南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (8): 1682-1696.doi: 10.12122/j.issn.1673-4254.2025.08.13
收稿日期:2024-11-28
出版日期:2025-08-20
发布日期:2025-09-05
通讯作者:
浦春
E-mail:823545914@qq.com;philipcpu@163.com
作者简介:马思源,硕士,检验师,E-mail: 823545914@qq.com
基金资助:
Siyuan MA1,2( ), Bochao ZHANG3, Chun PU2(
), Bochao ZHANG3, Chun PU2( )
)
Received:2024-11-28
Online:2025-08-20
Published:2025-09-05
Contact:
Chun PU
E-mail:823545914@qq.com;philipcpu@163.com
摘要:
目的 探讨环状RNA(circ_0000437)对乳腺癌细胞功能的影响及其分子机制。 方法 体外培养人乳腺细胞(MCF-10A)和人乳腺癌细胞(MCF-7和MDA-MB-231),乳腺癌细胞经过分别转染后设置sh-circ_0000437组、mimics组、inhibitor组、si-CTPS1组及其阴性对照组、sh-NC+inhibitor-NC组、sh-circ_0000437+inhibitor-NC组、sh-circ_0000437+inhibitor组、sh-NC+pcDNA-NC组、sh-circ_0000437+pcDNA-NC组、sh-circ_0000437+pcDNA-CTPS1组。qRT-PCR检测乳腺癌细胞株或组织中circ_0000437、let-7b-5p、CTPS1、Notch1、Hes1和Numb的基因表达水平,RNase R检测鉴定circ_0000437的环状结构,亚细胞定位检测显示circ_0000437在乳腺癌细胞中的分布,CCK-8检测细胞增殖能力,Transwell实验检测细胞侵袭能力,划痕实验检测细胞迁移能力,双荧光素酶报告基因和RNA免疫共沉淀实验探究circ_0000437/let-7b-5p/CTPS1间的结合位点,Western blotting检测细胞中CTPS1,E-cadherin、N-cadherin和Vimentin的蛋白表达,小鼠体内成瘤实验验证circ_0000437与CTPS1在体内促癌的作用机制。 结果 Circ_0000437和CTPS1在乳腺癌组织和细胞系中的表达均上调,let-7b-5p在乳腺癌组织和细胞系中的表达下调(P<0.01),敲低circ_0000437或CTPS1均可抑制乳腺癌细胞的增殖、侵袭、迁移和上皮间质转化(P<0.05),过表达let-7b-5p同样抑制乳腺癌的恶性生物学行为,而抑制let-7b-5p则相反(P<0.05),sh-circ_0000437+pcDNA-NC组的乳腺癌细胞在小鼠体内的生长速度低于sh-NC+pcDNA-NC组,sh-circ_0000437+pcDNA-CTPS1组生长速度高于sh-circ_0000437+pcDNA-NC组(P<0.01),circ_0000437和let-7b-5p、let-7b-5p和CTPS1之间存在结合位点(P<0.01),circ_0000437,let-7b-5p和CTPS1在乳腺癌细胞中存在相互作用(P<0.05)。 结论 Circ_0000437在乳腺癌组织和细胞中表达上调,且circ_0000437可以通过let-7b-5p/CTPS1轴促进乳腺癌细胞的增殖,侵袭,迁移以及上皮间质转化。
马思源, 张博超, 浦春. Circ_0000437通过靶向let-7b-5p/CTPS1轴促进乳腺癌细胞的增殖、侵袭、迁移及上皮间质转化[J]. 南方医科大学学报, 2025, 45(8): 1682-1696.
Siyuan MA, Bochao ZHANG, Chun PU. Circ_0000437 promotes proliferation, invasion, migration and epithelial-mesenchymal transition of breast cancer cells by targeting the let-7b-5p/CTPS1 axis[J]. Journal of Southern Medical University, 2025, 45(8): 1682-1696.
| Gene name | Primer sequence (s) |
|---|---|
| Circ_0000437 | F:5'-AATCCCCGTACGTCCACTAC-3' |
| R:5'-AGGGTCATAGAAAGGCAGCA-3' | |
| CORO1C | F:5'-ATGAGGCGGCACATATAC-3' |
| R:5'-ATCCCAGGTCACACGAGAAAC-3' | |
| CTPS1 | F:5'-CAGTGTGGGCACAATACTCAA-3' |
| R:5'-CGCTCATAGTTACCCAGGTCA-3' | |
| Notch1 | F:5'-CGAACCCGTGCCAGAA-3' |
| R:5'-CAGATGCCCAGTGAAGC-3' | |
| Hes1 | F:5'-CAGTGCCTTTGAGAAGCAGG-3' |
| R:5'-CAGATAACGGGCAACTTCGG-3' | |
| Numb | F:5'-CACAACTGCCACTGAGCAAG-3' |
| R:5'-GTTGCCAGGAGCCACTGAT-3' | |
| GAPDH | F:5'-CATCAAGAAGGTGGTGAAGCAG-3' |
| R:5'-GTGTCGCTGTTGAAGTCAGAG-3' | |
| let-7b-5p | 5'-GCTGAGGTAGTAGGTTGTGTGGG-3' |
| U6 | 5'-CGCTTCGGCACATATAC-3' |
表1 引物序列
Tab.1 Primer sequences for qRT-PCR
| Gene name | Primer sequence (s) |
|---|---|
| Circ_0000437 | F:5'-AATCCCCGTACGTCCACTAC-3' |
| R:5'-AGGGTCATAGAAAGGCAGCA-3' | |
| CORO1C | F:5'-ATGAGGCGGCACATATAC-3' |
| R:5'-ATCCCAGGTCACACGAGAAAC-3' | |
| CTPS1 | F:5'-CAGTGTGGGCACAATACTCAA-3' |
| R:5'-CGCTCATAGTTACCCAGGTCA-3' | |
| Notch1 | F:5'-CGAACCCGTGCCAGAA-3' |
| R:5'-CAGATGCCCAGTGAAGC-3' | |
| Hes1 | F:5'-CAGTGCCTTTGAGAAGCAGG-3' |
| R:5'-CAGATAACGGGCAACTTCGG-3' | |
| Numb | F:5'-CACAACTGCCACTGAGCAAG-3' |
| R:5'-GTTGCCAGGAGCCACTGAT-3' | |
| GAPDH | F:5'-CATCAAGAAGGTGGTGAAGCAG-3' |
| R:5'-GTGTCGCTGTTGAAGTCAGAG-3' | |
| let-7b-5p | 5'-GCTGAGGTAGTAGGTTGTGTGGG-3' |
| U6 | 5'-CGCTTCGGCACATATAC-3' |
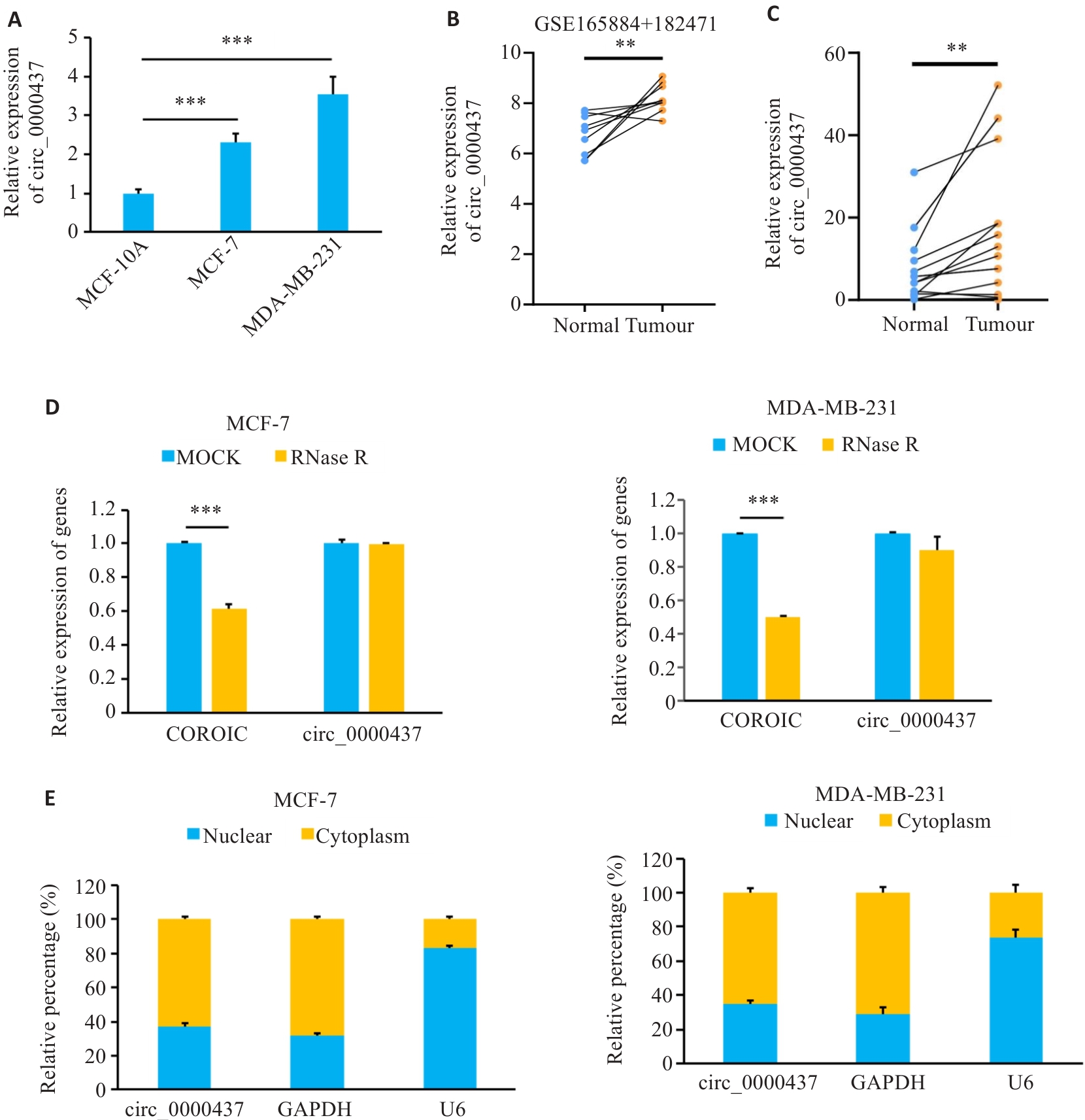
图1 乳腺癌组织和细胞中circ_0000437的表达特征
Fig.1 Expression levels of circ_0000437 in breast cancer cells and tissues. A: Expression of circ_0000437 in breast cancer cell line. B: Expression of circ_0000437 in datasets GSE165884 and 182471. C: Expression of circ_0000437 in breast cancer tissues. D: Expression of circ_0000437 and linear CORO1C in two breast cancer cell lines treated with RNase R. E: Subcellular localization of circ_0000437 in breast cancer cells. **P<0.01, ***P<0.001.

图2 敲减circ_0000437对乳腺癌细胞增殖、侵袭、迁移和EMT的影响
Fig.2 Effects of circ_0000437 knockdown on proliferation, invasion, migration and EMT of breast cancer cells. A, B: Gene knockout efficiency and fluorescence staining of the cells after lentivirus transfection (Original magnification: ×100). C: CCK-8 assay for assessing the effect of circ_0000437 on cell proliferation. D: Transwell assay for assessing the effect of circ_0000437 on cell invasion (×400). E: Scratch assay for assessing the effect of circ_0000437 on cell migration (×100). F: Western blotting for assessing the effect of circ_0000437 on EMT in breast cancer cells. *P<0.05, **P<0.01, ***P<0.001.
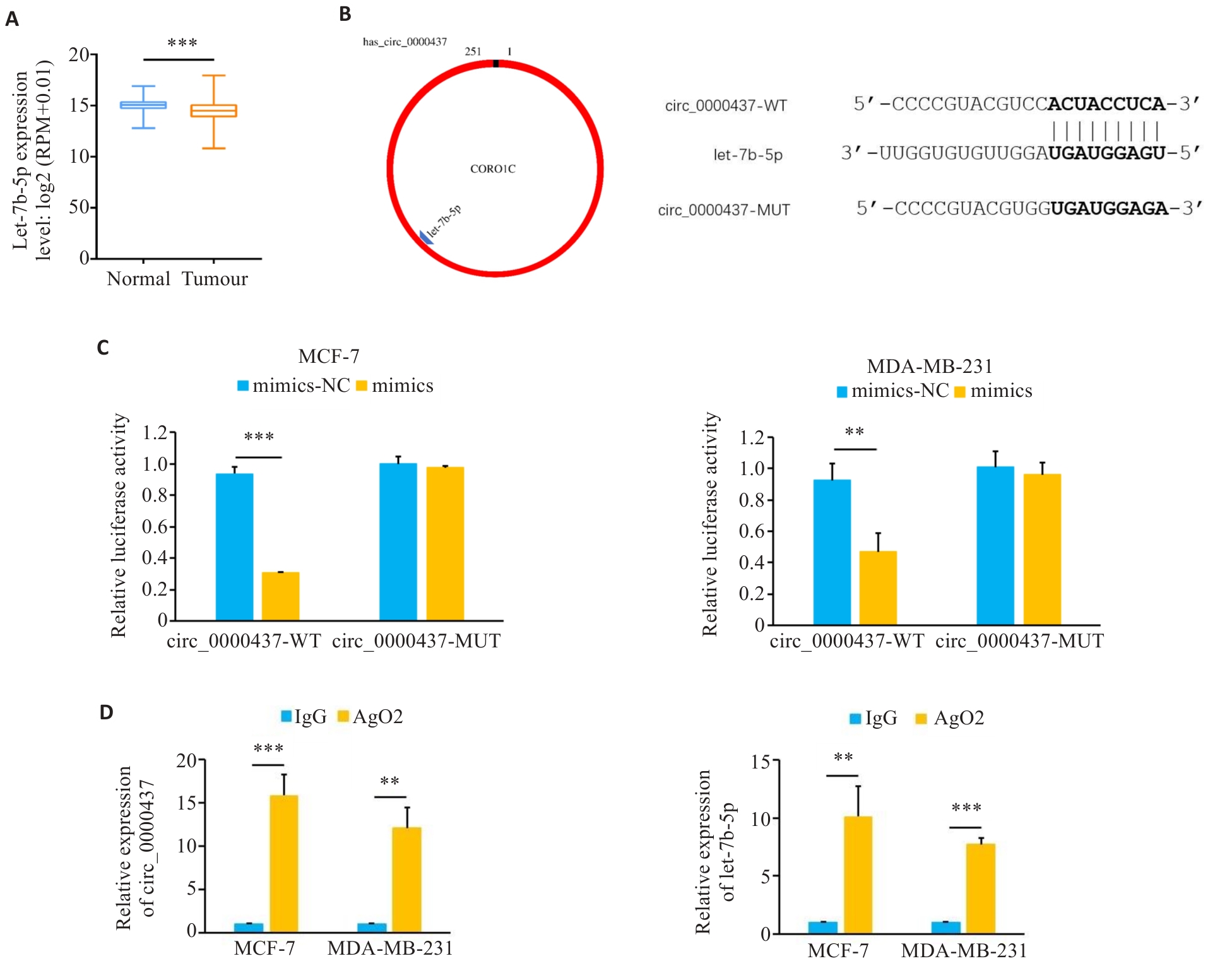
图3 Circ_0000437与let-7b-5p存在结合位点
Fig.3 Circ_0000437 has a binding site with let-7b-5p. A: Expression of let-7b-5p in breast cancer in TCGA database. B: The predicted binding diagram and binding sequence of let-7b-5p and circ_0000437. C: Luciferase activity in the cells co-transfected with mimics or mimics-NC and circ_0000437 WT plasmid or MUT plasmid. D: RIP analysis showing enrichment of circ_0000437 and let-7b-5p in the AGO2 part. **P<0.01, ***P<0.001.
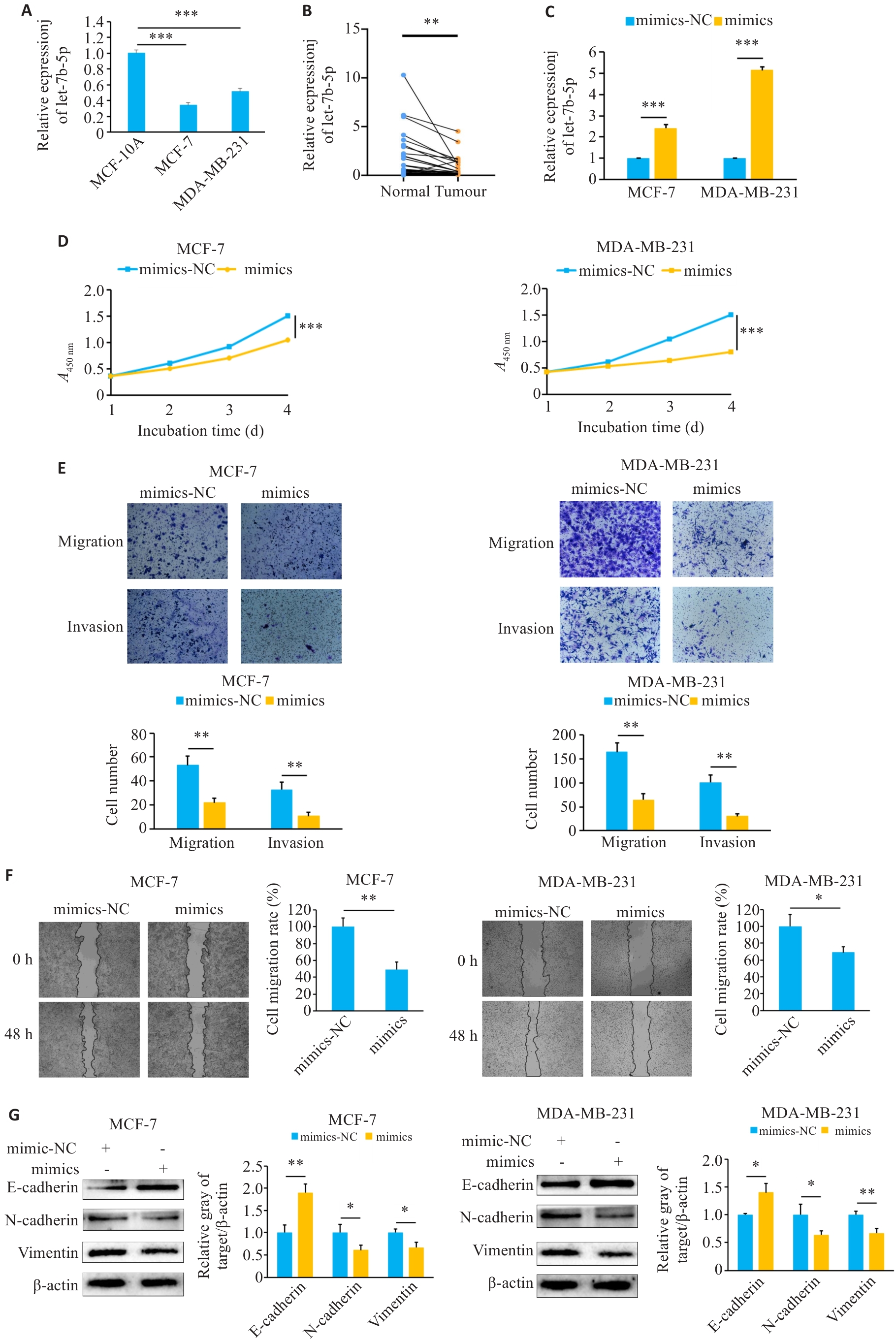
图4 过表达let-7b-5p抑制乳腺癌细胞的增殖、侵袭、迁移和EMT
Fig.4 Overexpression of let-7b-5p inhibits proliferation, invasion, migration and EMT of breast cancer cells. A, B: Expression of let-7b-5p in breast cancer tissues and cell lines. C: Transfection efficiency of let-7b-5p mimics. D: CCK-8 analysis to evaluate the effects of mimics on cell proliferation. E: Transwell assay to verify the effect of mimics on cell invasion (×400). F: Effect of mimics on cell migration determined using scratch assay (×100). G: Effect of mimics on EMT in breast cancer cells. *P<0.05, **P<0.01, ***P<0.001.

图5 抑制let-7b-5p抑制乳腺癌细胞的增殖、侵袭、迁移和EMT
Fig.5 Inhibition of let-7b-5p suppresses proliferation, invasion, migration and EMT of breast cancer cells. A: Transfection efficiency of let-7b-5p inhibitor. B: CCK-8 analysis to evaluate the effects of inhibitor on cell proliferation. C: Transwell assay to verify the effect of inhibitor on cell invasion ability (×400). D: Effect of the inhibitor on cell migration determined using scratch assay (×100). E: Effect of the inhibitor on EMT in breast cancer cells. *P<0.05, **P<0.01, ***P<0.001.
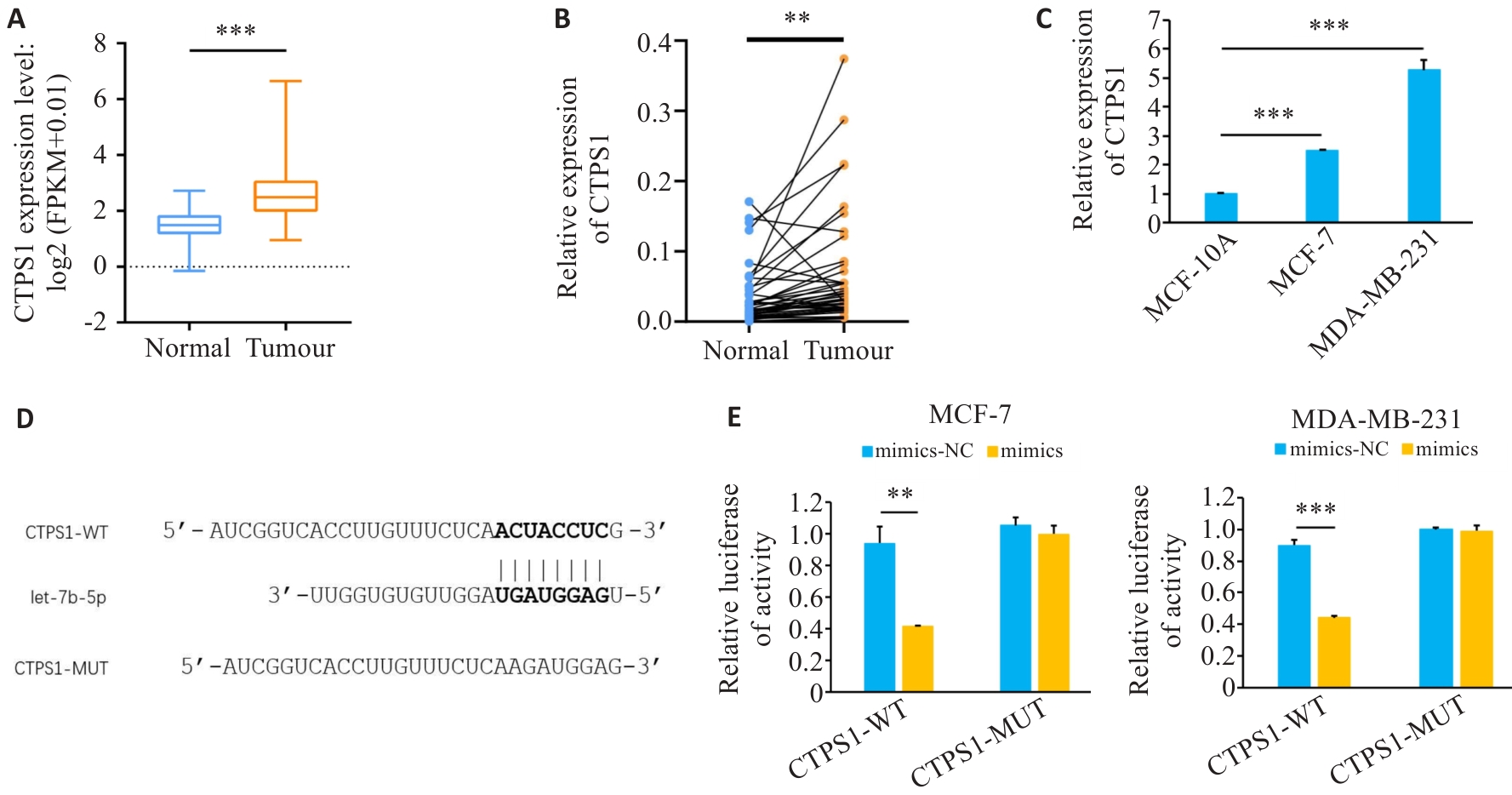
图6 CTPS1在乳腺癌中表达上调
Fig.6 Expression of CTPS1 up-regulated in breast cancer. A: Expression of CTPS1 in breast cancer in TCGA database. B, C: Expression of CTPS1 in breast cancer tissues and cell lines. D: Predicted CTPS1 and let-7b-5p binding sites. E: Mimics or mimics-NC were co-transfected with CTPS1 WT plasmid or MUT plasmid, respectively, and luciferase activity was measured in each group. **P<0.01, ***P<0.001.
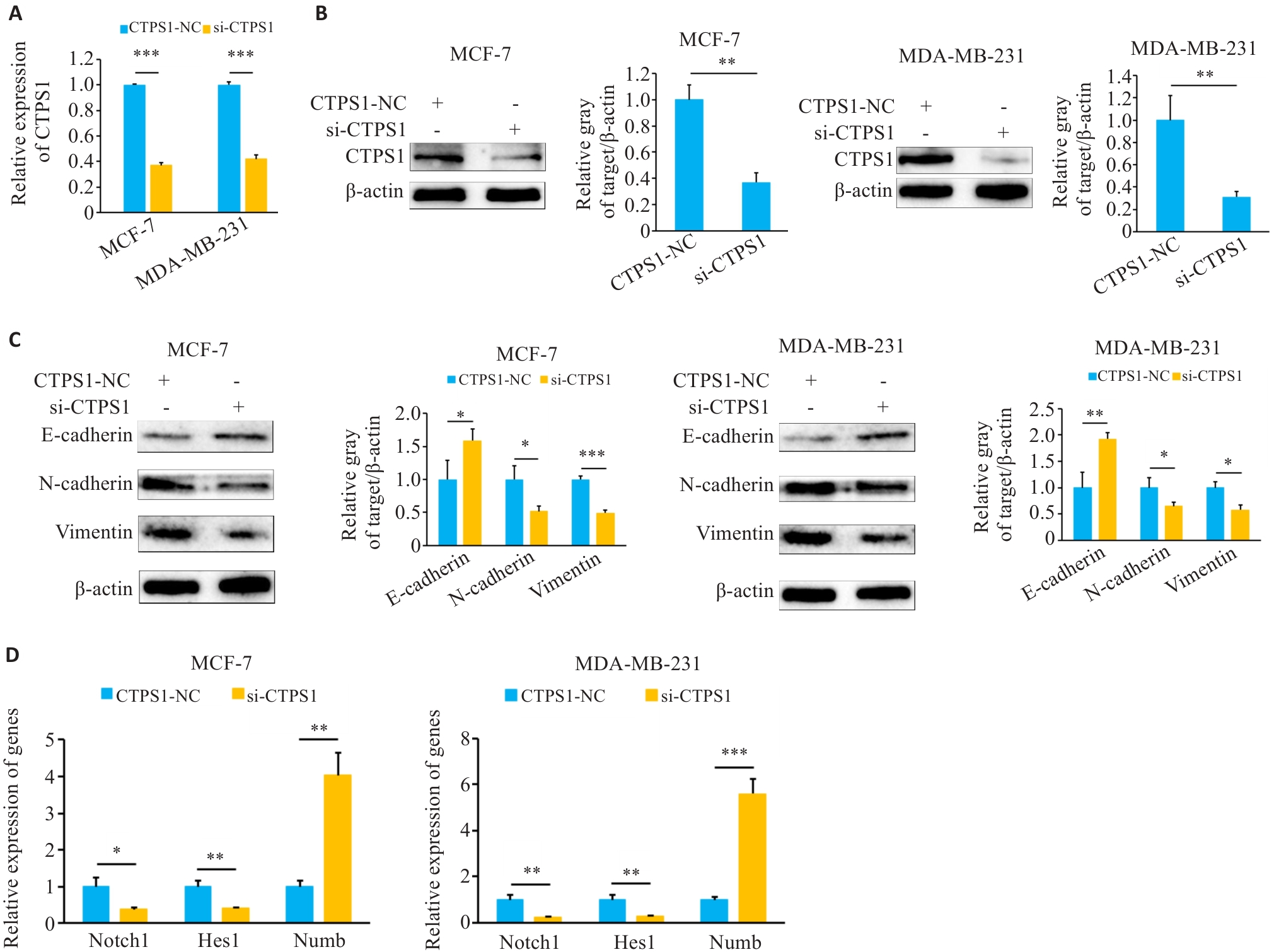
图7 敲低CTPS1抑制乳腺癌细胞EMT并影响Notch信号通路
Fig.7 CTPS1 knockdown inhibits EMT and regulates the Notch signaling pathway in breast cancer cells. A: Knockdown efficiency after transfection with si-CTPS1. B: Western blotting for assessing the effect of si-CTPS1 on expression of CTPS1 protein in breast cancer cells. C: Western blotting for assessing the effect of CTPS1 on EMT of breast cancer cells. D: qRT-PCR for assessing the effect of si-CTPS1 on the expression of genes related to the Notch signaling pathway. *P<0.05, **P<0.01, ***P<0.001.

图8 敲低CTPS1抑制乳腺癌细胞增殖、侵袭和迁移
Fig.8 CTPS1 knockdown inhibits proliferation, invasion and migration of breast cancer cells. A: CCK-8 assay for evaluating the effect of transfection with si-CTPS1 on cell proliferation. B: Transwell assay for assessing the effect of transfection with si-CTPS1 on cell invasion ability (×400). C: Effect of transfection with si-CTPS1 on cell migration determined using scratch assay (×100). *P<0.05, **P<0.01, ***P<0.001.
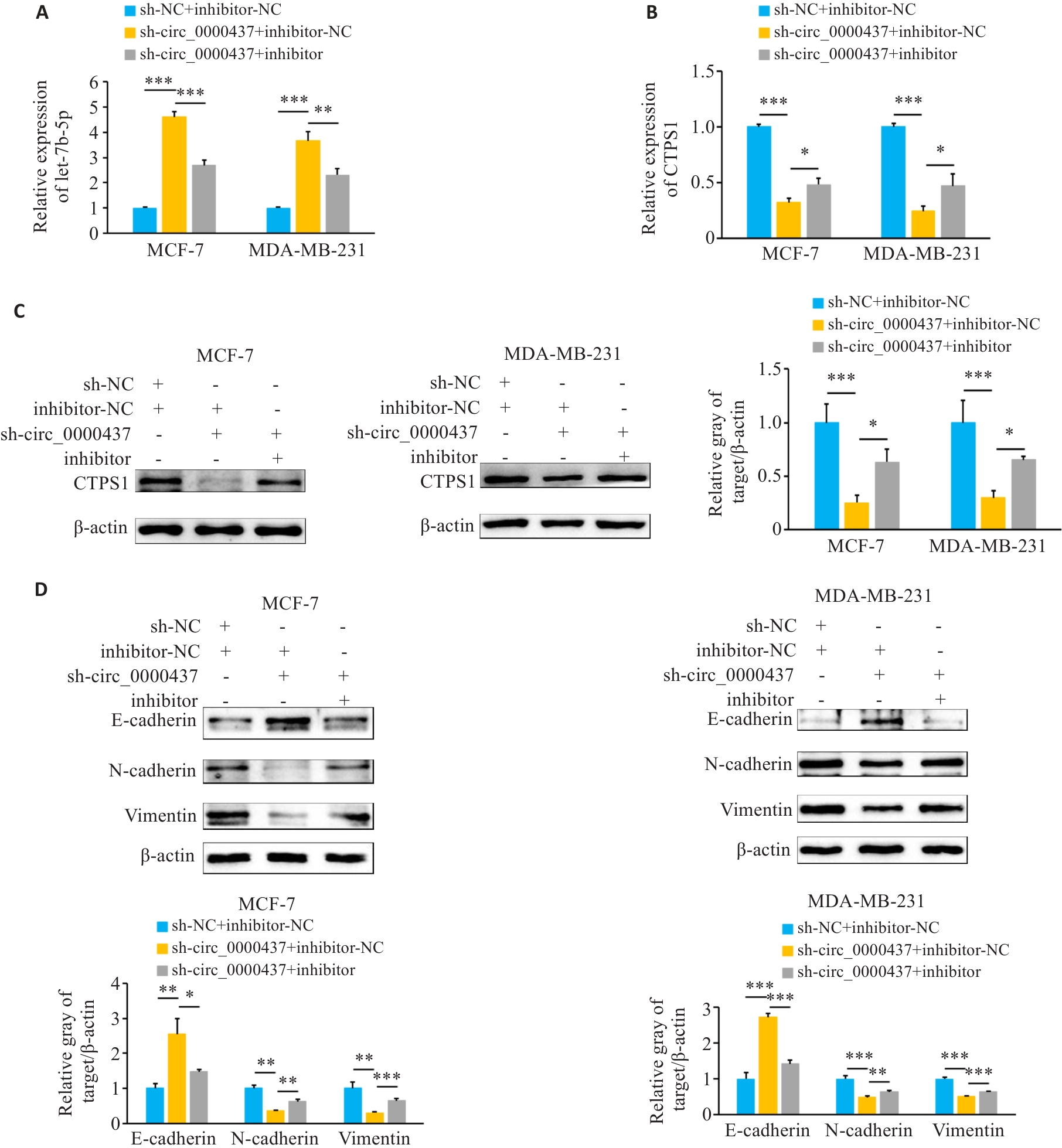
图10 Circ_0000437可以通过let-7b-5p/CTPS1轴促进乳腺癌细胞EMT
Fig.10 Circ_0000437 promotes EMT in breast cancer cells by regulating the let-7b-5p/CTPS1 axis. A: Expression of let-7b-5p in each group (sh-NC+inhibitor-NC, sh-circ_0000437+inhibitor-NC, and sh-circ_0000437+inhibitor) detected by qRT-PCR. B: qRT-PCR for detecting CTPS1 mRNA expression in breast cancer cells in each group. C: Western blotting for detecting the expression of CTPS1 protein in breast cancer cells in each group. D: Western blotting for detecting the expression of EMT-related proteins in breast cancer cells in each group. *P<0.05, **P<0.01, ***P<0.001.

图11 Circ_0000437 作为 let-7b-5p 的分子海绵,促进乳腺癌细胞的增殖、侵袭和迁移
Fig.11 Circ_0000437 promotes proliferation, invasion, and migration of breast cancer cells as a molecular sponge for let-7b-5p. A: CCK-8 assay of the breast cancer cells. B: Transwell assay of the breast cancer cells (×400). C: Scratch assay of the breast cancer cells (×100). *P<0.05, **P<0.01, ***P<0.001.
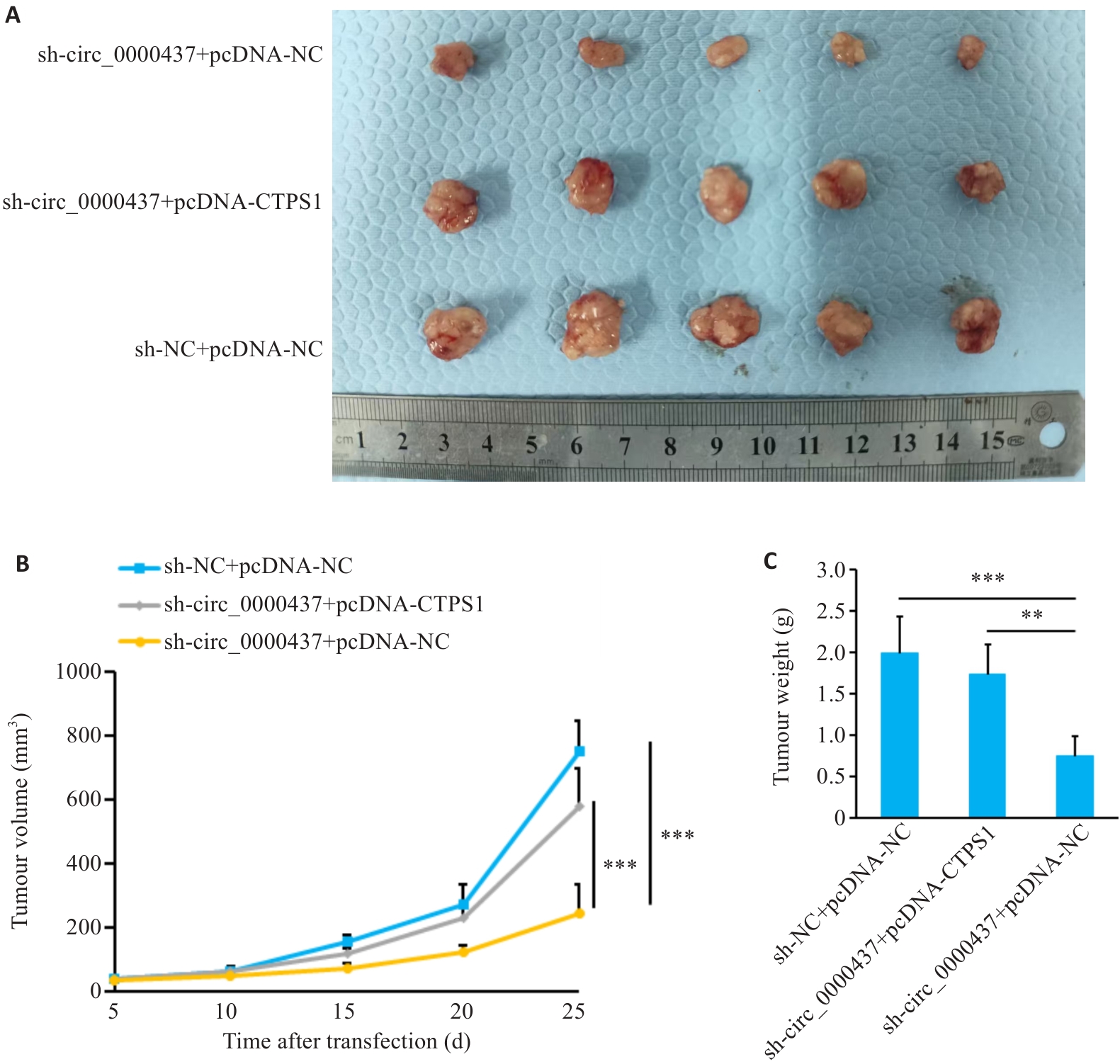
图12 小鼠体内肿瘤模型的构建
Fig.12 Construction of the tumor-bearing model in mice. A: Comparison of tumor volume 25 days after cell inoculation. B: Tumor growth curve in the mice. C: Comparison of tumor weight. **P<0.01, ***P<0.001.
| [1] | Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics[J]. CA Cancer J Clin, 2023, 73(1): 17-48. doi:10.3322/caac.21763 |
| [2] | Waks AG, Winer EP. Breast cancer treatment: a review[J]. JAMA, 2019, 321(3): 288-300. doi:10.1001/jama.2018.19323 |
| [3] | He X, Xu T, Hu WJ, et al. Circular RNAs: their role in the pathogenesis and orchestration of breast cancer[J]. Front Cell Dev Biol, 2021, 9: 647736. doi:10.3389/fcell.2021.647736 |
| [4] | DeSantis CE, Bray F, Ferlay J, et al. International variation in female breast cancer incidence and mortality rates[J]. Cancer Epidemiol Biomarkers Prev, 2015, 24(10): 1495-506. doi:10.1158/1055-9965.epi-15-0535 |
| [5] | Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-91. doi:10.1038/s41576-019-0158-7 |
| [6] | Sharma AR, Bhattacharya M, Bhakta S, et al. Recent research progress on circular RNAs: biogenesis, properties, functions, and therapeutic potential[J]. Mol Ther Nucleic Acids, 2021, 25: 355-71. doi:10.1016/j.omtn.2021.05.022 |
| [7] | Qu SB, Liu ZC, Yang XS, et al. The emerging functions and roles of circular RNAs in cancer[J]. Cancer Lett, 2018, 414: 301-9. doi:10.1016/j.canlet.2017.11.022 |
| [8] | Ruan Y, Li Z, Shen YJ, et al. Functions of circular RNAs and their potential applications in gastric cancer[J]. Expert Rev Gastroenterol Hepatol, 2020, 14(2): 85-92. doi:10.1080/17474124.2020.1715211 |
| [9] | Lei B, Tian ZQ, Fan WP, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers[J]. Int J Med Sci, 2019, 16(2): 292-301. doi:10.7150/ijms.28047 |
| [10] | Zhang ML, Bai X, Zeng XM, et al. circRNA-miRNA-mRNA in breast cancer[J]. Clin Chim Acta, 2021, 523: 120-30. doi:10.1016/j.cca.2021.09.013 |
| [11] | Salmena L, Poliseno L, Tay Y, et al. A CeRNA hypothesis: the Rosetta Stone of a hidden RNA language?[J]. Cell, 2011, 146(3): 353-8. doi:10.1016/j.cell.2011.07.014 |
| [12] | Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis[J]. Trends Cell Biol, 2019, 29(3): 212-26. doi:10.1016/j.tcb.2018.12.001 |
| [13] | Liu SY, Li LY, Ren DM. Anti-cancer potential of phytochemicals: the regulation of the epithelial-mesenchymal transition[J]. Molecules, 2023, 28(13): 5069. doi:10.3390/molecules28135069 |
| [14] | Cao ZQ, Wang Z, Leng P. Aberrant N-cadherin expression in cancer[J]. Biomed Pharmacother, 2019, 118: 109320. doi:10.1016/j.biopha.2019.109320 |
| [15] | Péglion F, Etienne-Manneville S. N-cadherin expression level as a critical indicator of invasion in non-epithelial tumors[J]. Cell Adh Migr, 2012, 6(4): 327-32. doi:10.4161/cam.20855 |
| [16] | Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways[J]. Cancer Res, 2008, 68(10): 3645-54. doi:10.1158/0008-5472.can-07-2938 |
| [17] | Liu P, Wang ZH, Ou XQ, et al. The FUS/circEZH2/KLF5/feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition[J]. Mol Cancer, 2022, 21(1): 198. doi:10.1186/s12943-022-01653-2 |
| [18] | Tian XL, Yang H, Fang Q, et al. Circ_ZFR affects FABP7 expression to regulate breast cancer progression by acting as a sponge for miR-223-3p[J]. Thorac Cancer, 2022, 13(9): 1369-80. doi:10.1111/1759-7714.14401 |
| [19] | Yang L, Chen YX. Circ_0008717 sponges miR-326 to elevate GATA6 expression to promote breast cancer tumorigenicity[J]. Biochem Genet, 2023, 61(2): 578-96. doi:10.1007/s10528-022-10270-z |
| [20] | Wu F, Sun GQ, Zheng WB, et al. circCORO1C promotes the proliferation and metastasis of hepatocellular carcinoma by enhancing the expression of PD-L1 through NF-κB pathway[J]. J Clin Lab Anal, 2021, 35(12): e24003. doi:10.1002/jcla.24003 |
| [21] | Shen XJ, Kong S, Ma S, et al. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis[J]. Oncogene, 2022, 41(42): 4724-35. doi:10.1038/s41388-022-02449-w |
| [22] | Zhu LW, Wang ZF, Sun LB, et al. Hsa_circ_0000437 upregulates and promotes disease progression in rheumatic valvular heart disease[J]. J Clin Lab Anal, 2022, 36(2): e24197. doi:10.1002/jcla.24197 |
| [23] | Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013, 19(2): 141-57. doi:10.1261/rna.035667.112 |
| [24] | Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells[J]. Cell Mol Life Sci, 2018, 75(6): 1071-98. doi:10.1007/s00018-017-2688-5 |
| [25] | Huang XJ, Dong HX, Liu Y, et al. Silencing of let-7b-5p inhibits ovarian cancer cell proliferation and stemness characteristics by Asp-Glu-Ala-Asp-box helicase 19A[J]. Bioengineered, 2021, 12(1): 7666-77. doi:10.1080/21655979.2021.1982276 |
| [26] | Dai YY, Liu JS, Li XY, et al. Let-7b-5p inhibits colon cancer progression by prohibiting APC ubiquitination degradation and the Wnt pathway by targeting NKD1[J]. Cancer Sci, 2023, 114(5): 1882-97. doi:10.1111/cas.15678 |
| [27] | Zheng ST, Liu Q, Ma R, et al. Let-7b-5p inhibits proliferation and motility in squamous cell carcinoma cells through negative modulation of KIAA1377[J]. Cell Biol Int, 2019, 43(6): 634-41. doi:10.1002/cbin.11136 |
| [28] | Sun Z, Zhang ZH, Wang QQ, et al. Combined inactivation of CTPS1 and ATR is synthetically lethal to MYC-overexpressing cancer cells[J]. Cancer Res, 2022, 82(6): 1013-24. doi:10.1158/0008-5472.can-21-1707 |
| [29] | Traut TW. Physiological concentrations of purines and pyrimidines[J]. Mol Cell Biochem, 1994, 140(1): 1-22. doi:10.1007/bf00928361 |
| [30] | Williams JC, Kizaki H, Weber G, et al. Increased CTP synthetase activity in cancer cells[J]. Nature, 1978, 271(5640): 71-3. doi:10.1038/271071a0 |
| [31] | Chitrakar I, Kim-Holzapfel DM, Zhou WJ, et al. Higher order structures in purine and pyrimidine metabolism[J]. J Struct Biol, 2017, 197(3): 354-64. doi:10.1016/j.jsb.2017.01.003 |
| [32] | Wu FH, Mao YD, Ma T, et al. CTPS1 inhibition suppresses proliferation and migration in colorectal cancer cells[J]. Cell Cycle, 2022, 21(24): 2563-74. doi:10.1080/15384101.2022.2105084 |
| [33] | Ohmine K, Kawaguchi K, Ohtsuki S, et al. Quantitative targeted proteomics of pancreatic cancer: deoxycytidine kinase protein level correlates to progression-free survival of patients receiving gemcitabine treatment[J]. Mol Pharm, 2015, 12(9): 3282-91. doi:10.1021/acs.molpharmaceut.5b00282 |
| [34] | Zhang C, Tian CX, Zhu RZ, et al. CircSATB1 promotes colorectal cancer liver metastasis through facilitating FKBP8 degradation via RNF25-mediated ubiquitination[J]. Adv Sci (Weinh), 2025, 12(13): e2406962. doi:10.1002/advs.202406962 |
| [1] | 张兆君, 吴琼, 谢苗苗, 叶洳吟, 耿晨晨, 石纪雯, 杨清玲, 王文锐, 石玉荣. 层状双氢氧化物负载si-NEAT1通过miR-133b/PD-L1轴调控乳腺癌紫杉醇耐药及巨噬细胞极化[J]. 南方医科大学学报, 2025, 45(8): 1718-1731. |
| [2] | 王子良, 陈孝华, 杨晶晶, 严晨, 张志郅, 黄炳轶, 赵萌, 刘嵩, 葛思堂, 左芦根, 陈德利. 高表达SURF4通过抑制紧密连接蛋白表达促进胃癌细胞的恶性生物学行为[J]. 南方医科大学学报, 2025, 45(8): 1732-1742. |
| [3] | 吴璇, 方家敏, 韩玮玮, 陈琳, 孙菁, 金齐力. 高表达PRELID1促进胃癌细胞上皮间质转化并与不良预后相关[J]. 南方医科大学学报, 2025, 45(7): 1535-1542. |
| [4] | 李嘉豪, 冼瑞婷, 李荣. 下调ACADM介导的脂毒性抑制雌激素受体阳性乳腺癌细胞的侵袭与转移[J]. 南方医科大学学报, 2025, 45(6): 1163-1173. |
| [5] | 马振南, 刘福全, 赵雪峰, 张晓微. DTX2促进奥沙利铂耐药的结直肠癌细胞增殖、侵袭和上皮间质转化[J]. 南方医科大学学报, 2025, 45(4): 829-836. |
| [6] | 陶露, 韦卓利, 王月月, 项平. CEACAM6通过调控上皮间质转化抑制鼻咽癌细胞的增殖和迁移[J]. 南方医科大学学报, 2025, 45(3): 566-576. |
| [7] | 蔡蕊, 黄卓, 贺文霞, 艾添红, 宋晓伟, 胡淑婷. 剪接因子HNRNPH1通过调控Circ-MYOCD的反向剪接影响心肌肥厚的发生[J]. 南方医科大学学报, 2025, 45(3): 587-594. |
| [8] | 陈镝, 吕莹, 郭怡欣, 张怡荣, 王蕊璇, 周小若, 陈雨欣, 武晓慧. 双氢青蒿素可显著增强阿霉素诱导的三阴性乳腺癌细胞凋亡:基于负向调控STAT3/HIF-1α通路[J]. 南方医科大学学报, 2025, 45(2): 254-260. |
| [9] | 褚乔, 王小娜, 续佳颖, 彭荟林, 赵裕琳, 张静, 陆国玉, 王恺. 白头翁皂苷D通过多靶点和多途径抑制三阴性乳腺癌侵袭转移[J]. 南方医科大学学报, 2025, 45(1): 150-161. |
| [10] | 薛良军, 谈秋瑜, 许静文, 冯璐, 李文锦, 颜亮, 李玉磊. MiR-6838-5p过表达下调DDR1基因表达抑制乳腺癌MCF-7细胞的增殖[J]. 南方医科大学学报, 2024, 44(9): 1677-1684. |
| [11] | 欧阳明子, 崔佳琦, 王慧, 梁正, 皮大锦, 陈利国, 陈前军, 吴迎朝. 开心散通过减轻前额叶皮质铁死亡缓解小鼠的阿霉素化疗性抑郁[J]. 南方医科大学学报, 2024, 44(8): 1441-1449. |
| [12] | 朱梦云, 王剑锋. 康柏西普可逆转TGF-β2诱导的晶状体上皮细胞发生上皮间质转化:基于调节TGF-β/Smad信号通路[J]. 南方医科大学学报, 2024, 44(8): 1459-1466. |
| [13] | 房锦存, 刘立威, 林俊豪, 陈逢生. CDHR2过表达通过抑制PI3K/Akt通路抑制乳腺癌细胞增殖[J]. 南方医科大学学报, 2024, 44(6): 1117-1125. |
| [14] | 崔芝, 马萃娇, 王倩茹, 陈金豪, 严子阳, 杨建林, 吕亚丰, 曹春雨. 表达 TGF-βⅡ受体的腺相关病毒载体抑制小鼠三阴性乳腺癌4T1细胞的增殖和肺转移[J]. 南方医科大学学报, 2024, 44(5): 818-826. |
| [15] | 张富星, 刘国庆, 董锐, 高磊, 陆伟晨, 高连霞, 赵忠扩, 陆飞, 刘牧林. 高表达CRTAC1通过调控PI3K信号通路促进胃癌细胞增殖、迁移及免疫浸润[J]. 南方医科大学学报, 2024, 44(12): 2421-2433. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||