Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (2): 269-284.doi: 10.12122/j.issn.1673-4254.2025.02.08
Previous Articles Next Articles
Jinhua ZOU( ), Hui WANG(
), Hui WANG( ), Dongyan ZHANG(
), Dongyan ZHANG( )
)
Received:2024-10-11
Online:2025-02-20
Published:2025-03-03
Contact:
Dongyan ZHANG
E-mail:360722772@qq.com;635137884@qq.com;15013091401@163.com
Supported by:Jinhua ZOU, Hui WANG, Dongyan ZHANG. SLC1A5 overexpression accelerates progression of hepatocellular carcinoma by promoting M2 polarization of macrophages[J]. Journal of Southern Medical University, 2025, 45(2): 269-284.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.02.08
| Products name | Target sequence |
|---|---|
| SLC1A5(siRNA# 1) | 5'-GTACCGTCCTCAATGTAGA-3' |
| SLC1A5(siRNA# 2) | 5'- GAAGCACAGAGCCTGAGTT-3' |
Tab.1 siRNA sequences for SLC1A5
| Products name | Target sequence |
|---|---|
| SLC1A5(siRNA# 1) | 5'-GTACCGTCCTCAATGTAGA-3' |
| SLC1A5(siRNA# 2) | 5'- GAAGCACAGAGCCTGAGTT-3' |
| Primer | Sequence | |
|---|---|---|
| SLC1A5 | Sense Antisense | 5'-GAGGCTTTCTCTGGCTGGTAA-3' 5'-GTCTTGGACACTGAGGGCTG-3' |
TGFB1 IL-10 CSF1R CXCL1 | Sense Antisense Sense Antisense Sense Antisense Sense Antisense | 5'-CAAGTGGACATCAACGGGTTC-3' 5'-GCAGCAGTTCTTCTCCGTGG-3' 5'-GAGGAGGTGATGCCCCAAGC-3' 5'-GGGAAGAAATCGATGACAGC-3' 5'-CCTCGCTTCCAAGAATTGCA-3' 5'-CCCAATCTTGGCCACATGA-3' 5'-CTCTTCCGCTCCTCTCACAG-3' 5'-CACGGACGCTCCTGCTGC-3' |
Tab.2 PCR primer sequences
| Primer | Sequence | |
|---|---|---|
| SLC1A5 | Sense Antisense | 5'-GAGGCTTTCTCTGGCTGGTAA-3' 5'-GTCTTGGACACTGAGGGCTG-3' |
TGFB1 IL-10 CSF1R CXCL1 | Sense Antisense Sense Antisense Sense Antisense Sense Antisense | 5'-CAAGTGGACATCAACGGGTTC-3' 5'-GCAGCAGTTCTTCTCCGTGG-3' 5'-GAGGAGGTGATGCCCCAAGC-3' 5'-GGGAAGAAATCGATGACAGC-3' 5'-CCTCGCTTCCAAGAATTGCA-3' 5'-CCCAATCTTGGCCACATGA-3' 5'-CTCTTCCGCTCCTCTCACAG-3' 5'-CACGGACGCTCCTGCTGC-3' |
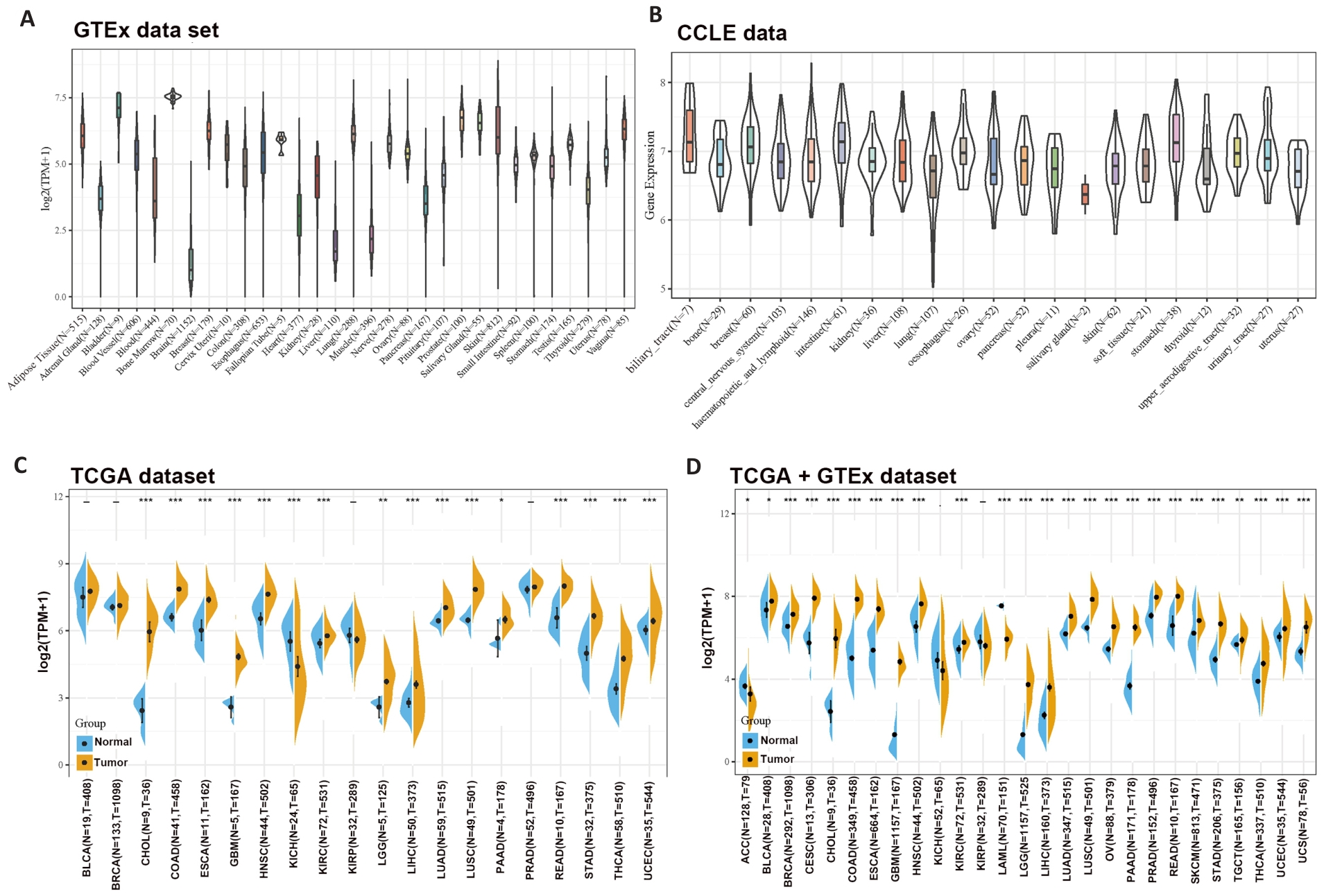
Fig1 Differential expressions of SLC1A5 across different cancer types. A: SLC1A5 expression in normal tissues. B: SLC1A5 expression in tumor cell lines. C: Expression status of SLC1A5 gene in paired tumor and non-tumor tissues across 22 cancer types from TCGA. D: Analysis of SLC1A5 expression in 33 cancer types with their corresponding normal tissues in GTEx dataset as controls. *P<0.05, **P<0.01, *** P<0.001.
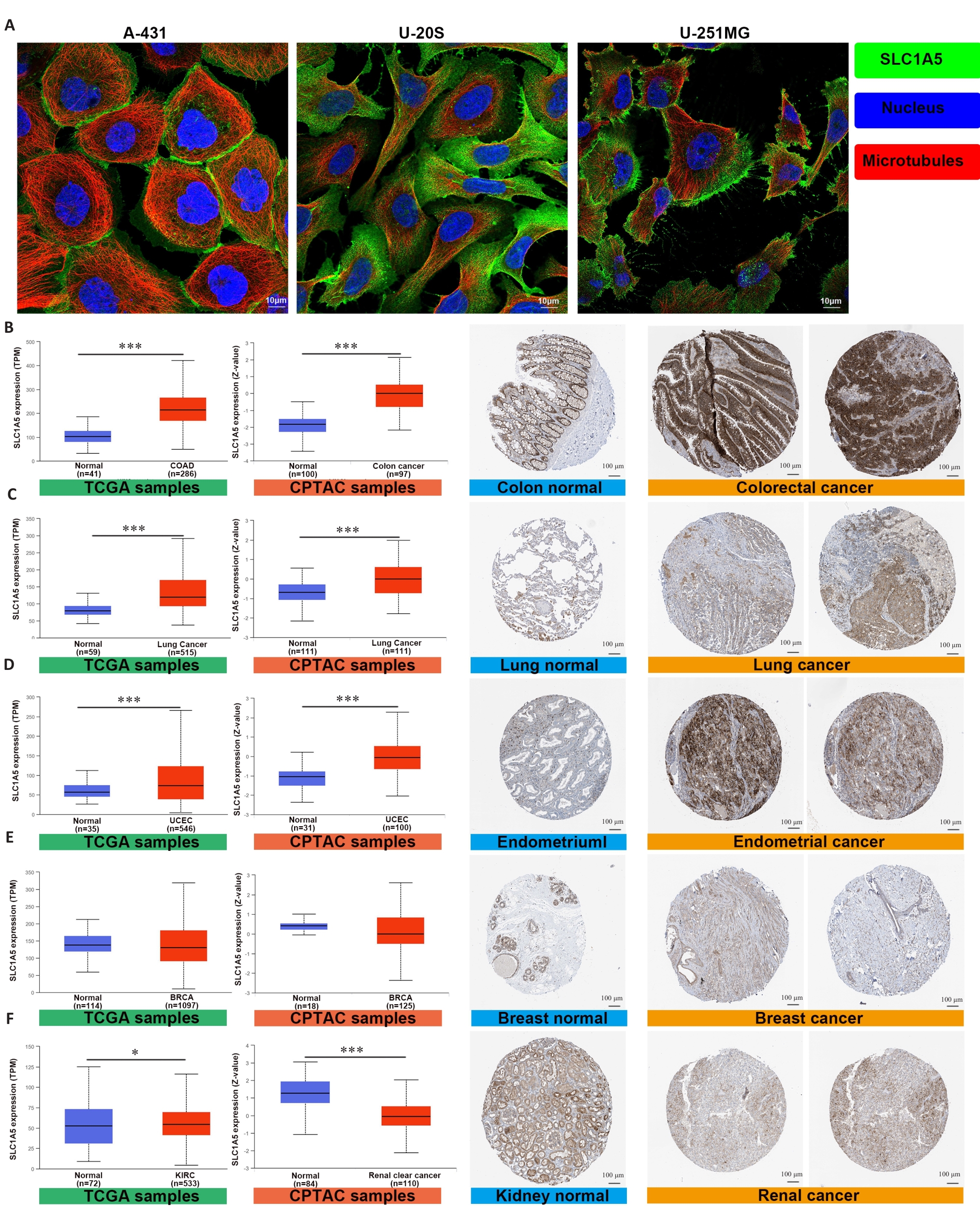
Fig.2 SLC1A5 is highly expressed in various cancers. A: Expression and distribution of SLC1A5 in different cancer cell lines (Scale bar=10 μm); B-F: Comparison of SLC1A5 gene expression between normal and tumor tissues from TCGA databases (left), SLC1A5 protein expression between normal and tumor tissues from CPTAC (middle), and immunohistochemistry images of normal and tumor tissues from HPA (right, Scale bar=100 μm). SLC1A5 expression at both the mRNA and protein levels was significantly higher in colorectal cancer (B), lung cancer (C), and endometrial cancer (D). There was no significant difference in SLC1A5 expression between tumor and non-tumor in breast cancer (E). SLC1A5 gene expression was slightly upregulated in renal cancer, but was significantly downregulated in the tumor tissues at the protein level (F). *P<0.05, ***P<0.001.
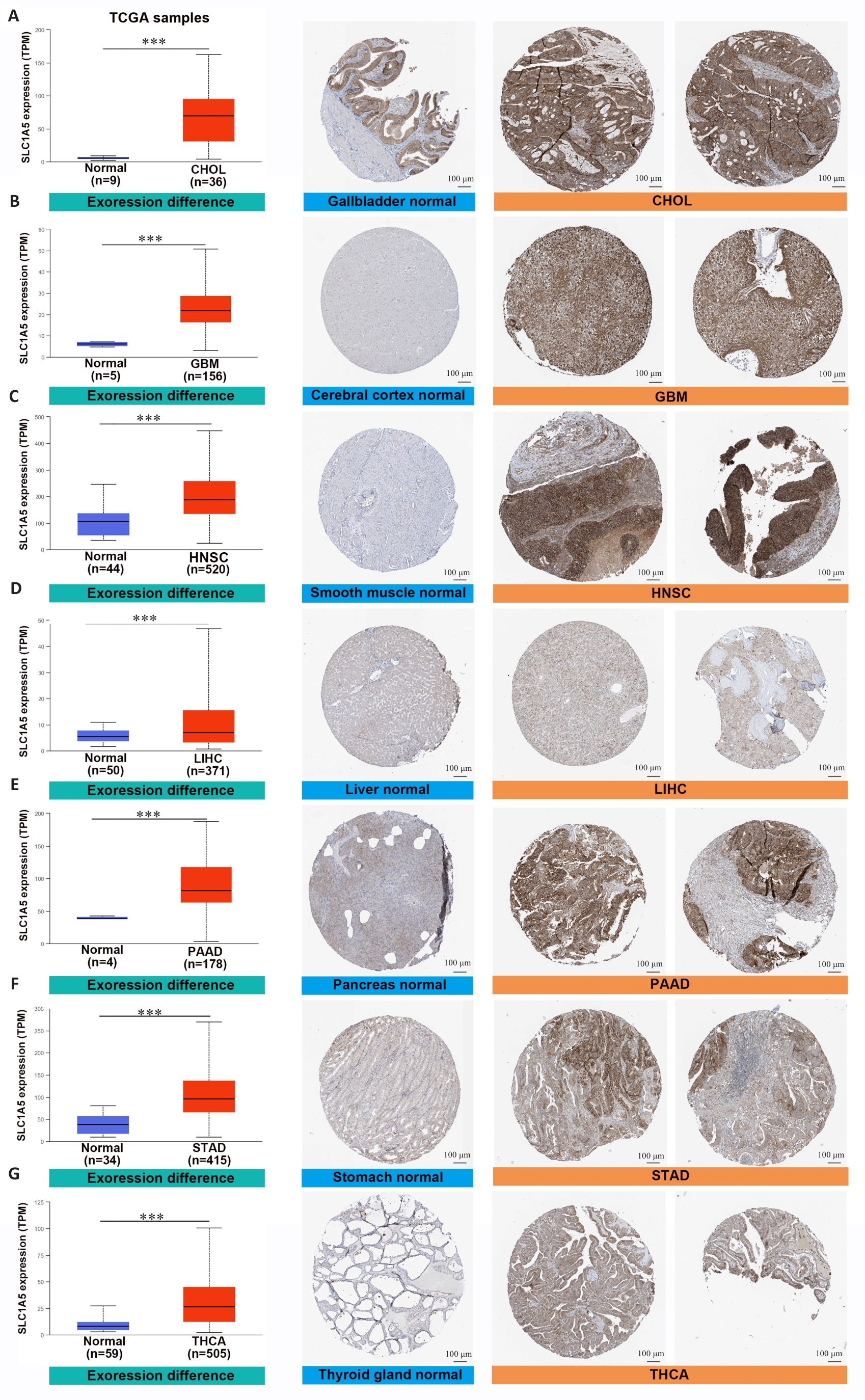
Fig.3 SLC1A5 is highly expressed in various cancer types. Comparison of SLC1A5 gene expression between normal and tumor tissues from TCGA databases (left), SLC1A5 protein expression between normal and tumor tissues from CPTAC (middle), and immunohistochemistry images of normal and tumor tissues from HPA (right, Scale bar=100 μm). ***P<0.001.
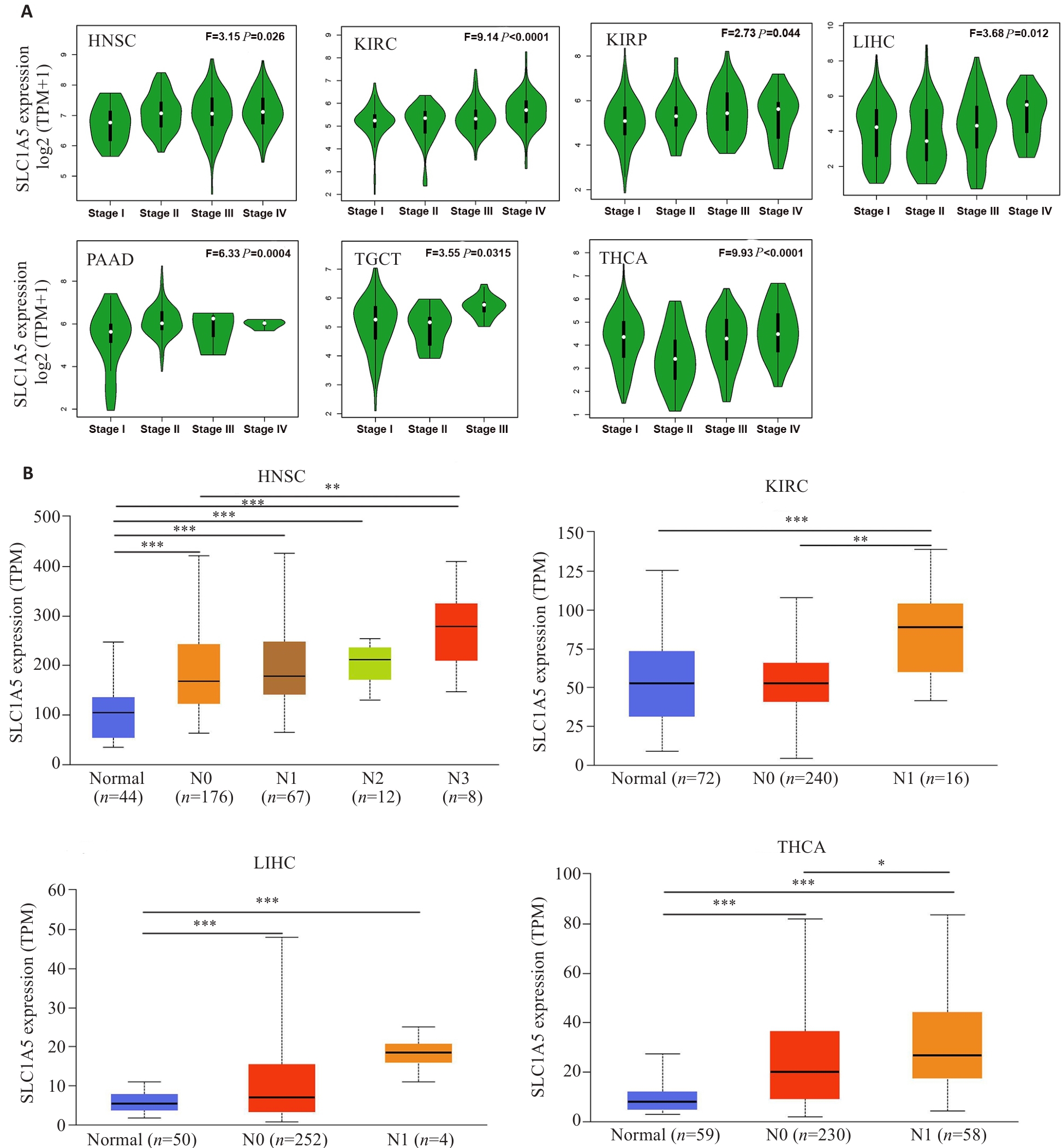
Fig.4 Expression levels of SLC1A5 gene in cancers in realtion to pathological stages and status of lymphatic metastasis. A: Expression levels of SLC1A5 mRNA in different pathological stages of HNSC, KIRC, KIRP, LIHC, PAAD, TGCT, and THCA. B: Expression levels of SLC1A5 mRNA analyzed in relation to lymphatic metastasis in HNSC, KIRC, LIHC, and THCA. *P<0.05, **P<0.01, ***P<0.001.
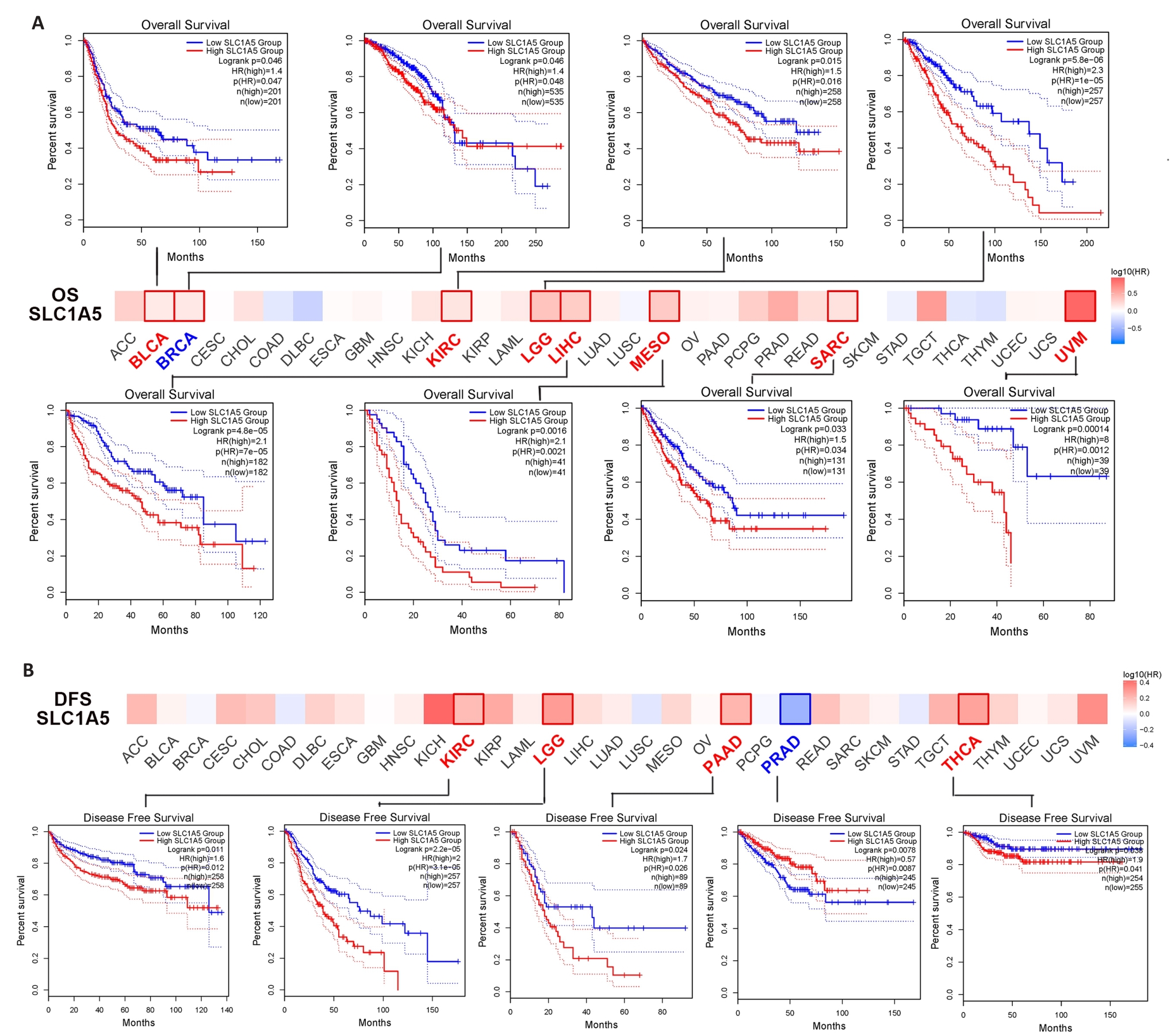
Fig.5 Correlation between SLC1A5 expression and survival outcomes of different cancers. A: GEPIA2 tool was used for analysis of overall survival (A) and disease-survival (B) for 33 cancers in TCGA in relation to SLC1A5 expression. The survival map and survival curves with statistical significance are shown.
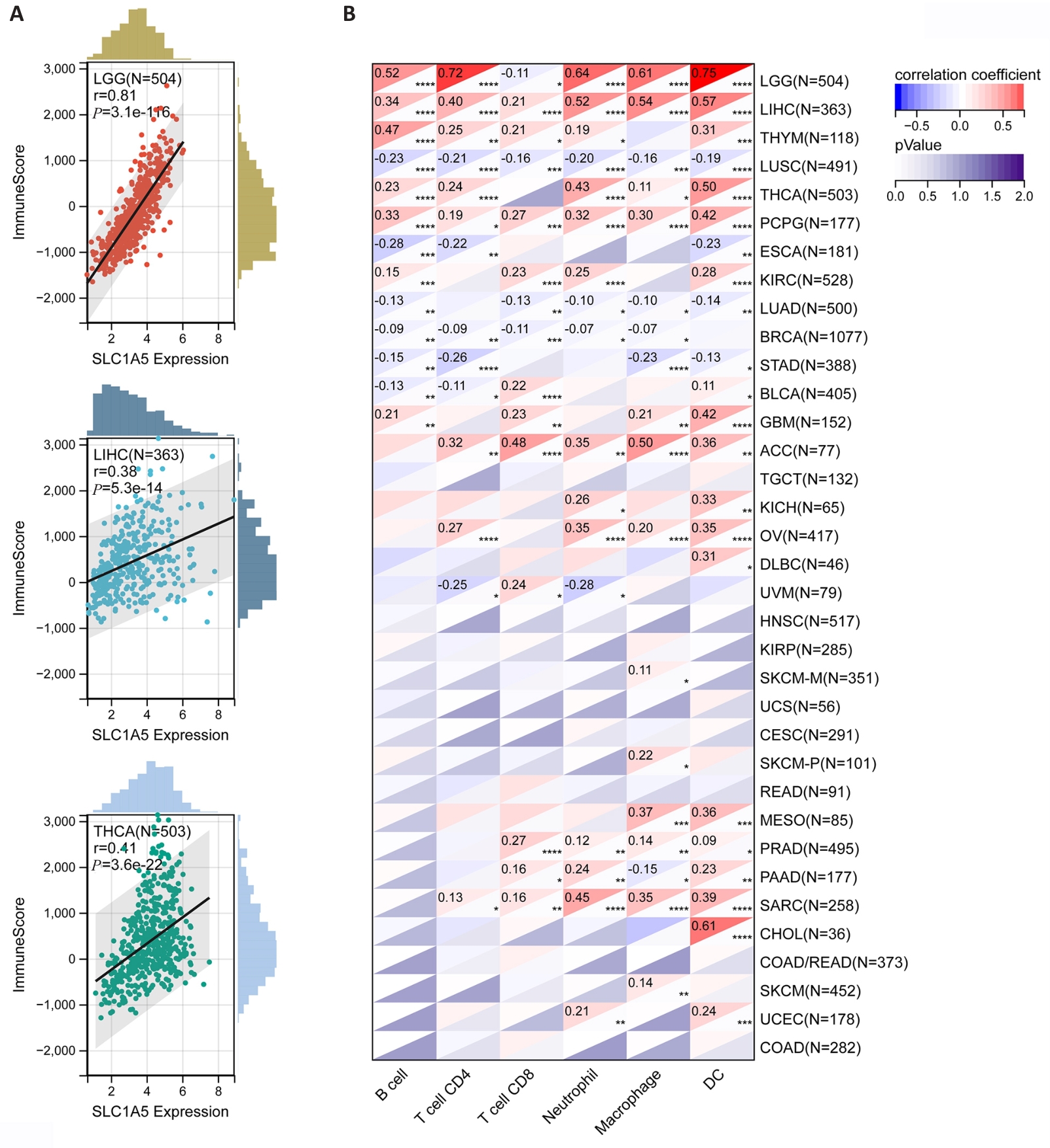
Fig.6 Correlation of SLC1A5 expression levels with tumor microenvironment and levels of tumor immune cell infiltration. A: Three tumors with the highest correlation coefficients between SLC1A5 expression and the tumor microenvironment. B: Correlation coefficients between SLC1A5 expression and tumor infiltration level of different immune cells in 33 cancer types. *P<0.05; **P<0.01, ***P<0.001, ****P<0.0001.
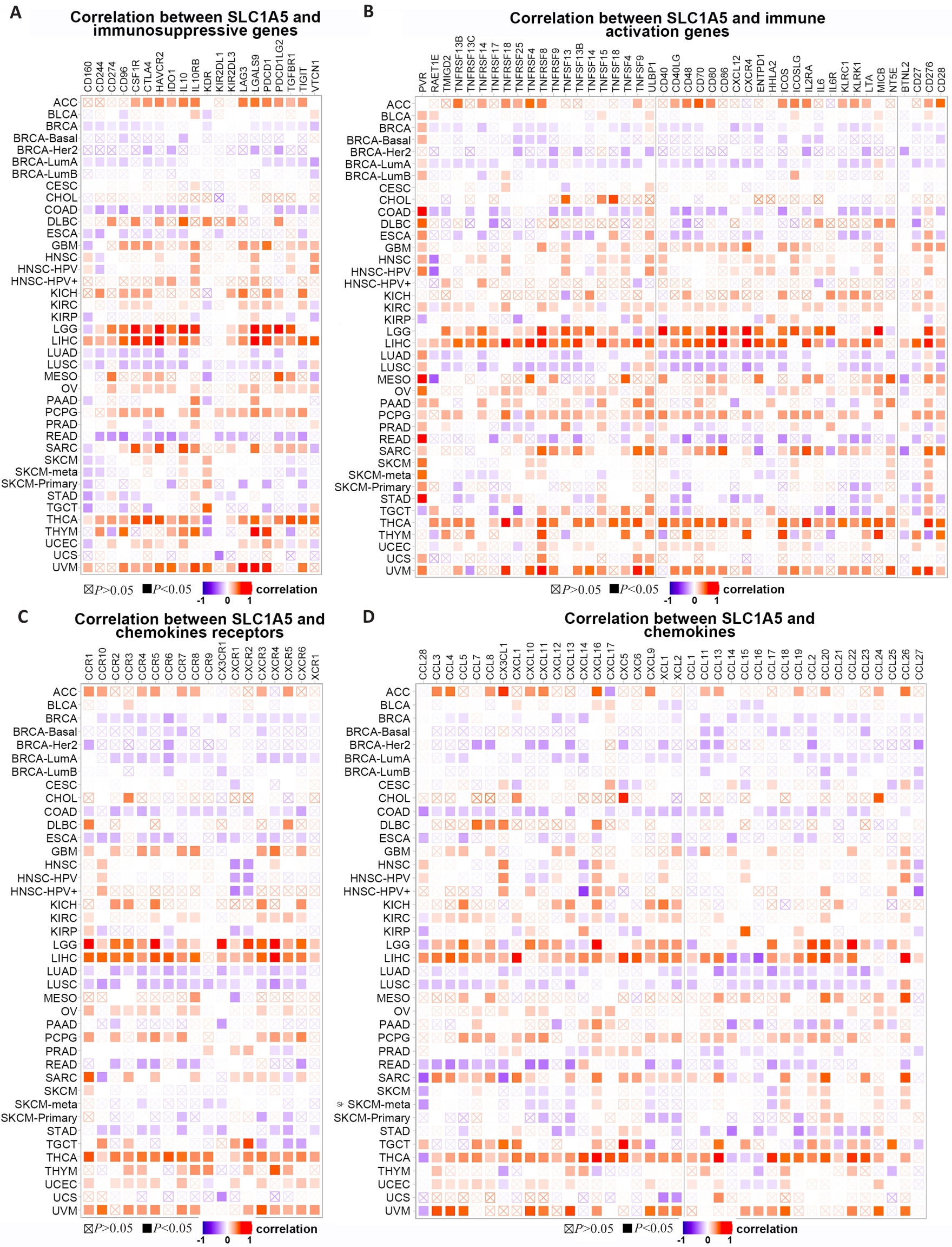
Fig.7 Co-expression of SLC1A5 expression and immune-related genes. Correlations of SLC1A5 expression with immunosuppressive genes (A), immune activation genes (B), chemokines receptor (C), and chemokines (D) are presented.
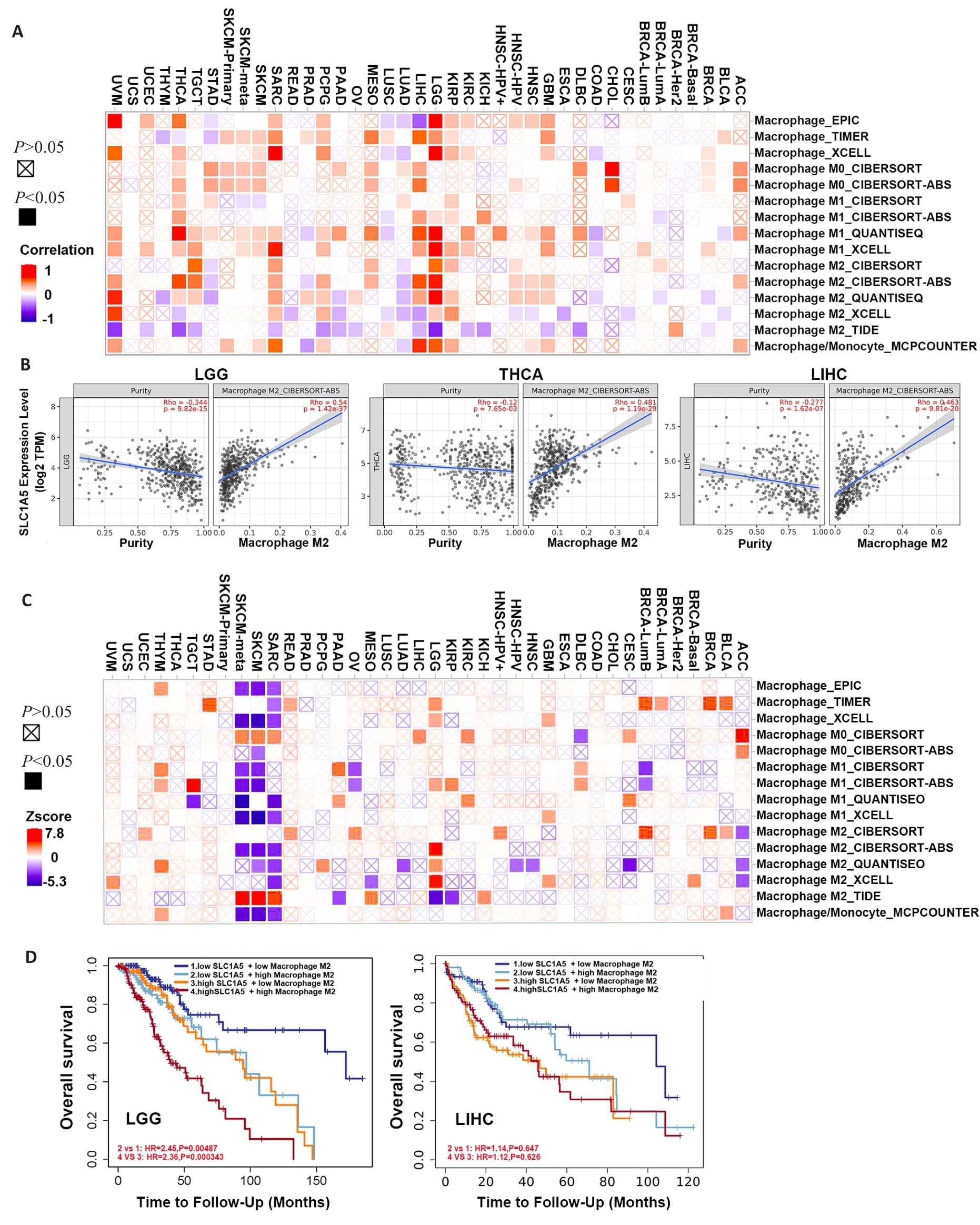
Fig.8 Correlation between SLC1A5 expression and different subtypes of macrophage. A: Relationship between SLC1A5 expression and different subtypes of macrophage. B: Three tumors with the highest correlation coefficients between SLC1A5 expression and M2 macrophages. C: Survival map illustrating the correlation between different subtypes of macrophage and overall survival in different cancers. D: Kaplan-Meier survival analysis of LGG (left) and LIHC (right) patients with different SLC1A5 expression levels stratified by levels of infiltrating M2 macrophages.
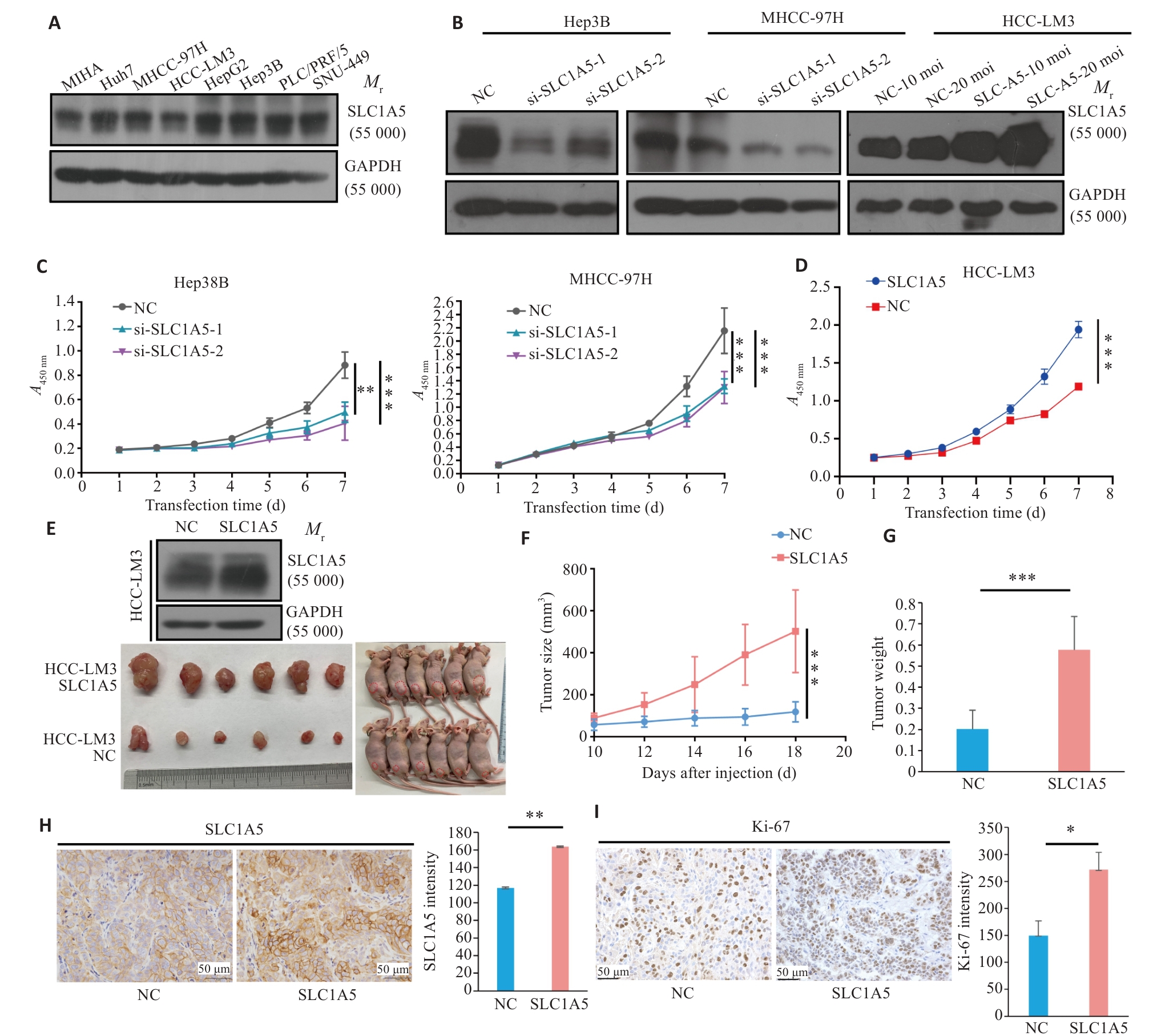
Fig.9 Overexpression of SLC1A5 promotes proliferation of HCC cells. A: Expression of SLC1A5 in HCC cell lines detected by Western blotting. LE: Long exposure; SE: Short exposure. B: Verification of SLC1A5 knockout and overexpression efficiency in different HCC cell lines using Western blotting. C: Effect of RNA interference of SLC1A5 on HCC cell proliferation assessed using CCK-8 assay. D: Effect of SLC1A5 overexpression on HCC cell proliferation assessed using by CCK-8 assay. E: Overexpression efficiency of SLC1A5 in HCC-LM3 cells examined by Western blotting (upper). Examination of subcutaneous tumors in nude mice (bottom). F, G: Proliferation curve of subcutaneous tumor (F) and tumor weight (G) in nude mice. H, I: Expressions of SLC1A5 and Ki-67 in subcutaneous tumors detected by immunohistochemistry. *P<0.05, **P<0.01, ***P<0.001.
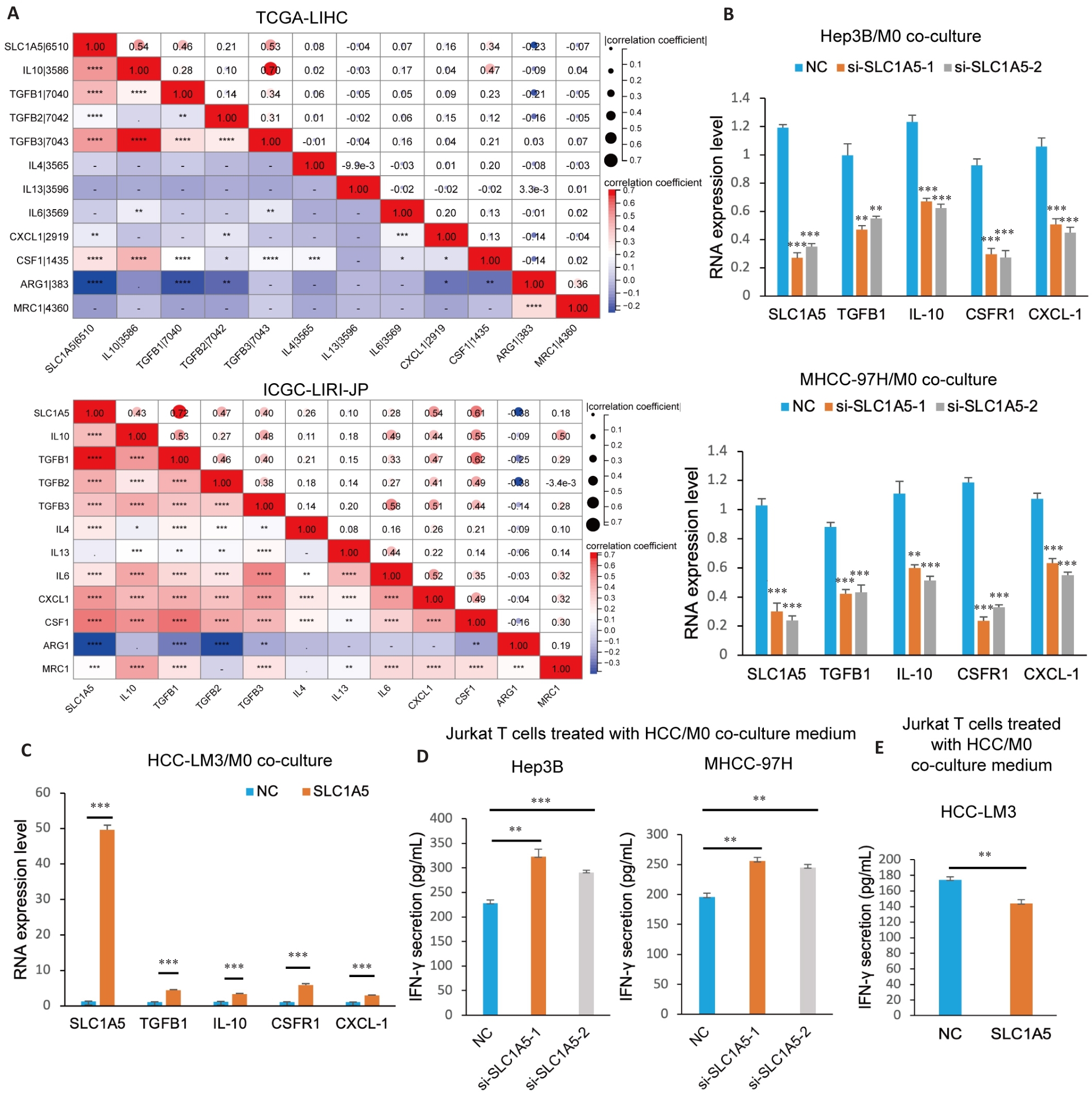
Fig.10 SLC1A5 promotes M2 polarization of macrophages and induces CD8+ T cell dysfunction. A: Correlation analysis between SLC1A5 expression and molecular markers of M2 macrophages. B: M0 cells co-cultured with HCC cells transfected with SLC1A5-specific siRNA or negative control siRNA and induced to polarize into M2 phenotype using IL-4 (20 ng/mL). Interference efficiency of SLC1A5 and expressions of M2 markers (IL-10, TGFB1, CSF1R and CXCL-1) detected using qRT-PCR. C: M0 cells co-cultured with SLC1A5-overpressing HCC-LM3 cells and induced to polarize into M2 phenotype using IL-4 (20 ng/mL). The overpression efficiency of SLC1A5 and the expression of M2 markers (IL-10, TGFB1, CSF1R and CXCL-1) were detected using qRT-PCR. D, E: M0 cells co-cultured with HCC cells with SLC1A5 knockdown (D) and overexpression (E) and induced into the M2 phenotype using IL-4 (20 ng/mL). The culture medium was used to treat activated Jurkat T cells for 48 h, and IFN-γ secretion was detected by ELISA. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
| 1 | Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors in cancer therapy: a focus on T-regulatory cells[J]. Immunol Cell Biol, 2018, 96(1): 21-33. |
| 2 | Sharma P, Goswami S, Raychaudhuri D, et al. Immune checkpoint therapy-current perspectives and future directions[J]. Cell, 2023, 186(8): 1652-69. |
| 3 | Wang QY, Shen XF, Chen G, et al. How to overcome resistance to immune checkpoint inhibitors in colorectal cancer: from mechanisms to translation[J]. Int J Cancer, 2023, 153(4): 709-22. |
| 4 | Bou-Dargham MJ, Sha LL, Sarker DB, et al. TCGA RNA-seq and tumor-infiltrating lymphocyte imaging data reveal cold tumor signatures of invasive ductal carcinomas and estrogen receptor-positive human breast tumors[J]. Int J Mol Sci, 2023, 24(11): 9355. |
| 5 | He Y, Wang XS. Identifying biomarkers associated with immunotherapy response in melanoma by multi-omics analysis[J]. Comput Biol Med, 2023, 167: 107591. |
| 6 | Osman I, He XQ, Liu JH, et al. TEAD1 (TEA domain transcription factor 1) promotes smooth muscle cell proliferation through upregulating SLC1A5 (solute carrier family 1 member 5)-mediated glutamine uptake[J]. Circ Res, 2019, 124(9): 1309-22. |
| 7 | Hassanein M, Hoeksema MD, Shiota M, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival[J]. Clin Cancer Res, 2013, 19(3): 560-70. |
| 8 | Huang F, Zhao YC, Zhao JZ, et al. Upregulated SLC1A5 promotes cell growth and survival in colorectal cancer[J]. Int J Clin Exp Pathol, 2014, 7(9): 6006-14. |
| 9 | van Geldermalsen M, Wang Q, Nagarajah R, et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer[J]. Oncogene, 2016, 35(24): 3201-8. |
| 10 | Wang Q, Hardie RA, Hoy AJ, et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development[J]. J Pathol, 2015, 236(3): 278-89. |
| 11 | Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression[J]. Cancer Res, 2019, 79(18): 4557-66. |
| 12 | Hui LL, Chen Y. Tumor microenvironment: sanctuary of the devil[J]. Cancer Lett, 2015, 368(1): 7-13. |
| 13 | Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing[J]. Signal Transduct Target Ther, 2020, 5(1): 166. |
| 14 | Nakaya M, Xiao YC, Zhou XF, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation[J]. Immunity, 2014, 40(5): 692-705. |
| 15 | Masle-Farquhar E, Bröer A, Yabas M, et al. ASCT2 (SLC1A5)-deficient mice have normal B-cell development, proliferation, and antibody production[J]. Front Immunol, 2017, 8: 549. |
| 16 | Ansari RE, Craze ML, Althobiti M, et al. Enhanced glutamine uptake influences composition of immune cell infiltrates in breast cancer[J]. Br J Cancer, 2020, 122(1): 94-101. |
| 17 | Han LY, Zhou JP, Li LY, et al. SLC1A5 enhances malignant phenotypes through modulating ferroptosis status and immune microenvironment in glioma[J]. Cell Death Dis, 2022, 13(12): 1071. |
| 18 | Zheng ST, Liu T, Li L, et al. SLC1A5, unrelated to prognosis, was associated with CD8+ T-cell exclusion in the tumor micro-environment of squamous cell carcinoma[J]. Heliyon, 2023, 9(3): e14571. |
| 19 | Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors[J]. PLoS One, 2017, 12(7): e0179726. |
| 20 | Zhou R, Zhang JW, Zeng DQ, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer[J]. Cancer Immunol Immunother, 2019, 68(3): 433-42. |
| 21 | Csanadi A, Oser A, Aumann K, et al. Overexpression of SLC1a5 in lymph node metastases outperforms assessment in the primary as a negative prognosticator in non-small cell lung cancer[J]. Pathology, 2018, 50(3): 269-75. |
| 22 | Liu YD, Yang L, An HM, et al. High expression of Solute Carrier Family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma[J]. Sci Rep, 2015, 5: 16954. |
| 23 | Sun HW, Yu XJ, Wu WC, et al. GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma[J]. PLoS One, 2016, 11(12): e0168907. |
| 24 | Kaira K, Sunose Y, Arakawa K, et al. Clinicopathological significance of ASC amino acid transporter-2 expression in panc-reatic ductal carcinoma[J]. Histopathology, 2015, 66(2): 234-43. |
| 25 | Lu J, Chen M, Tao ZH, et al. Effects of targeting SLC1A5 on inhibiting gastric cancer growth and tumor development in vitro and in vivo [J]. Oncotarget, 2017, 8(44): 76458-67. |
| 26 | Liao ZX, Hsu SH, Tang SC, et al. Potential targeting of the tumor microenvironment to improve cancer virotherapy[J]. Pharmacol Ther, 2023, 250: 108521. |
| 27 | Wang CX, Zheng XC, Zhang JL, et al. CD300ld on neutrophils is required for tumour-driven immune suppression[J]. Nature, 2023, 621(7980): 830-9. |
| 28 | Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023 [J]. CA Cancer J Clin, 2023, 73(1): 17-48. |
| 29 | Xia CF, Dong XS, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants[J]. Chin Med J, 2022, 135(5): 584-90. |
| 30 | Li YM, Liu F. The extracellular vesicles targeting tumor microenvironment: a promising therapeutic strategy for melanoma[J]. Front Immunol, 2023, 14: 1200249. |
| 31 | Vitale I, Manic G, Coussens LM, et al. Macrophages and metabolism in the tumor microenvironment[J]. Cell Metab, 2019, 30(1): 36-50. |
| 32 | Guilliams M, Scott CL. Liver macrophages in health and disease[J]. Immunity, 2022, 55(9): 1515-29. |
| 33 | Chen YN, Hu MR, Wang L, et al. Macrophage M1/M2 polarization[J]. Eur J Pharmacol, 2020, 877: 173090. |
| 34 | Mantovani A, Sozzani S, Locati M, et al. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes[J]. Trends Immunol, 2002, 23(11): 549-55. |
| [1] | Ting XIE, Yunyun WANG, Ting GUO, Chunhua YUAN. The peptide toxin components and nucleotide metabolites in Macrothele raveni venom synergistically inhibit cancer cell proliferation by activating the pro-apoptotic pathways [J]. Journal of Southern Medical University, 2025, 45(7): 1460-1470. |
| [2] | Yumei ZENG, Jike LI, Zhongxi HUANG, Yibo ZHOU. Villin-like protein VILL suppresses proliferation of nasopharyngeal carcinoma cells by interacting with LMO7 protein [J]. Journal of Southern Medical University, 2025, 45(5): 954-961. |
| [3] | Tianli SONG, Yimin WANG, Tong SUN, Xu LIU, Sheng HUANG, Yun RAN. Zheng Gan Decoction inhibits diethylnitrosamine-induced hepatocellular carcinoma in rats by activating the Hippo/YAP signaling pathway [J]. Journal of Southern Medical University, 2025, 45(4): 799-809. |
| [4] | Yaqing YUE, Zhaoxia MU, Xibo WANG, Yan LIU. Aurora-A overexpression promotes cervical cancer cell invasion and metastasis by activating the NF-κBp65/ARPC4 signaling axis [J]. Journal of Southern Medical University, 2025, 45(4): 837-843. |
| [5] | Yi ZHANG, Yu SHEN, Zhiqiang WAN, Song TAO, Yakui LIU, Shuanhu WANG. High expression of CDKN3 promotes migration and invasion of gastric cancer cells by regulating the p53/NF-κB signaling pathway and inhibiting cell apoptosis [J]. Journal of Southern Medical University, 2025, 45(4): 853-861. |
| [6] | Shunjie QING, Zhiyong SHEN. High expression of hexokinase 2 promotes proliferation, migration and invasion of colorectal cancer cells by activating the JAK/STAT pathway and regulating tumor immune microenvironment [J]. Journal of Southern Medical University, 2025, 45(3): 542-553. |
| [7] | Qingqing HUANG, Wenjing ZHANG, Xiaofeng ZHANG, Lian WANG, Xue SONG, Zhijun GENG, Lugen ZUO, Yueyue WANG, Jing LI, Jianguo HU. High MYO1B expression promotes proliferation, migration and invasion of gastric cancer cells and is associated with poor patient prognosis [J]. Journal of Southern Medical University, 2025, 45(3): 622-631. |
| [8] | Yu BIN, Ziwen LI, Suwei ZUO, Sinuo SUN, Min LI, Jiayin SONG, Xu LIN, Gang XUE, Jingfang WU. High expression of apolipoprotein C1 promotes proliferation and inhibits apoptosis of papillary thyroid carcinoma cells by activating the JAK2/STAT3 signaling pathway [J]. Journal of Southern Medical University, 2025, 45(2): 359-370. |
| [9] | Zhoufang CAO, Yuan WANG, Mengna WANG, Yue SUN, Feifei LIU. LINC00837/miR-671-5p/SERPINE2 functional axis promotes pathological processes of fibroblast-like synovial cells in rheumatoid arthritis [J]. Journal of Southern Medical University, 2025, 45(2): 371-378. |
| [10] | Yaobin WANG, Liuyan CHEN, Yiling LUO, Jiqing SHEN, Sufang ZHOU. Predictive value of NUF2 for prognosis and immunotherapy responses in pan-cancer [J]. Journal of Southern Medical University, 2025, 45(1): 137-149. |
| [11] | Meng XU, Lina CHEN, Jinyu WU, Lili LIU, Mei SHI, Hao ZHOU, Guoliang ZHANG. Mechanism of Hedyotis diffusa-Scutellaria barbata D. Don for treatment of primary liver cancer: analysis with network pharmacology, molecular docking and in vitro validation [J]. Journal of Southern Medical University, 2025, 45(1): 80-89. |
| [12] | Xiaohua CHEN, Hui LU, Ziliang WANG, Lian WANG, Yongsheng XIA, Zhijun GENG, Xiaofeng ZHANG, Xue SONG, Yueyue WANG, Jing LI, Jianguo HU, Lugen ZUO. Role of Abelson interactor 2 in progression and prognosis of gastric cancer and its regulatory mechanisms [J]. Journal of Southern Medical University, 2024, 44(9): 1653-1661. |
| [13] | Liangjun XUE, Qiuyu TAN, Jingwen XU, Lu FENG, Wenjin LI, Liang YAN, Yulei LI. MiR-6838-5p overexpression inhibits proliferation of breast cancer MCF-7 cells by downregulating DDR1 expression [J]. Journal of Southern Medical University, 2024, 44(9): 1677-1684. |
| [14] | Liping ZHANG, Xijuan LIU, Xiao HU, Jiali WANG, Xihe YU, Guoliang LI, Haimin YOU, Qizhou ZHANG, Haibo ZHANG. Efficacy and safety of transcatheter arterial chemoembolization followed by hepatic arterial infusion chemotherapy combined with TKI and PD-1 inhibitors as first-line treatment for advanced hepatocellular carcinoma [J]. Journal of Southern Medical University, 2024, 44(9): 1831-1838. |
| [15] | Kai JI, Guanyu YU, Leqi ZHOU, Tianshuai ZHANG, Qianlong LING, Wenjiang MAN, Bing ZHU, Wei ZHANG. HNRNPA1 gene is highly expressed in colorectal cancer: its prognostic implications and potential as a therapeutic target [J]. Journal of Southern Medical University, 2024, 44(9): 1685-1695. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||