1ITK, HSP90AB1, GRIA2, GRIN1, C5AR1, HSP90AA1, MMP1, FPR2, CFTR, ACACB, HDAC2, TLR8, S1PR5, SLC9A1, PSMB1, AURKB, NTSR2, HDAC7, LTA4H, F13A1, SOAT1, ATG4B, NFE2L2, CACNA1B, CHUK, NR2E3, CDK1, ADORA1, CYSLTR2, AR, CDC25B, MDM4, NR4A1, SLC40A1, SLC1A3, SCN4A, AKR1C3, CAPN1, KIF11, WDR5, TERT, PRCP, HDAC5, MAP3K14, CHRM1, TACR2, QRFPR, NR1I2, GABRA1, PSMB9, ABL1, PTGER2, CBX4, PRMT1, ABCB1, CACNA1H, DCUN1D1, FAAH, FPR1, ACACA, MC4R, RXFP1, ADK, CDK5, PDGFRB, SLC1A2, METAP2, NOX1, PROC, PRSS1, PLA2G2A, MME, PTK2B, RECQ2, PTP1B, APE, AOF2, COX1, BTEB2, TOP2, RNF82, GLYT2, KUZ, CPSD, PDGFR2, TRKC, MCR, CK2N, HBA, PTPT, FRAP, LACI, KIAA1356, GRB1, VHR, CYP3A3, EMT, HSP90B, GLUR2, NMDAR1, C5AR, HSP90A, CLG, FPRH1, ABCC7, ACC2, UNQ249/PRO286, EDG8, APNH1, PSC5, AIK2, HDAC7A, LTA4, F13A, ACACT, APG4B, NRF2, CACH5, IKKA, PNR, CDC2, CYSLT2, DHTR, CDC25HU2, MDMX, GFRP1, FPN, EAAT1, DDH1, CANPL1, EG5, BIG3, EST2, PCP, KIAA0600, NIK, NK2R, GPR103, PXR, LMP2, ABL, HMT2, MDR1, DCN1, FAAH1, ACAC, LGR7, CDKN5, PDGFR, EAAT2, MNPEP, MOX1, TRP1, PLA2B, EPN, FAK2, RECQL3, APE1, KDM1, CKLF, TIF1, NET1, MADM, RHEPDGFRA, MLR, G5A, NAC2, FRAP1, TFPI1, NAC3, LYK, HSPC2, C5R1, HSPC1, FPRL1, ACCB, NHE1, AIM1, ACACT1, AUTL1, CACNL1A5, TCF16, RNR, CDC28A, CYSLT2R, NR3C4, HMR, FPN1, GLAST, HSD17B5, PIG30, KNSL1, TCS1, NKNAR, PSMB6i, JTK7, HRMT1L2, PGY1, DCUN1L1, ACC1, PSSALRE, PDGFR1, GLT1, P67EIF2, NOH1, TRY1, PLA2L, PYK2, APEX, KIAA0601, IKLF, TIF1A, SCN2A1, FRAP2, HSPCB, HSPCA, LXA4R, AIRK2, ACAT, KIAA0943, CDKN1, PSEC0146, NAK1, IREG1, GLAST1, KIAA0119, TRIP5, TRT, TAC2R, RING12, IR1B4, RP42, ACCA, TRYP1, RASF-A, RAFTK, SCCRO, APX, LSD1, HAP1, REF1, RAPT1, SCN2A2, RAFT1, STK12, STK5, ARK2, STAT, ACAT1, P34CDC2, MSTP079, SLC11A3, PGFS, STK1, SOAT | 285 |
表1 PSD的潜在作用靶点
Tab.1 Potential targets of Pulsatilla saponin D (PSD)
| Compound | Target genes | Number of targets |
|---|---|---|
| PSD | BLM, PTPN1, GPR6, APEX1, KDM1A, GPR55, PTGS1, KLF5, TOP2A, TRIM24, NFKB1, SLC6A5, CLK4, ADAM10, CTSD, PDGFRA, NTRK3, NR3C2, CSNK2B, SCN2A, PTPN2, MTOR, TFPI, TLR4, SCN3A, PIK3R1, DUSP3, CYP3A4, PDE3A, GLRA1ITK, HSP90AB1, GRIA2, GRIN1, C5AR1, HSP90AA1, MMP1, FPR2, CFTR, ACACB, HDAC2, TLR8, S1PR5, SLC9A1, PSMB1, AURKB, NTSR2, HDAC7, LTA4H, F13A1, SOAT1, ATG4B, NFE2L2, CACNA1B, CHUK, NR2E3, CDK1, ADORA1, CYSLTR2, AR, CDC25B, MDM4, NR4A1, SLC40A1, SLC1A3, SCN4A, AKR1C3, CAPN1, KIF11, WDR5, TERT, PRCP, HDAC5, MAP3K14, CHRM1, TACR2, QRFPR, NR1I2, GABRA1, PSMB9, ABL1, PTGER2, CBX4, PRMT1, ABCB1, CACNA1H, DCUN1D1, FAAH, FPR1, ACACA, MC4R, RXFP1, ADK, CDK5, PDGFRB, SLC1A2, METAP2, NOX1, PROC, PRSS1, PLA2G2A, MME, PTK2B, RECQ2, PTP1B, APE, AOF2, COX1, BTEB2, TOP2, RNF82, GLYT2, KUZ, CPSD, PDGFR2, TRKC, MCR, CK2N, HBA, PTPT, FRAP, LACI, KIAA1356, GRB1, VHR, CYP3A3, EMT, HSP90B, GLUR2, NMDAR1, C5AR, HSP90A, CLG, FPRH1, ABCC7, ACC2, UNQ249/PRO286, EDG8, APNH1, PSC5, AIK2, HDAC7A, LTA4, F13A, ACACT, APG4B, NRF2, CACH5, IKKA, PNR, CDC2, CYSLT2, DHTR, CDC25HU2, MDMX, GFRP1, FPN, EAAT1, DDH1, CANPL1, EG5, BIG3, EST2, PCP, KIAA0600, NIK, NK2R, GPR103, PXR, LMP2, ABL, HMT2, MDR1, DCN1, FAAH1, ACAC, LGR7, CDKN5, PDGFR, EAAT2, MNPEP, MOX1, TRP1, PLA2B, EPN, FAK2, RECQL3, APE1, KDM1, CKLF, TIF1, NET1, MADM, RHEPDGFRA, MLR, G5A, NAC2, FRAP1, TFPI1, NAC3, LYK, HSPC2, C5R1, HSPC1, FPRL1, ACCB, NHE1, AIM1, ACACT1, AUTL1, CACNL1A5, TCF16, RNR, CDC28A, CYSLT2R, NR3C4, HMR, FPN1, GLAST, HSD17B5, PIG30, KNSL1, TCS1, NKNAR, PSMB6i, JTK7, HRMT1L2, PGY1, DCUN1L1, ACC1, PSSALRE, PDGFR1, GLT1, P67EIF2, NOH1, TRY1, PLA2L, PYK2, APEX, KIAA0601, IKLF, TIF1A, SCN2A1, FRAP2, HSPCB, HSPCA, LXA4R, AIRK2, ACAT, KIAA0943, CDKN1, PSEC0146, NAK1, IREG1, GLAST1, KIAA0119, TRIP5, TRT, TAC2R, RING12, IR1B4, RP42, ACCA, TRYP1, RASF-A, RAFTK, SCCRO, APX, LSD1, HAP1, REF1, RAPT1, SCN2A2, RAFT1, STK12, STK5, ARK2, STAT, ACAT1, P34CDC2, MSTP079, SLC11A3, PGFS, STK1, SOAT | 285 |
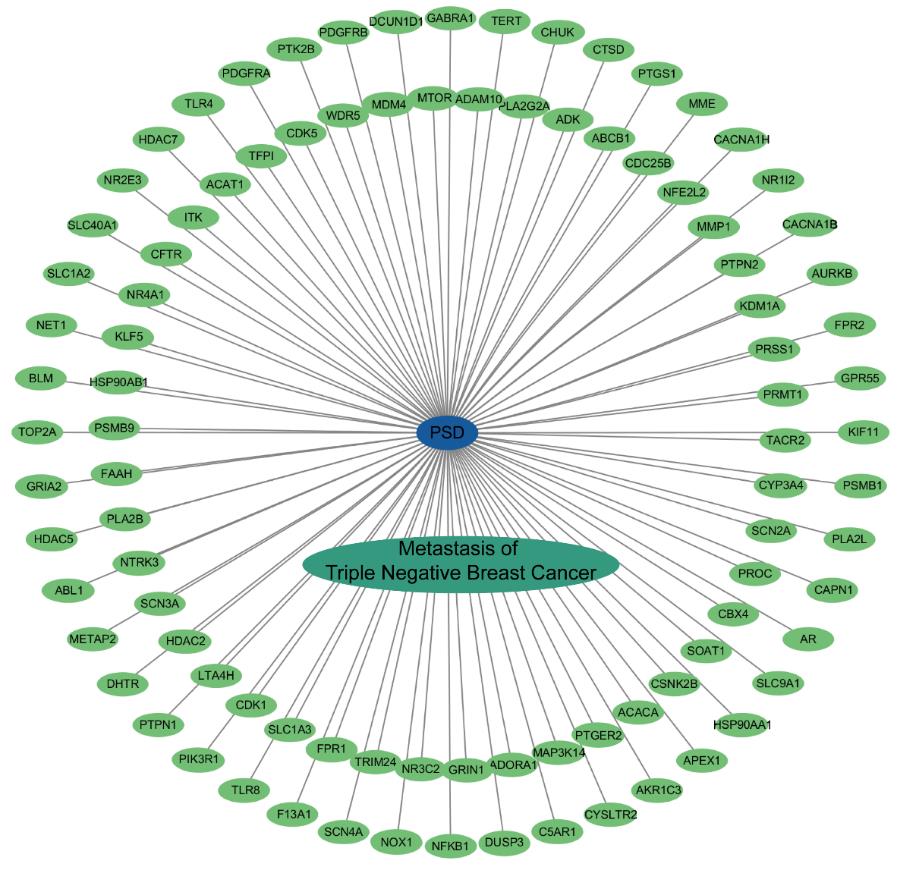
图2 “PSD-靶点-疾病”互作网络
Fig.2 "PSD-target-disease" interaction network. Blue oval nodes represent compound PSD, green oval nodes represent crossover genes, cyan oval nodes denote triple-negative breast cancer metastasis (disease), and gray lines stand for interactions.
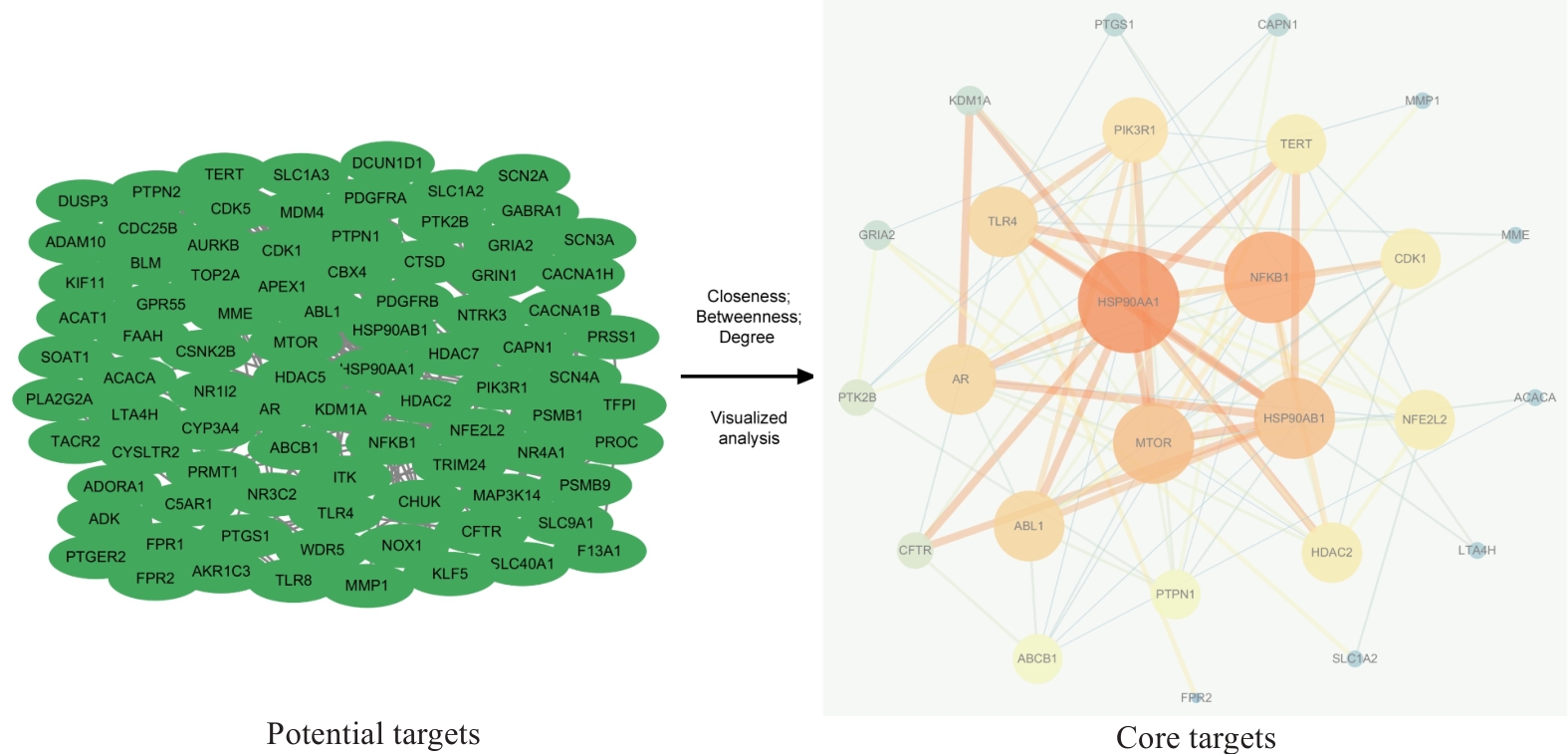
图3 PSD治疗TNBC侵袭转移的PPI网络可视化图
Fig.3 Visualization of PPI network for PSD treatment of TNBC invasion and metastasis. Green oval nodes on the left side represent 94 potential targets and the circular nodes on the right side represent 26 core targets. The size and color shades of the nodes are proportional to their importance, and the thickness and color shades of the lines are proportional to the importance between nodes.
| Core targets | Degree unDir | PDB ID |
|---|---|---|
| HSP90AA1 | 39 | 6GR5 |
| HSP90AB1 | 33 | 6N8Y |
| NFKB1 | 27 | 1SVC |
| MTOR | 24 | 4JSN |
| HDAC2 | 23 | 7JS8 |
| ABL1 | 20 | 5HU9 |
| CDK1 | 19 | 6GU2 |
| TLR4 | 18 | 2Z62 |
| TERT | 17 | 5UGW |
| AR | 17 | 8E1A |
| PIK3R1 | 17 | 7PG5 |
| NFE2L2 | 16 | 7X5G |
| ABCB1 | 12 | 7A69 |
| KDM1A | 12 | 7E0G |
| CFTR | 11 | 5TFJ |
| PTPN1 | 11 | 7MN9 |
| PTGS1 | 10 | 6Y3C |
| GRIA2 | 9 | 7F3O |
| MME | 8 | 6SUK |
| PTK2B | 7 | 3CC6 |
| ACACA | 6 | 2YL2 |
| FPR2 | 6 | 6LW5 |
| CAPN1 | 6 | 7W7O |
| SLC1A2 | 5 | 7VR7 |
| LTA4H | 4 | 3U9W |
| MMP1 | 4 | 966C |
表2 PSD治疗TNBC侵袭转移的核心靶点
Tab.2 Core targets of PSD for the treatment of TNBC invasion and metastasis
| Core targets | Degree unDir | PDB ID |
|---|---|---|
| HSP90AA1 | 39 | 6GR5 |
| HSP90AB1 | 33 | 6N8Y |
| NFKB1 | 27 | 1SVC |
| MTOR | 24 | 4JSN |
| HDAC2 | 23 | 7JS8 |
| ABL1 | 20 | 5HU9 |
| CDK1 | 19 | 6GU2 |
| TLR4 | 18 | 2Z62 |
| TERT | 17 | 5UGW |
| AR | 17 | 8E1A |
| PIK3R1 | 17 | 7PG5 |
| NFE2L2 | 16 | 7X5G |
| ABCB1 | 12 | 7A69 |
| KDM1A | 12 | 7E0G |
| CFTR | 11 | 5TFJ |
| PTPN1 | 11 | 7MN9 |
| PTGS1 | 10 | 6Y3C |
| GRIA2 | 9 | 7F3O |
| MME | 8 | 6SUK |
| PTK2B | 7 | 3CC6 |
| ACACA | 6 | 2YL2 |
| FPR2 | 6 | 6LW5 |
| CAPN1 | 6 | 7W7O |
| SLC1A2 | 5 | 7VR7 |
| LTA4H | 4 | 3U9W |
| MMP1 | 4 | 966C |
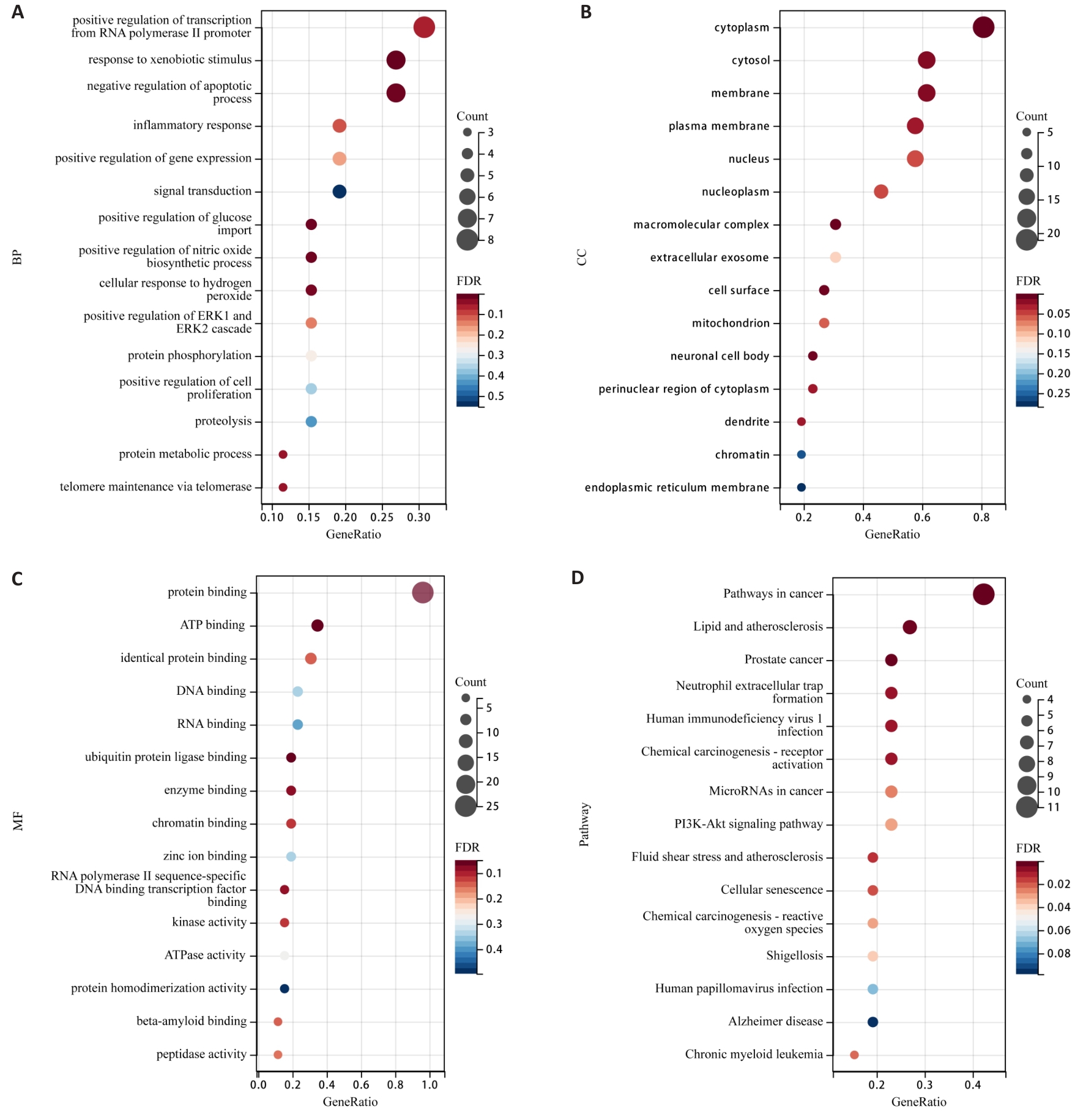
图4 PSD抑制TNBC侵袭转移的GO功能和KEGG通路富集气泡图
Fig.4 Bubble plots of GO function and KEGG pathway enrichment for PSD inhibition of TNBC invasion and metastasis. A: Enrichment analysis of BP. B: Enrichment analysis of CC. C: Enrichment analysis of MF. D: KEGG pathway enrichment analysis.
| ID | Pathways | P | Number of related genes |
|---|---|---|---|
| hsa05200 | Pathways in cancer | 0.000000974 | 11 |
| hsa05417 | Lipid and atherosclerosis | 0.000026000 | 7 |
| hsa05215 | Prostate cancer | 0.000007060 | 6 |
| hsa04613 | Neutrophil extracellular trap formation | 0.000184000 | 6 |
| hsa05170 | Human immunodeficiency virus 1 infection | 0.000298000 | 6 |
| hsa05207 | Chemical carcinogenesis - receptor activation | 0.000298000 | 6 |
| hsa05206 | MicroRNAs in cancer | 0.001673574 | 6 |
| hsa04151 | PI3K-Akt signaling pathway | 0.003182088 | 6 |
| hsa05418 | Fluid shear stress and atherosclerosis | 0.000618000 | 5 |
| hsa04218 | Cellular senescence | 0.000953000 | 5 |
| hsa05208 | Chemical carcinogenesis-reactive oxygen species | 0.003535389 | 5 |
| hsa05131 | Shigellosis | 0.005091764 | 5 |
| hsa05165 | Human papillomavirus infection | 0.014031422 | 5 |
| hsa05010 | Alzheimer disease | 0.022984313 | 5 |
| hsa05220 | Chronic myeloid leukemia | 0.001299397 | 4 |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 0.002047819 | 4 |
| hsa04657 | IL-17 signaling pathway | 0.002394257 | 4 |
| hsa04914 | Progesterone-mediated oocyte maturation | 0.003020588 | 4 |
| hsa04931 | Insulin resistance | 0.003550859 | 4 |
| hsa04659 | Th17 cell differentiation | 0.003550859 | 4 |
| hsa04066 | HIF-1 signaling pathway | 0.003644426 | 4 |
| hsa04152 | AMPK signaling pathway | 0.004886673 | 4 |
| hsa04910 | Insulin signaling pathway | 0.006900929 | 4 |
| hsa05135 | Yersinia infection | 0.006900929 | 4 |
| hsa05226 | Gastric cancer | 0.008693554 | 4 |
| hsa04217 | Necroptosis | 0.010557294 | 4 |
| hsa05161 | Hepatitis B | 0.010919274 | 4 |
| hsa05225 | Hepatocellular carcinoma | 0.012048446 | 4 |
| hsa04141 | Protein processing in endoplasmic reticulum | 0.012439336 | 4 |
| hsa04621 | NOD-like receptor signaling pathway | 0.015830764 | 4 |
| hsa05203 | Viral carcinogenesis | 0.020215918 | 4 |
| hsa05163 | Human cytomegalovirus infection | 0.026104939 | 4 |
| hsa04024 | cAMP signaling pathway | 0.026104939 | 4 |
| hsa05171 | Coronavirus disease - COVID-19 | 0.028568364 | 4 |
| hsa05132 | Salmonella infection | 0.033858771 | 4 |
| hsa05016 | Huntington disease | 0.056576135 | 4 |
| hsa04213 | Longevity regulating pathway - multiple species | 0.013188318 | 3 |
| hsa05221 | Acute myeloid leukemia | 0.015766996 | 3 |
| hsa05212 | Pancreatic cancer | 0.020005321 | 3 |
| hsa04012 | ErbB signaling pathway | 0.02466712 | 3 |
| hsa04211 | Longevity regulating pathway | 0.026868984 | 3 |
| hsa05146 | Amoebiasis | 0.034548353 | 3 |
| hsa05142 | Chagas disease | 0.034548353 | 3 |
| hsa04620 | Toll-like receptor signaling pathway | 0.038349814 | 3 |
| hsa04722 | Neurotrophin signaling pathway | 0.045714487 | 3 |
| hsa04919 | Thyroid hormone signaling pathway | 0.047106228 | 3 |
表3 KEGG通路富集
Tab.3 KEGG pathway enrichment
| ID | Pathways | P | Number of related genes |
|---|---|---|---|
| hsa05200 | Pathways in cancer | 0.000000974 | 11 |
| hsa05417 | Lipid and atherosclerosis | 0.000026000 | 7 |
| hsa05215 | Prostate cancer | 0.000007060 | 6 |
| hsa04613 | Neutrophil extracellular trap formation | 0.000184000 | 6 |
| hsa05170 | Human immunodeficiency virus 1 infection | 0.000298000 | 6 |
| hsa05207 | Chemical carcinogenesis - receptor activation | 0.000298000 | 6 |
| hsa05206 | MicroRNAs in cancer | 0.001673574 | 6 |
| hsa04151 | PI3K-Akt signaling pathway | 0.003182088 | 6 |
| hsa05418 | Fluid shear stress and atherosclerosis | 0.000618000 | 5 |
| hsa04218 | Cellular senescence | 0.000953000 | 5 |
| hsa05208 | Chemical carcinogenesis-reactive oxygen species | 0.003535389 | 5 |
| hsa05131 | Shigellosis | 0.005091764 | 5 |
| hsa05165 | Human papillomavirus infection | 0.014031422 | 5 |
| hsa05010 | Alzheimer disease | 0.022984313 | 5 |
| hsa05220 | Chronic myeloid leukemia | 0.001299397 | 4 |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 0.002047819 | 4 |
| hsa04657 | IL-17 signaling pathway | 0.002394257 | 4 |
| hsa04914 | Progesterone-mediated oocyte maturation | 0.003020588 | 4 |
| hsa04931 | Insulin resistance | 0.003550859 | 4 |
| hsa04659 | Th17 cell differentiation | 0.003550859 | 4 |
| hsa04066 | HIF-1 signaling pathway | 0.003644426 | 4 |
| hsa04152 | AMPK signaling pathway | 0.004886673 | 4 |
| hsa04910 | Insulin signaling pathway | 0.006900929 | 4 |
| hsa05135 | Yersinia infection | 0.006900929 | 4 |
| hsa05226 | Gastric cancer | 0.008693554 | 4 |
| hsa04217 | Necroptosis | 0.010557294 | 4 |
| hsa05161 | Hepatitis B | 0.010919274 | 4 |
| hsa05225 | Hepatocellular carcinoma | 0.012048446 | 4 |
| hsa04141 | Protein processing in endoplasmic reticulum | 0.012439336 | 4 |
| hsa04621 | NOD-like receptor signaling pathway | 0.015830764 | 4 |
| hsa05203 | Viral carcinogenesis | 0.020215918 | 4 |
| hsa05163 | Human cytomegalovirus infection | 0.026104939 | 4 |
| hsa04024 | cAMP signaling pathway | 0.026104939 | 4 |
| hsa05171 | Coronavirus disease - COVID-19 | 0.028568364 | 4 |
| hsa05132 | Salmonella infection | 0.033858771 | 4 |
| hsa05016 | Huntington disease | 0.056576135 | 4 |
| hsa04213 | Longevity regulating pathway - multiple species | 0.013188318 | 3 |
| hsa05221 | Acute myeloid leukemia | 0.015766996 | 3 |
| hsa05212 | Pancreatic cancer | 0.020005321 | 3 |
| hsa04012 | ErbB signaling pathway | 0.02466712 | 3 |
| hsa04211 | Longevity regulating pathway | 0.026868984 | 3 |
| hsa05146 | Amoebiasis | 0.034548353 | 3 |
| hsa05142 | Chagas disease | 0.034548353 | 3 |
| hsa04620 | Toll-like receptor signaling pathway | 0.038349814 | 3 |
| hsa04722 | Neurotrophin signaling pathway | 0.045714487 | 3 |
| hsa04919 | Thyroid hormone signaling pathway | 0.047106228 | 3 |
| Compound | Targets | PDB ID | MM-GBSA dG Bind (kcal/mol) |
|---|---|---|---|
| PSD | MTOR | 4JSN | -32.00 |
| HDAC2 | 7JS8 | -17.49 | |
| ABL1 | 5HU9 | -30.56 | |
| CDK1 | 6GU2 | -32.77 | |
| TLR4 | 2Z62 | -19.87 | |
| TERT | 5UGW | -39.09 | |
| PIK3R1 | 7PG5 | -31.43 | |
| NFE2L2 | 7X5G | -43.75 | |
| KDM1A | 7E0G | -43.68 | |
| PTPN1 | 7MN9 | -20.59 |
表4 PSD与核心靶蛋白的结合能
Tab.4 Free energy of PSD binding to the core target proteins
| Compound | Targets | PDB ID | MM-GBSA dG Bind (kcal/mol) |
|---|---|---|---|
| PSD | MTOR | 4JSN | -32.00 |
| HDAC2 | 7JS8 | -17.49 | |
| ABL1 | 5HU9 | -30.56 | |
| CDK1 | 6GU2 | -32.77 | |
| TLR4 | 2Z62 | -19.87 | |
| TERT | 5UGW | -39.09 | |
| PIK3R1 | 7PG5 | -31.43 | |
| NFE2L2 | 7X5G | -43.75 | |
| KDM1A | 7E0G | -43.68 | |
| PTPN1 | 7MN9 | -20.59 |
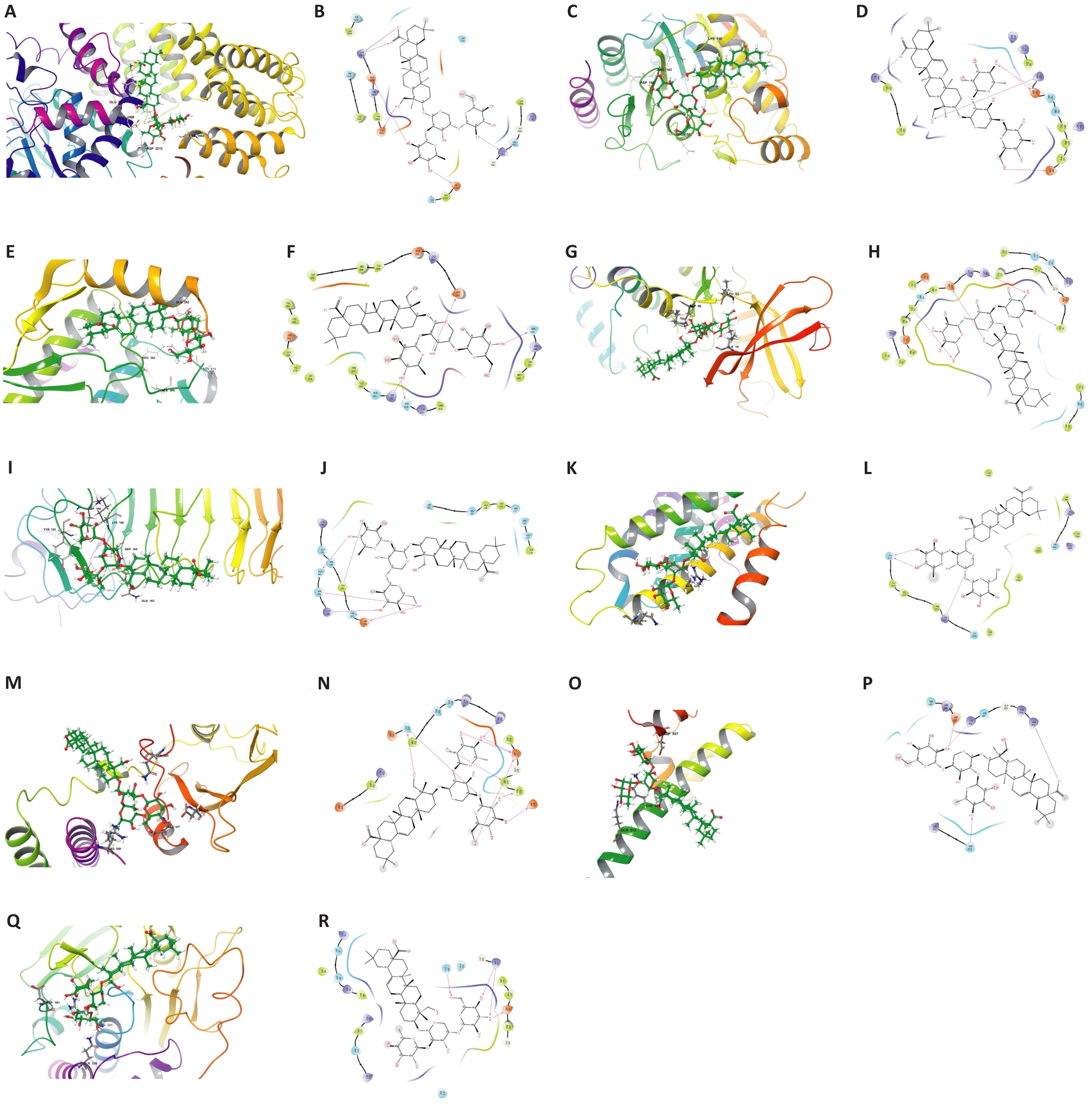
图5 PSD与关键靶蛋白的2D和3D结合模式图
Fig.5 2D and 3D binding pattern maps of PSD to the core target proteins. A: 3D binding model of PSD and MTOR. B: 2D binding model of PSD and MTOR. C: 3D binding model of PSD and HDAC2. D: 2D binding model of PSD and HDAC2. E: 3D binding model of PSD and ABL1. F: 2D binding model of PSD and ABL1. G: 3D binding model of PSD and CDK1. H: 2D binding model of PSD and CDK1. I: 3D binding model of PSD and TLR4. J: 2D binding model of PSD and TLR4. K: 3D binding model of PSD and TERT. L: 2D binding model of PSD and TERT. M: 3D binding model of PSD and PIK3R1. N: 2D binding model of PSD and PIK3R1. O: 3D binding model of PSD and NFE2L2. P: 2D binding model of PSD and NFE2L2. Q: 3D binding model of PSD and PTPN1. R: 2D binding model of PSD and PTPN1.
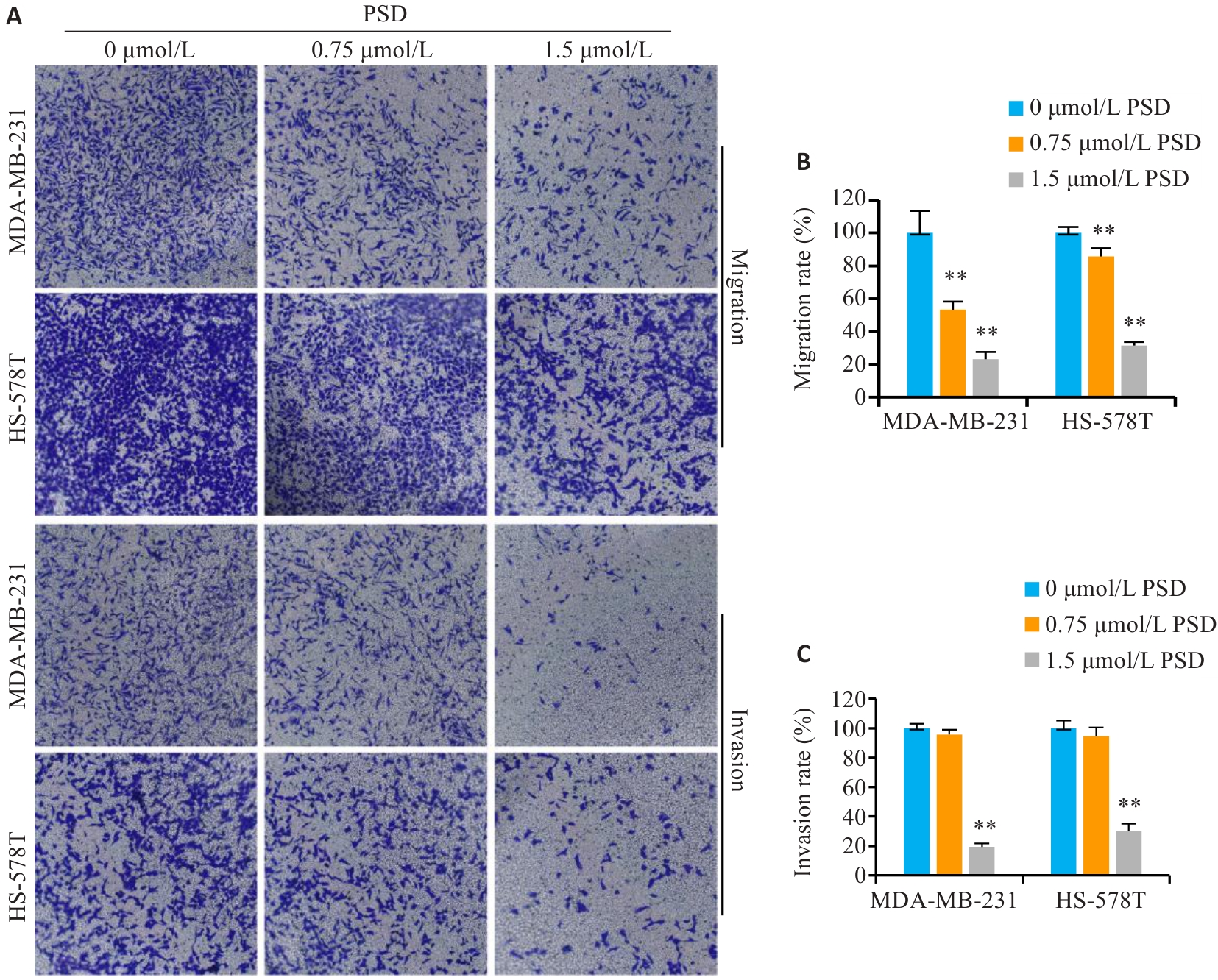
图6 PSD抑制TNBC细胞侵袭和迁移
Fig.6 PSD inhibits invasion and migration of TNBC cells. A: Crystal violet staining for detecting migration and invasion of MDA-MB-231 and HS-578T cells (Original magnification: ×200). B, C: Quantitative analysis of the results. **P<0.01 vs 0 μmol/L PSD group.
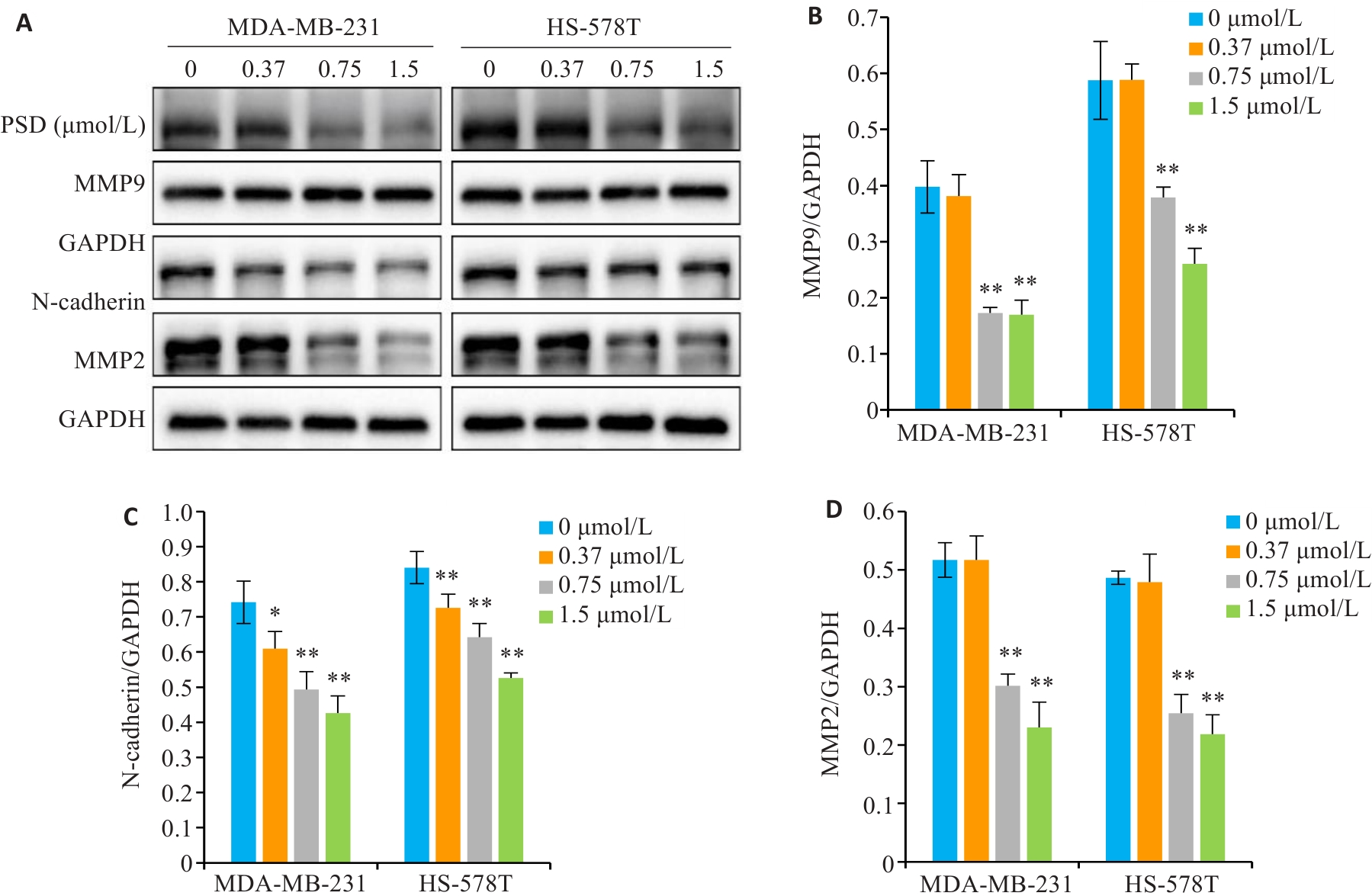
图7 PSD抑制侵袭迁移相关蛋白表达
Fig.7 PSD inhibits the expression of invasion and migration related proteins. A: MMP9, N-cadherin, and MMP2 protein levels were detected by Western blotting. B-D: Quantitative analysis of the results. *P<0.05, **P<0.01 vs 0 μmol/L group.

图8 PSD抑制TNBC细胞中9种核心靶蛋白表达
Fig.8 PSD inhibits the expression of 9 core target proteins in TNBC cells. A: p-mTOR, mTOR, CDK1, ABL1, TERT, PTPN1, HDAC2, PIK3R1, TLR4 and nuclear NRF2 protein levels detected by Western blotting. B-J: Quantitative analysis of the results. NRF2: NFE2L2. *P<0.05, **P<0.01 vs 0 μmol/L PSD group.
| 1 | Adrada BE, Moseley TW, Kapoor MM, et al. Triple-negative breast cancer: histopathologic features, genomics, and treatment[J]. Radiographics, 2023, 43(10): e230034. |
| 2 | Lin SY, Mo HN, Li YQ, et al. Clinicopathological characteristics and survival outcomes in patients with synchronous lung metastases upon initial metastatic breast cancer diagnosis in Han population[J]. BMC Cancer, 2021, 21(1): 1330. |
| 3 | 尹海庆, 苏 莉, 陈万贞, 等. 术后首发肺转移的三阴性乳腺癌临床病理特点及预后因素分析[J]. 现代肿瘤医学, 2019, 27(16): 2872-6. |
| 4 | Yang R, Shi YY, Han XH, et al. The impact of platinum-containing chemotherapies in advanced triple-negative breast cancer: meta-analytical approach to evaluating its efficacy and safety[J]. Oncol Res Treat, 2021, 44(6): 333-43. |
| 5 | Mason SR, Willson ML, Egger SJ, et al. Platinum-based chemotherapy for early triple-negative breast cancer[J]. Cochrane Database Syst Rev, 2023, 9(9): CD014805. |
| 6 | McGuinness JE, Kalinsky K. Antibody-drug conjugates in metastatic triple negative breast cancer: a spotlight on sacituzumab govitecan, ladiratuzumab vedotin, and trastuzumab deruxtecan[J]. Expert Opin Biol Ther, 2021, 21(7): 903-13. |
| 7 | Mediratta K, El-Sahli S, D’Costa V, et al. Current progresses and challenges of immunotherapy in triple-negative breast cancer[J]. Cancers, 2020, 12(12): 3529. |
| 8 | Li LY, Zhang F, Liu ZY, et al. Immunotherapy for triple-negative breast cancer: combination strategies to improve outcome[J]. Cancers, 2023, 15(1): 321. |
| 9 | Ye F, Dewanjee S, Li YH, et al. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer[J]. Mol Cancer, 2023, 22(1): 105. |
| 10 | Lev S. Targeted therapy and drug resistance in triple-negative breast cancer: the EGFR axis[J]. Biochem Soc Trans, 2020, 48(2): 657-65. |
| 11 | Li YH, Zou M, Han Q, et al. Therapeutic potential of triterpenoid saponin anemoside B4 from Pulsatilla chinensis [J]. Pharmacol Res, 2020, 160: 105079. |
| 12 | Fang YY, Hu DY, Li HL, et al. Synthesis, biological evaluation, and mode of action of Pulsatilla saponin D derivatives as promising anticancer agents[J]. Front Pharmacol, 2019, 10: 1208. |
| 13 | Zhang YL, Bao JL, Wang K, et al. Pulsatilla saponin D inhibits autophagic flux and synergistically enhances the anticancer activity of chemotherapeutic agents against HeLa cells[J]. Am J Chin Med, 2015, 43(8): 1657-70. |
| 14 | Hong SW, Jung KH, Lee HS, et al. SB365, Pulsatilla saponin D, targets c-Met and exerts antiangiogenic and antitumor activities[J]. Carcinogenesis, 2013, 34(9): 2156-69. |
| 15 | Lu YY, He WB, Huang XX, et al. Pulsatilla saponin D regulates ras-related C3 botulinum toxin substrate 3 (RAC3) to overcome resistance to paclitaxel in lung adenocarcinoma cells[J]. BMC Cancer, 2024, 24(1): 55. |
| 16 | Lim JH, Jung KH, Kim MS, et al. SB365 induces apoptosis and suppresses proliferation of glioblastoma cells[J]. Indian J Pharmacol, 2020, 52(2): 102-7. |
| 17 | Wang K, Tu YB, Wan JB, et al. Synergistic anti-breast cancer effect of pulsatilla saponin D and camptothecin through interrupting autophagic-lysosomal function and promoting p62-mediated ubiquitinated protein aggregation[J]. Carcinogenesis, 2020, 41(6): 804-16. |
| 18 | Zhong JM, Tan LH, Chen MW, et al. Pharmacological activities and molecular mechanisms of Pulsatilla saponins[J]. Chin Med, 2022, 17(1): 59. |
| 19 | Bryan S, Witzel I, Borgmann K, et al. Molecular mechanisms associated with brain metastases in HER2-positive and triple negative breast cancers[J]. Cancers, 2021, 13(16): 4137. |
| 20 | 江远玲, 冯 楠, 邵欣宇, 等. 黄芪的现代药理作用研究进展[J]. 西南医科大学学报, 2023, 46(5): 456-60, 封3. |
| 21 | Tewari D, Patni P, Bishayee A, et al. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: a novel therapeutic strategy[J]. Semin Cancer Biol, 2022, 80: 1-17. |
| 22 | Zheng YY, Dai MR, Dong Y, et al. ZEB2/TWIST1/PRMT5/NuRD Multi complex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma[J]. Cancers, 2022, 14(14): 3426. |
| 23 | Bradley WD, Koleske AJ. Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts[J]. J Cell Sci, 2009, 122(Pt 19): 3441-54. |
| 24 | Ren LW, Yang YH, Li W, et al. CDK1 serves as a therapeutic target of adrenocortical carcinoma via regulating epithelial-mesenchymal transition, G2/M phase transition, and PANoptosis[J]. J Transl Med, 2022, 20(1): 444. |
| 25 | Kashani B, Zandi Z, Pourbagheri-Sigaroodi A, et al. The role of toll-like receptor 4 (TLR4) in cancer progression: a possible therapeutic target[J]? J Cell Physiol, 2021, 236(6): 4121-37. |
| 26 | Zhou Q, Xue CQ, Man JW, et al. Correlation of tumor-associated macrophage infiltration in glioblastoma with magnetic resonance imaging characteristics: a retrospective cross-sectional study[J]. Quant Imaging Med Surg, 2023, 13(9): 5958-73. |
| 27 | Lin L, Wu Q, Lu FF, et al. Nrf2 signaling pathway: current status and potential therapeutic targetable role in human cancers[J]. Front Oncol, 2023, 13: 1184079. |
| 28 | Sivaganesh V, Sivaganesh V, Scanlon C, et al. Protein tyrosine phosphatases: mechanisms in cancer[J]. Int J Mol Sci, 2021, 22(23): 12865. |
| 29 | Liu Q, Guan CC, Liu C, et al. Targeting hypoxia-inducible factor-1alpha: a new strategy for triple-negative breast cancer therapy[J]. Biomed Pharmacother, 2022, 156: 113861. |
| 30 | Xu ZW, Goel HL, Burkart C, et al. Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer[J]. Sci Transl Med, 2023, 15(694): eadf1128. |
| 31 | Wu TQ, Song HM, Xie D, et al. Silencing of ASPP2 promotes the proliferation, migration and invasion of triple-negative breast cancer cells via the PI3K/AKT pathway[J]. Int J Oncol, 2018, 52(6): 2001-10. |
| 32 | Liu X, Adorno-Cruz V, Chang YF, et al. EGFR inhibition blocks cancer stem cell clustering and lung metastasis of triple negative breast cancer[J]. Theranostics, 2021, 11(13): 6632-43. |
| 33 | Liu T, Li KD, Zhang ZX, et al. Tetrandrine inhibits cancer stem cell characteristics and epithelial to mesenchymal transition in triple-negative breast cancer via SOD1/ROS signaling pathway[J]. Am J Chin Med, 2023, 51(2): 425-44. |
| 34 | Fang H, Xie JP, Zhang M, et al. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN[J]. Am J Transl Res, 2017, 9(3): 953-61. |
| 35 | Xu JH, Zhao JX, Jiang MY, et al. MiR-193 promotes cell proliferation and invasion by ING5/PI3K/AKT pathway of triple-negative breast cancer[J]. Eur Rev Med Pharmacol Sci, 2020, 24(6): 3122-9. |
| 36 | Chen CH, Li SH, Xue JL, et al. PD-L1 tumor-intrinsic signaling and its therapeutic implication in triple-negative breast cancer[J]. JCI Insight, 2021, 6(8): e131458. |
| 37 | Bacci M, Lorito N, Ippolito L, et al. Reprogramming of amino acid transporters to support aspartate and glutamate dependency sustains endocrine resistance in breast cancer[J]. Cell Rep, 2019, 28(1): 104-18.e8. |
| [1] | 陈鑫源, 吴成挺, 李瑞迪, 潘雪芹, 张耀丹, 陶俊宇, 林才志. 双术汤通过P53/SLC7A11/GPX4通路诱导胃癌细胞铁死亡[J]. 南方医科大学学报, 2025, 45(7): 1363-1371. |
| [2] | 王立明, 陈宏睿, 杜燕, 赵鹏, 王玉洁, 田燕歌, 刘新光, 李建生. 益气滋肾方通过抑制PI3K/Akt/NF-κB通路改善小鼠慢性阻塞性肺疾病的炎症反应[J]. 南方医科大学学报, 2025, 45(7): 1409-1422. |
| [3] | 朱胤福, 李怡燃, 王奕, 黄颖而, 龚昆翔, 郝文波, 孙玲玲. 桂枝茯苓丸活性成分常春藤皂苷元通过抑制JAK2/STAT3通路抑制宫颈癌细胞的生长[J]. 南方医科大学学报, 2025, 45(7): 1423-1433. |
| [4] | 龚秀莹, 侯顺福, 赵苗苗, 王晓娜, 张致涵, 刘清华, 尹崇高, 李洪利. LncRNA SNHG15通过miR-30b-3p调控COX6B1轴促进肺腺癌细胞增殖、迁移和侵袭的分子机制[J]. 南方医科大学学报, 2025, 45(7): 1498-1505. |
| [5] | 何丽君, 陈晓菲, 闫陈昕, 师林. 扶正化积汤治疗非小细胞肺癌的分子机制:基于网络药理学及体外实验验证[J]. 南方医科大学学报, 2025, 45(6): 1143-1152. |
| [6] | 李嘉豪, 冼瑞婷, 李荣. 下调ACADM介导的脂毒性抑制雌激素受体阳性乳腺癌细胞的侵袭与转移[J]. 南方医科大学学报, 2025, 45(6): 1163-1173. |
| [7] | 李国永, 黎仁玲, 刘艺婷, 柯宏霞, 李菁, 王新华. 牛蒡子治疗小鼠病毒性肺炎后肺纤维化的机制:基于代谢组学、网络药理学和实验验证方法[J]. 南方医科大学学报, 2025, 45(6): 1185-1199. |
| [8] | 管丽萍, 颜燕, 卢心怡, 李智峰, 高晖, 曹东, 侯晨曦, 曾靖宇, 李欣怡, 赵洋, 王俊杰, 方会龙. 复方积雪草减轻小鼠日本血吸虫引起的肝纤维化:通过调控TLR4/MyD88通路抑制炎症-纤维化级联反应[J]. 南方医科大学学报, 2025, 45(6): 1307-1316. |
| [9] | 翁诺舟, 谭彬, 曾文涛, 古家宇, 翁炼基, 郑克鸿. 过表达RGL1通过激活CDC42/RAC1复合体上调运动型黏着斑组装促进结直肠癌转移[J]. 南方医科大学学报, 2025, 45(5): 1031-1038. |
| [10] | 唐培培, 谈勇, 殷燕云, 聂晓伟, 黄菁宇, 左文婷, 李玉玲. 调周滋阴方治疗早发性卵巢功能不全的疗效、安全性及作用机制[J]. 南方医科大学学报, 2025, 45(5): 929-941. |
| [11] | 梁晓涛, 熊一凡, 刘雪琪, 梁小珊, 朱晓煜, 谢炜. 活血疏风颗粒通过抑制TLR4/NF-κB通路改善慢性偏头痛小鼠的中枢敏化[J]. 南方医科大学学报, 2025, 45(5): 986-994. |
| [12] | 冉念东, 刘杰, 徐剑, 张永萍, 郭江涛. 黑骨藤正丁醇萃取成分治疗大鼠阿尔茨海默病的药效学及作用机制[J]. 南方医科大学学报, 2025, 45(4): 785-798. |
| [13] | 岳雅清, 牟召霞, 王希波, 刘艳. Aurora-A过表达通过激活NF-κBp65/ARPC4信号轴促进宫颈癌细胞的侵袭和转移[J]. 南方医科大学学报, 2025, 45(4): 837-843. |
| [14] | 庆顺杰, 沈智勇. 过表达己糖激酶2通过激活JAK/STAT途径促进结直肠癌细胞的增殖、迁移和侵袭并调节肿瘤免疫微环境[J]. 南方医科大学学报, 2025, 45(3): 542-553. |
| [15] | 陈镝, 吕莹, 郭怡欣, 张怡荣, 王蕊璇, 周小若, 陈雨欣, 武晓慧. 双氢青蒿素可显著增强阿霉素诱导的三阴性乳腺癌细胞凋亡:基于负向调控STAT3/HIF-1α通路[J]. 南方医科大学学报, 2025, 45(2): 254-260. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||