南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (2): 313-321.doi: 10.12122/j.issn.1673-4254.2025.02.12
• • 上一篇
收稿日期:2024-09-03
出版日期:2025-02-20
发布日期:2025-03-03
通讯作者:
吴竞
E-mail:631517383@qq.com;wjwjwj1973@126.com
作者简介:高俊杰,检验师,E-mail: 631517383@qq.com
基金资助:
Junjie GAO( ), Kai YE, Jing WU(
), Kai YE, Jing WU( )
)
Received:2024-09-03
Online:2025-02-20
Published:2025-03-03
Contact:
Jing WU
E-mail:631517383@qq.com;wjwjwj1973@126.com
Supported by:摘要:
目的 筛选槲皮素治疗肾透明细胞癌(ccRCC)的潜在分子靶点。 方法 运用网络药理学方法从多个数据库中筛选槲皮素的治疗靶点,运用孟德尔随机化方法从4907种血浆蛋白中筛选与ccRCC显著相关的靶点,构建药物-疾病网络模型,筛选出潜在的关键靶点,运用生物信息学技术评估这些靶点的作用,培养ccRCC细胞并用不用浓度槲皮素处理,行CCK-8实验、创伤愈合实验、RT-qPCR和Western blotting对筛选靶点进行验证。 结果 网络药理学和孟德尔随机化综合分析筛选出了TP53(OR=3.325,95% CI:1.805-6.124,P=0.0001)、ARF4(OR=0.173,95% CI:0.065-0.456,P=0.0003)和DPP4(OR=0.463,95% CI:0.302-0.711,P=0.0004)3个槲皮素治疗ccRCC的核心靶点。生物信息学分析表明TP53在肾透明细胞癌中表达增高(P<0.001);生存分析显示高表达TP53的肾癌患者预后更差(P<0.001);分子对接显示槲皮素与TP53的结合能为-5.83 kcal/mol;CCK-8实验和创伤愈合实验显示槲皮素在体外抑制786-O细胞的增殖和迁移(P<0.05);RT-qPCR和Western blotting显示槲皮素在体外抑制TP53 mRNA和蛋白的表达(P<0.05)。 结论 TP53可能是槲皮素治疗ccRCC的关键靶点,为槲皮素治疗肾透明细胞癌的分子机制研究奠定了基础。
高俊杰, 叶开, 吴竞. 槲皮素通过调控TP53基因抑制肾透明细胞癌的增殖和迁移[J]. 南方医科大学学报, 2025, 45(2): 313-321.
Junjie GAO, Kai YE, Jing WU. Quercetin inhibits proliferation and migration of clear cell renal cell carcinoma cells by regulating TP53 gene[J]. Journal of Southern Medical University, 2025, 45(2): 313-321.
| Exposure | Outcome | Gen | Method | Nsnp | b | Se | P |
|---|---|---|---|---|---|---|---|
| 9816_37_ISOC1_ISOC1 | KIRC | ISOC1 | IVW | 8 | 0.736746 | 0.171048 | 1.65E-05 |
| 6354_13_HEPHL1_HPHL1 | KIRC | HEPHL1 | IVW | 5 | -1.28851 | 0.310833 | 3.39E-05 |
| 17742_2_RRAS_RRAS | KIRC | RRAS | IVW | 5 | -1.33358 | 0.325935 | 4.29E-05 |
| 12621_55_PPP2R1A_PP2AAA | KIRC | PPP2R1A | IVW | 4 | -0.9648 | 0.243754 | 7.56E-05 |
| 5345_51_CKAP2_CKAP2 | KIRC | CKAP2 | IVW | 15 | -0.66094 | 0.169567 | 9.71E-05 |
| 12807_89_ARHGAP30_RHG30 | KIRC | ARHGAP30 | IVW | 3 | 1.721247 | 0.446005 | 0.000114 |
| 6123_69_TP53_p53 | KIRC | TP53 | IVW | 7 | 1.201512 | 0.311593 | 0.000115 |
| 9511_61_NXPH2_NXPH2 | KIRC | NXPH2 | IVW | 18 | -0.39638 | 0.103952 | 0.000137 |
| 6388_21_CCDC126_CC126 | KIRC | CCDC126 | IVW | 18 | -0.46547 | 0.124775 | 0.000191 |
| 13673_21_CCT7_TCPH | KIRC | CCT7 | IVW | 4 | -1.0039 | 0.269598 | 0.000196 |
| 7083_74_MATN4_MATN4 | KIRC | MATN4 | IVW | 7 | 0.634937 | 0.174364 | 0.000271 |
| 4568_17_SLITRK5_SLIK5 | KIRC | SLITRK5 | IVW | 12 | 0.821988 | 0.226306 | 0.000281 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | IVW | 3 | -1.75547 | 0.495302 | 0.000394 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | IVW | 8 | -0.76964 | 0.218311 | 0.000423 |
| 8040_9_LEMD1_LEMD1 | KIRC | LEMD1 | IVW | 3 | -2.26505 | 0.647584 | 0.000469 |
| 6578_29_TNMD_TNMD | KIRC | TNMD | IVW | 3 | -1.94913 | 0.558892 | 0.000488 |
| 9012_1_PRPF6_PRP6 | KIRC | PRPF6 | IVW | 2 | -2.88761 | 0.828756 | 0.000493 |
| 8535_102_DMKN_Dermokine | KIRC | DMKN | IVW | 2 | -1.63853 | 0.479425 | 0.000632 |
| 8748_45_CD74_HG2A | KIRC | CD74 | IVW | 3 | -2.22233 | 0.652226 | 0.000656 |
| 10085_25_STAR_STAR | KIRC | STAR | IVW | 5 | -1.39378 | 0.418791 | 0.000874 |
| 18198_51_HBQ1_HBAT | KIRC | HBQ1 | IVW | 8 | -0.86326 | 0.261832 | 0.000977 |
| 18380_78_ALB_Albumin | KIRC | ALB | IVW | 11 | -0.41249 | 0.13172 | 0.001739 |
| 12587_65_ARL2_ARL2 | KIRC | ARL2 | IVW | 12 | -0.47391 | 0.152486 | 0.001884 |
| 12041_33_HSPA1L_HS71L | KIRC | HSPA1L | IVW | 11 | -0.55241 | 0.178001 | 0.001913 |
| 10842_7_GCNT4_GCNT4 | KIRC | GCNT4 | IVW | 4 | -1.6224 | 0.541404 | 0.00273 |
| 11160_56_RNF122_RN122 | KIRC | RNF122 | IVW | 6 | -0.87475 | 0.299384 | 0.00348 |
表1 血浆蛋白和肾透明细胞癌孟德尔随机化分析结果
Tab.1 Result of Mendelian randomization between plasma proteins and ccRCC
| Exposure | Outcome | Gen | Method | Nsnp | b | Se | P |
|---|---|---|---|---|---|---|---|
| 9816_37_ISOC1_ISOC1 | KIRC | ISOC1 | IVW | 8 | 0.736746 | 0.171048 | 1.65E-05 |
| 6354_13_HEPHL1_HPHL1 | KIRC | HEPHL1 | IVW | 5 | -1.28851 | 0.310833 | 3.39E-05 |
| 17742_2_RRAS_RRAS | KIRC | RRAS | IVW | 5 | -1.33358 | 0.325935 | 4.29E-05 |
| 12621_55_PPP2R1A_PP2AAA | KIRC | PPP2R1A | IVW | 4 | -0.9648 | 0.243754 | 7.56E-05 |
| 5345_51_CKAP2_CKAP2 | KIRC | CKAP2 | IVW | 15 | -0.66094 | 0.169567 | 9.71E-05 |
| 12807_89_ARHGAP30_RHG30 | KIRC | ARHGAP30 | IVW | 3 | 1.721247 | 0.446005 | 0.000114 |
| 6123_69_TP53_p53 | KIRC | TP53 | IVW | 7 | 1.201512 | 0.311593 | 0.000115 |
| 9511_61_NXPH2_NXPH2 | KIRC | NXPH2 | IVW | 18 | -0.39638 | 0.103952 | 0.000137 |
| 6388_21_CCDC126_CC126 | KIRC | CCDC126 | IVW | 18 | -0.46547 | 0.124775 | 0.000191 |
| 13673_21_CCT7_TCPH | KIRC | CCT7 | IVW | 4 | -1.0039 | 0.269598 | 0.000196 |
| 7083_74_MATN4_MATN4 | KIRC | MATN4 | IVW | 7 | 0.634937 | 0.174364 | 0.000271 |
| 4568_17_SLITRK5_SLIK5 | KIRC | SLITRK5 | IVW | 12 | 0.821988 | 0.226306 | 0.000281 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | IVW | 3 | -1.75547 | 0.495302 | 0.000394 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | IVW | 8 | -0.76964 | 0.218311 | 0.000423 |
| 8040_9_LEMD1_LEMD1 | KIRC | LEMD1 | IVW | 3 | -2.26505 | 0.647584 | 0.000469 |
| 6578_29_TNMD_TNMD | KIRC | TNMD | IVW | 3 | -1.94913 | 0.558892 | 0.000488 |
| 9012_1_PRPF6_PRP6 | KIRC | PRPF6 | IVW | 2 | -2.88761 | 0.828756 | 0.000493 |
| 8535_102_DMKN_Dermokine | KIRC | DMKN | IVW | 2 | -1.63853 | 0.479425 | 0.000632 |
| 8748_45_CD74_HG2A | KIRC | CD74 | IVW | 3 | -2.22233 | 0.652226 | 0.000656 |
| 10085_25_STAR_STAR | KIRC | STAR | IVW | 5 | -1.39378 | 0.418791 | 0.000874 |
| 18198_51_HBQ1_HBAT | KIRC | HBQ1 | IVW | 8 | -0.86326 | 0.261832 | 0.000977 |
| 18380_78_ALB_Albumin | KIRC | ALB | IVW | 11 | -0.41249 | 0.13172 | 0.001739 |
| 12587_65_ARL2_ARL2 | KIRC | ARL2 | IVW | 12 | -0.47391 | 0.152486 | 0.001884 |
| 12041_33_HSPA1L_HS71L | KIRC | HSPA1L | IVW | 11 | -0.55241 | 0.178001 | 0.001913 |
| 10842_7_GCNT4_GCNT4 | KIRC | GCNT4 | IVW | 4 | -1.6224 | 0.541404 | 0.00273 |
| 11160_56_RNF122_RN122 | KIRC | RNF122 | IVW | 6 | -0.87475 | 0.299384 | 0.00348 |
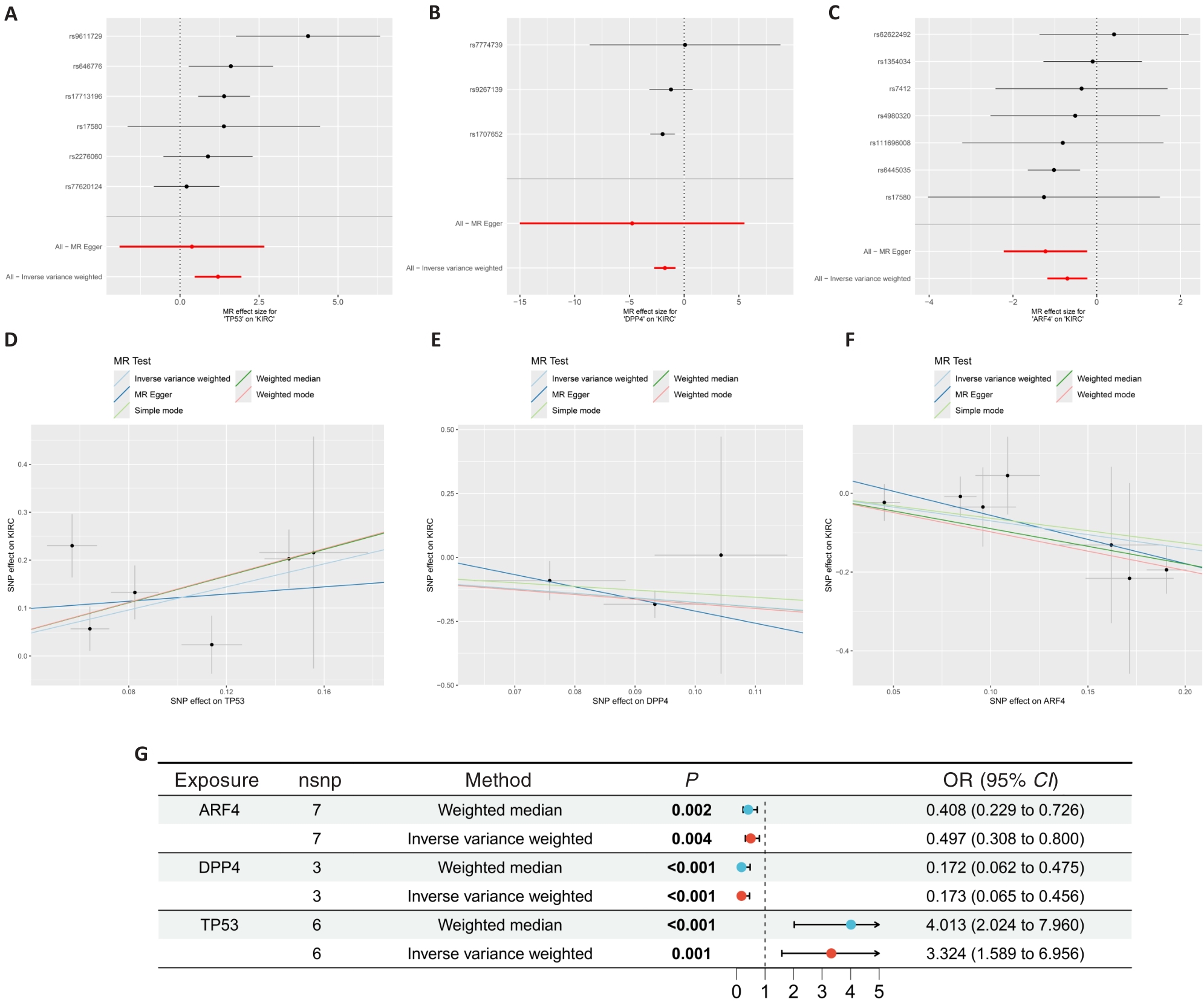
图3 肾透明细胞癌的孟德尔随机化结果
Fig.3 Mendelian randomization results on ccRCC. A-C: Forest plot showing the results of Mendelian randomization of the core genes. D-F: Scatter plots of Mendelian randomization results on core targets and ccRCC. G: Forest plot showing Mendelian randomization results of the core genes.
| Id.exposure | Outcome | Exposure | Method | Q | Q_pval |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | MR Egger | 2.46079 | 0.782388313 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | Inverse variance weighted | 3.836625 | 0.698774273 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | MR Egger | 0.27862 | 0.597607084 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | Inverse variance weighted | 0.608893 | 0.737531351 |
| 6123_69_TP53_p53 | KIRC | TP53 | MR Egger | 9.021383 | 0.060567227 |
| 6123_69_TP53_p53 | KIRC | TP53 | Inverse variance weighted | 10.29088 | 0.06740046 |
表2 血浆蛋白质与肾透明细胞癌孟德尔随机化的异质性分析
Tab. 2 Heterogeneity analysis of Mendelian randomization between plasma proteins and ccRCC
| Id.exposure | Outcome | Exposure | Method | Q | Q_pval |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | MR Egger | 2.46079 | 0.782388313 |
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | Inverse variance weighted | 3.836625 | 0.698774273 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | MR Egger | 0.27862 | 0.597607084 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | Inverse variance weighted | 0.608893 | 0.737531351 |
| 6123_69_TP53_p53 | KIRC | TP53 | MR Egger | 9.021383 | 0.060567227 |
| 6123_69_TP53_p53 | KIRC | TP53 | Inverse variance weighted | 10.29088 | 0.06740046 |
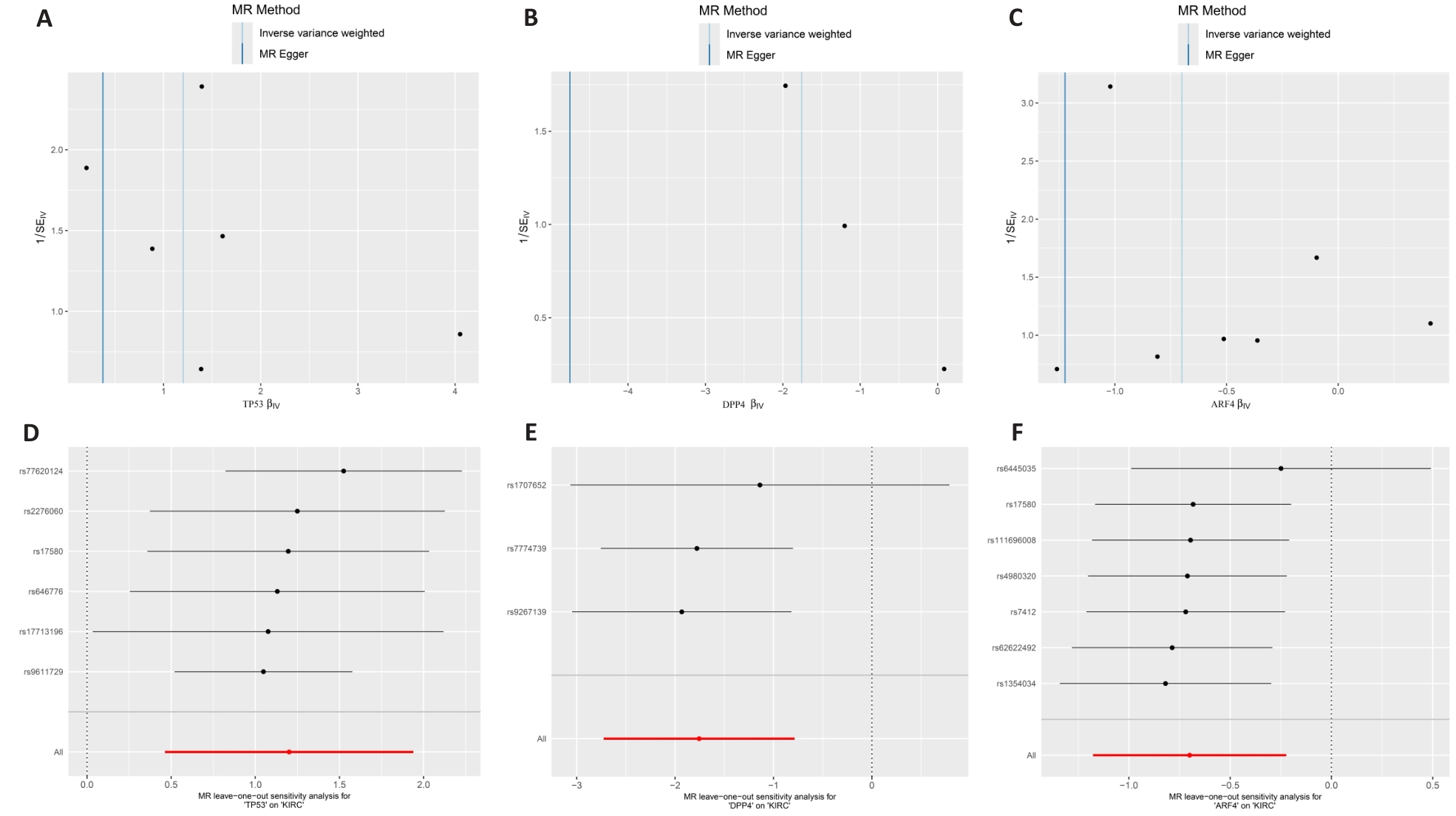
图4 孟德尔随机化的敏感性分析
Fig.4 Sensitivity analysis of Mendelian randomization. A-C: MR funnel plot for core targets and ccRCC. D-F: Leave-one-out sensitivity analysis for potential core genes and ccRCC.
| Id.exposure | Outcome | Exposure | Egger_intercept | Se | P |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | 0.066322526 | 0.056543 | 0.293645 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | 0.264971682 | 0.461066 | 0.667936 |
| 6123_69_TP53_p53 | KIRC | TP53 | 0.08422687 | 0.112264 | 0.494821 |
表3 血浆蛋白质和肾透明细胞癌之间孟德尔随机化的多效性分析
Tab.3 Pleiotropy analysis of Mendelian randomization between plasma proteins and ccRCC
| Id.exposure | Outcome | Exposure | Egger_intercept | Se | P |
|---|---|---|---|---|---|
| 18408_26_ARF4_ARF4 | KIRC | ARF4 | 0.066322526 | 0.056543 | 0.293645 |
| 15460_9_DPP4_CD26 | KIRC | DPP4 | 0.264971682 | 0.461066 | 0.667936 |
| 6123_69_TP53_p53 | KIRC | TP53 | 0.08422687 | 0.112264 | 0.494821 |
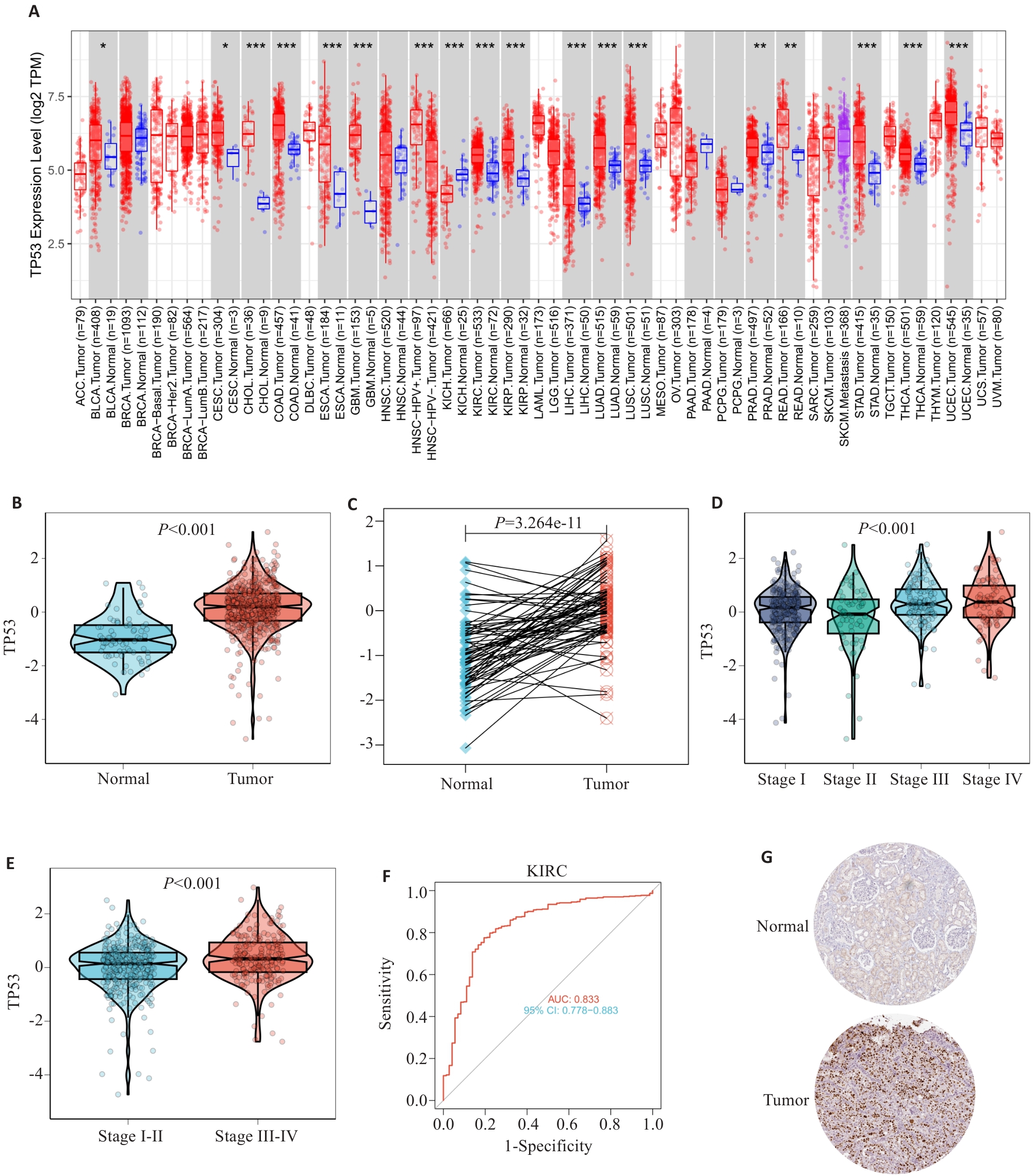
图5 TP53在肾透明细胞癌中的表达
Fig.5 TP53 expression in ccRCC. A: TP53 mRNA evpression in different tumor samples. B: TP53 mRNA expression in ccRCC samples and normal tissues. C: TP53 mRNA expression in ccRCC and paired adjacent normal tissues. D,E: TP53 mRNA expression levels in different ccRCC clinical stages. F: Receiver operating characteristic analysis (ROC) of TP53 in patients with ccRCC (AUC=0.833). G: Immunohistochemical staining of TP53 in renal cancer tissues and normal renal tissues (Original magnification: ×20).
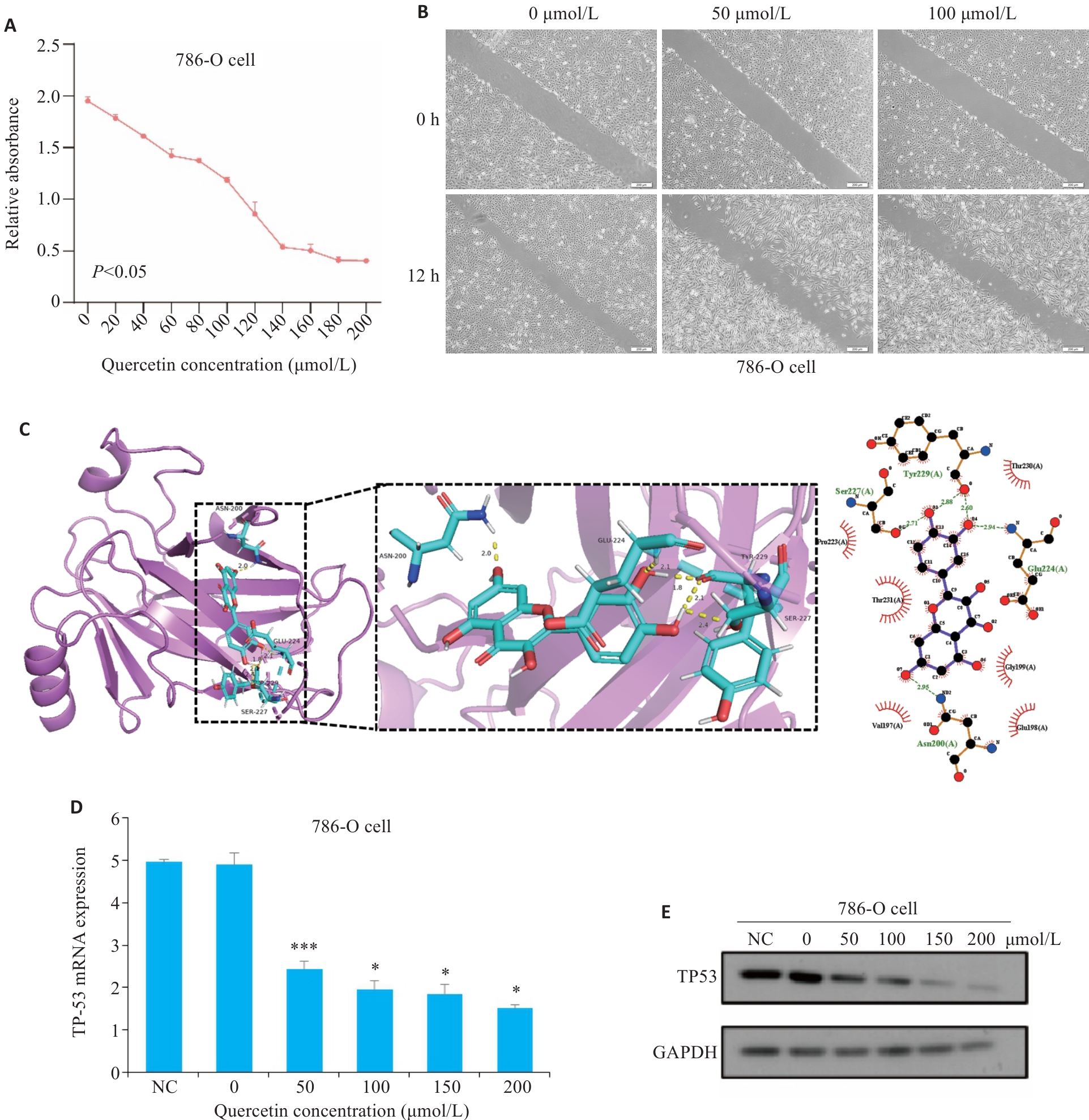
图6 细胞实验与分子对接验证
Fig.6 Cell experiments and molecular docking validation. A: Viability of 786-O cells measured using CCK-8 assay after 24 h of quercetin treatment. B: Wound healing assay for assessing migration capacity of 786-O cells after quercetin treatment for 12 h (scale bar=200 μm). C: Docking of quercetin with TP53 molecule. D: Effect of different concentrations of quercetin on TP53 mRNA expression in 786-O cells detected using RT-qPCR (*P<0.05,***P<0.001 vs 0 μmol/L). E: Effect of different concentrations of quercetin on TP53 protein expression in 786-O cells detected using Western blotting.
| 1 | Marston Linehan W, Schmidt LS, Crooks DR, et al. The metabolic basis of kidney cancer[J]. Cancer Discov, 2019, 9(8): 1006-21. |
| 2 | Singh D, Vignat J, Lorenzoni V, et al. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative[J]. Lancet Glob Health, 2023, 11(2): e197-206. |
| 3 | Capitanio U, Montorsi F. Renal cancer[J]. Lancet, 2016, 387(10021): 894-906. |
| 4 | Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European association of urology guidelines on renal cell carcinoma: the 2022 update[J]. Eur Urol, 2022, 82(4): 399-410. |
| 5 | Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma[J]. N Engl J Med, 2019, 380(12): 1103-15. |
| 6 | Panossian A, Wikman G. Pharmacology of Schisandra chinensis bail.: an overview of Russian research and uses in medicine[J]. J Ethnopharmacol, 2008, 118(2): 183-212. |
| 7 | Li Y, Yao JY, Han CY, et al. Quercetin, inflammation and immunity[J]. Nutrients, 2016, 8(3): 167. |
| 8 | Lim H, Min DS, Yun HE, et al. Impressic acid from Acanthopanax koreanum, possesses matrix metalloproteinase-13 down-regulating capacity and protects cartilage destruction[J]. J Ethnopharmacol, 2017, 209: 73-81. |
| 9 | Wong CK. Simplifying electrocardiographic assessment in STEMI reperfusion management: Pros and cons[J]. Int J Cardiol, 2017, 227: 30-6. |
| 10 | Vandemoortele A, Babat P, Yakubu M, et al. Reactivity of free malondialdehyde during in vitro simulated gastrointestinal digestion[J]. J Agric Food Chem, 2017, 65(10): 2198-204. |
| 11 | Ru JL, Li P, Wang JN, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines[J]. J Cheminform, 2014, 6: 13. |
| 12 | Wang X, Shen YH, Wang SW, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database[J]. Nucleic Acids Res, 2017, 45(W1): W356-60. |
| 13 | Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules[J]. Nucleic Acids Res, 2019, 47(W1): W357-64. |
| 14 | Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018[J]. Nucleic Acids Res, 2018, 46(D1): D1074-82. |
| 15 | Gilson MK, Liu TQ, Baitaluk M, et al. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology[J]. Nucleic Acids Res, 2016, 44(D1): D1045-53. |
| 16 | Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility[J]. J Comput Chem, 2009, 30(16): 2785-91. |
| 17 | Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina[J]. J Comput Aided Mol Des, 2010, 24(5): 417-22. |
| 18 | Larson AJ, David Symons J, Jalili T. Therapeutic potential of quercetin to decrease blood pressure: review of efficacy and mechanisms[J]. Adv Nutr, 2012, 3(1): 39-46. |
| 19 | Russo M, Spagnuolo C, Tedesco I, et al. Phytochemicals in cancer prevention and therapy: truth or dare[J]? Toxins, 2010, 2(4): 517-51. |
| 20 | Tang SM, Deng XT, Zhou J, et al. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects[J]. Biomed Pharmacother, 2020, 121: 109604. |
| 21 | Levine AJ. p53: 800 million years of evolution and 40 years of discovery[J]. Nat Rev Cancer, 2020, 20(8): 471-80. |
| 22 | Network CGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma[J]. Nature, 2013, 499(7456): 43-9. |
| 23 | Wang B, Song Q, Wei YA, et al. Comprehensive investigation into cuproptosis in the characterization of clinical features, molecular characteristics, and immune situations of clear cell renal cell carcinoma[J]. Front Immunol, 2022, 13: 948042. |
| 24 | Paul JY, Harding R, Tushemereirwe W, et al. Banana21: from gene discovery to deregulated golden bananas[J]. Front Plant Sci, 2018, 9: 558. |
| 25 | D’Andrea G. Quercetin: a flavonol with multifaceted therapeutic applications[J]? Fitoterapia, 2015, 106: 256-71. |
| 26 | Li FZ, Aljahdali IAM, Zhang RY, et al. Kidney cancer biomarkers and targets for therapeutics: survivin (BIRC5), XIAP, MCL-1, HIF1α, HIF2α, NRF2, MDM2, MDM4, p53, KRAS and AKT in renal cell carcinoma[J]. J Exp Clin Cancer Res, 2021, 40(1): 254. |
| 27 | Uhlman DL, Nguyen PL, Manivel JC, et al. Association of immunohistochemical staining for p53 with metastatic progression and poor survival in patients with renal cell carcinoma[J]. J Natl Cancer Inst, 1994, 86(19): 1470-5. |
| 28 | Shvarts O, Seligson D, Lam J, et al. p53 is an independent predictor of tumor recurrence and progression after nephrectomy in patients with localized renal cell carcinoma[J]. J Urol, 2005, 173(3): 725-8. |
| 29 | Noon AP, Vlatković N, Polański R, et al. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets[J]? Cancer, 2010, 116(4): 780-90. |
| [1] | 徐皓男, 张放, 黄钰莹, 姚其盛, 管悦琴, 陈浩. 百蕊草通过调节肠道菌群和调控EGFR/PI3K/Akt信号通路改善小鼠抗生素相关性腹泻[J]. 南方医科大学学报, 2025, 45(2): 285-295. |
| [2] | 黄鹏伟, 陈洁, 邹金虎, 高雪锋, 曹虹. 槲皮素促进应激颗粒G3BP1解聚改善HIV-1 gp120诱导的星形胶质细胞神经毒性[J]. 南方医科大学学报, 2025, 45(2): 304-312. |
| [3] | 陈晓睿, 魏青政, 张宗亮, 原江水, 宋卫青. 过表达带电多泡体蛋白2B基因抑制肾透明细胞癌细胞的增殖[J]. 南方医科大学学报, 2025, 45(1): 126-136. |
| [4] | 褚乔, 王小娜, 续佳颖, 彭荟林, 赵裕琳, 张静, 陆国玉, 王恺. 白头翁皂苷D通过多靶点和多途径抑制三阴性乳腺癌侵袭转移[J]. 南方医科大学学报, 2025, 45(1): 150-161. |
| [5] | 龙秀鹏, 陶顺, 阳绅, 李素云, 饶利兵, 李莉, 张哲. 槲皮素通过抑制MAPK信号通路改善心力衰竭[J]. 南方医科大学学报, 2025, 45(1): 187-196. |
| [6] | 徐朦, 陈丽娜, 吴金玉, 刘丽丽, 施美, 周灏, 张国梁. “白花蛇舌草-半枝莲”治疗原发性肝癌的机制研究:基于网络药理学、分子对接及体外实验验证[J]. 南方医科大学学报, 2025, 45(1): 80-89. |
| [7] | 刘青, 刘敬, 郑逸航, 雷金, 黄建华, 刘思妤, 刘芳, 彭群龙, 张远芳, 王俊杰, 李玉娟. 积雪草活性成分槲皮素通过介导STAT3磷酸化抑制IL-23/IL-17A炎症轴发挥抗银屑病作用[J]. 南方医科大学学报, 2025, 45(1): 90-99. |
| [8] | 姚辰, 李文佳, 庞瑞明, 周继红. 臀肌腱炎、原发性髋关节病可能导致髂胫束综合征—一项孟德尔随机化研究[J]. 南方医科大学学报, 2024, 44(9): 1821-1830. |
| [9] | 姜一凡, 李小荣, 耿嘉逸, 陈永锋, 唐碧, 康品方. 槲皮素通过抑制HMGB1/RAGE/NF-κB信号通路减轻糖尿病引起的大鼠肾脏损伤[J]. 南方医科大学学报, 2024, 44(9): 1769-1775. |
| [10] | 陈星梅, 刘琴文, 李镱, 钟晓宇, 樊奇灵, 马柯, 罗柳婷, 官道刚, 朱志博. 茵陈蒿汤治疗肝纤维化的核心功能成分群以及潜在通路[J]. 南方医科大学学报, 2024, 44(8): 1508-1517. |
| [11] | 张珊苑, 蔡巧燕, 祁江晗, 殷恺馨, 何晨晨, 高铸烨, 张铃, 褚剑锋. 清心解瘀颗粒抗动脉粥样硬化的药效学及调控机制[J]. 南方医科大学学报, 2024, 44(8): 1518-1528. |
| [12] | 张钰明, 夏士程, 张淋淋, 陈梦茜, 刘晓婧, 高琴, 叶红伟. 金银花提取物对小鼠阿霉素肝脏损伤的保护作用[J]. 南方医科大学学报, 2024, 44(8): 1571-1581. |
| [13] | 钟帷韬, 李伟松, 李泽霖, 王强, 张旺明. 睡眠性状与特发性正常压力脑积水的因果关联:一项两样本双向孟德尔随机化研究[J]. 南方医科大学学报, 2024, 44(8): 1612-1619. |
| [14] | 王瑾瑾, 崔文飞, 窦雪伟, 尹冰磊, 牛钰琪, 牛羚, 闫国立. 鬼箭羽通过调节EGFR酪氨酸激酶抑制剂耐药信号通路延缓糖尿病肾病的进展[J]. 南方医科大学学报, 2024, 44(7): 1243-1255. |
| [15] | 王琳月, 戚文月, 高记华, 田茂生, 许建成. 痛痒消洗剂可促进大鼠肛瘘术后的创面愈合[J]. 南方医科大学学报, 2024, 44(7): 1256-1265. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||