Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (5): 827-840.doi: 10.12122/j.issn.1673-4254.2024.05.04
• Clinical Research • Previous Articles Next Articles
Pengcheng LIU( ), Lijuan LOU, Xia LIU, Jian WANG, Ying JIANG(
), Lijuan LOU, Xia LIU, Jian WANG, Ying JIANG( )
)
Received:2024-03-10
Online:2024-05-20
Published:2024-06-04
Contact:
Ying JIANG
E-mail:pc18782995020@163.com;jiangying@ncpsb.org.cn
Supported by:Pengcheng LIU, Lijuan LOU, Xia LIU, Jian WANG, Ying JIANG. A risk scoring model based on M2 macrophage-related genes for predicting prognosis of HBV-related hepatocellular carcinoma[J]. Journal of Southern Medical University, 2024, 44(5): 827-840.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.05.04
| Item | Overall | GSE10140 | TCGA-LIHC |
|---|---|---|---|
| n | 155 | 82 | 73 |
| Futime (median [IQR]) | 1613.00 [630.50, 2871.75] | 2854.00 [1368.00, 3662.50] | 682.50 [437.00, 1632.50] |
| Fustat =1 [n (%)] | 48 (30.9) | 32 (39.0) | 16 (21.9) |
| EGF [n (%)] | |||
| High | 9 (5.8) | 9 (11.0) | 0 (0.0) |
| Low | 73 (47.1) | 73 (89.0) | 0 (0.0) |
| NA | 73 (47.1) | 0 (0.0) | 73 (100.0) |
| Age (year, median [IQR]) | 52.00 [46.00, 62.75] | NA [NA, NA] | 52.00 [46.00, 62.75] |
| Gender [n (%)] | |||
| Female | 15 (9.7) | 0 (0.0) | 15 (20.5) |
| Male | 58 (37.4) | 0 (0.0) | 58 (79.5) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Grade (%) | |||
| G1 | 6 (3.8) | 0 (0.0) | 6 (8.2) |
| G2 | 27 (17.4) | 0 (0.0) | 27 (37.0) |
| G3 | 34 (21.9) | 0 (0.0) | 34 (46.6) |
| G4 | 6 (3.9) | 0 (0.0) | 6 (8.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Stage [n (%)] | |||
| Stage I | 49 (31.6) | 0 (0.0) | 49 (67.1) |
| Stage II | 15 (9.8) | 0 (0.0) | 15 (20.5) |
| Stage III | 5 (3.2) | 0 (0.0) | 5 (6.8) |
| Stage IV | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| Unknow | 3 (1.8) | 0 (0.0) | 3 (4.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 |
| T stage [n (%)] | |||
| T1 | 52(33.5) | 0 (0.0) | 52 (71.3) |
| T2 | 15 (9.8) | 0 (0.0) | 15 (20.5) |
| T3 | 5 (3.2) | 0 (0.0) | 5 (6.8) |
| T4 | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Metastasis [n (%)] | |||
| M0 | 70 (45.3) | 0 (0.0) | 70 (95.8) |
| M1 | 2 (1.2) | 0 (0.0) | 2 (2.8) |
| MX | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Nodes [n (%)] | |||
| N0 | 70 (45.3) | 0 (0.0) | 70 (95.8) |
| NX | 3 (1.8) | 0 (0.0) | 3 (4.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
Tab.1 Baseline characteristics of HCC patients in TCGA dataset and GEO dataset
| Item | Overall | GSE10140 | TCGA-LIHC |
|---|---|---|---|
| n | 155 | 82 | 73 |
| Futime (median [IQR]) | 1613.00 [630.50, 2871.75] | 2854.00 [1368.00, 3662.50] | 682.50 [437.00, 1632.50] |
| Fustat =1 [n (%)] | 48 (30.9) | 32 (39.0) | 16 (21.9) |
| EGF [n (%)] | |||
| High | 9 (5.8) | 9 (11.0) | 0 (0.0) |
| Low | 73 (47.1) | 73 (89.0) | 0 (0.0) |
| NA | 73 (47.1) | 0 (0.0) | 73 (100.0) |
| Age (year, median [IQR]) | 52.00 [46.00, 62.75] | NA [NA, NA] | 52.00 [46.00, 62.75] |
| Gender [n (%)] | |||
| Female | 15 (9.7) | 0 (0.0) | 15 (20.5) |
| Male | 58 (37.4) | 0 (0.0) | 58 (79.5) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Grade (%) | |||
| G1 | 6 (3.8) | 0 (0.0) | 6 (8.2) |
| G2 | 27 (17.4) | 0 (0.0) | 27 (37.0) |
| G3 | 34 (21.9) | 0 (0.0) | 34 (46.6) |
| G4 | 6 (3.9) | 0 (0.0) | 6 (8.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Stage [n (%)] | |||
| Stage I | 49 (31.6) | 0 (0.0) | 49 (67.1) |
| Stage II | 15 (9.8) | 0 (0.0) | 15 (20.5) |
| Stage III | 5 (3.2) | 0 (0.0) | 5 (6.8) |
| Stage IV | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| Unknow | 3 (1.8) | 0 (0.0) | 3 (4.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 |
| T stage [n (%)] | |||
| T1 | 52(33.5) | 0 (0.0) | 52 (71.3) |
| T2 | 15 (9.8) | 0 (0.0) | 15 (20.5) |
| T3 | 5 (3.2) | 0 (0.0) | 5 (6.8) |
| T4 | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Metastasis [n (%)] | |||
| M0 | 70 (45.3) | 0 (0.0) | 70 (95.8) |
| M1 | 2 (1.2) | 0 (0.0) | 2 (2.8) |
| MX | 1 (0.6) | 0 (0.0) | 1 (1.4) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| Nodes [n (%)] | |||
| N0 | 70 (45.3) | 0 (0.0) | 70 (95.8) |
| NX | 3 (1.8) | 0 (0.0) | 3 (4.2) |
| NA | 82 (52.9) | 82 (100.0) | 0 (0.0) |
| MRG | Forword primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| GAPDH | AGATCCCTCCAAAATCAAGTGG | GGCAGAGATGATGACCCTTTT |
| VTN | AAGCCCCAAGTGACTCGC | TTTTCTCCTCGCCATCGTCA |
| GCLC | ACTTCATTTCCCAGTACCTTAACA | GCAGCACTCAAAGCCATAACA |
| GMPR | GGGCCACATCATCTCTGATGGA | TCAGTCCCCCGAGAATATCCAG |
| PARVB | GGTTCACTTCTCCCTGGCTC | CGCTCCTCGTTCTCCTCAAG |
| TRIM27 | AGCATGAGTATCGCCTCCTG | CTGATTCTTTCAGCCCTGCTC |
Tab.2 RT-qPCR primer sequences
| MRG | Forword primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| GAPDH | AGATCCCTCCAAAATCAAGTGG | GGCAGAGATGATGACCCTTTT |
| VTN | AAGCCCCAAGTGACTCGC | TTTTCTCCTCGCCATCGTCA |
| GCLC | ACTTCATTTCCCAGTACCTTAACA | GCAGCACTCAAAGCCATAACA |
| GMPR | GGGCCACATCATCTCTGATGGA | TCAGTCCCCCGAGAATATCCAG |
| PARVB | GGTTCACTTCTCCCTGGCTC | CGCTCCTCGTTCTCCTCAAG |
| TRIM27 | AGCATGAGTATCGCCTCCTG | CTGATTCTTTCAGCCCTGCTC |
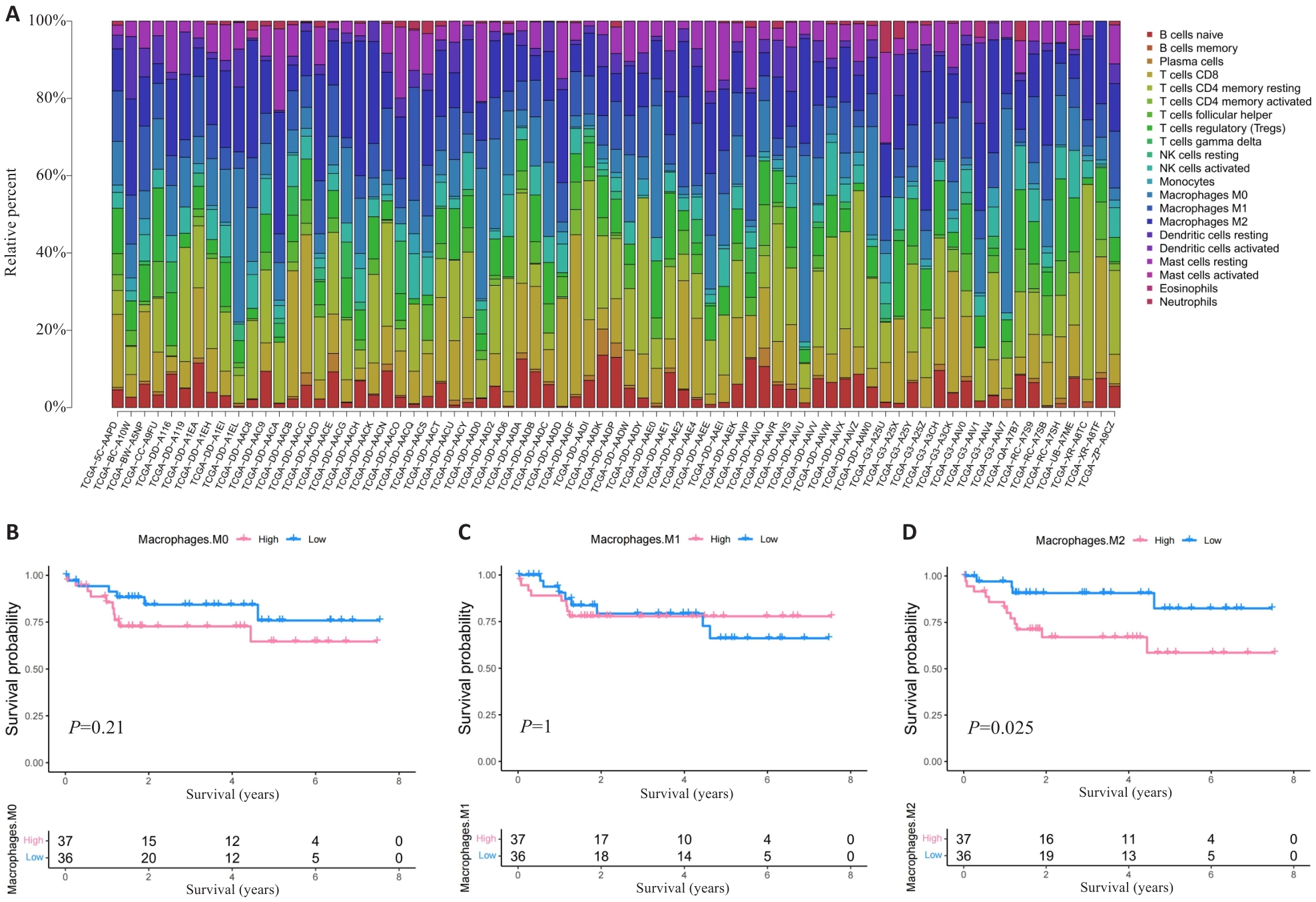
Fig.1 HCC patients with high M2 macrophage infiltration level have poor prognosis. A: Proportion of immune cells in HBV-related HCC patients. B-D: Prognosis of patients stratified by the number of M0 macrophage (B), M1 macrophage (C) and M2 macrophage (D).
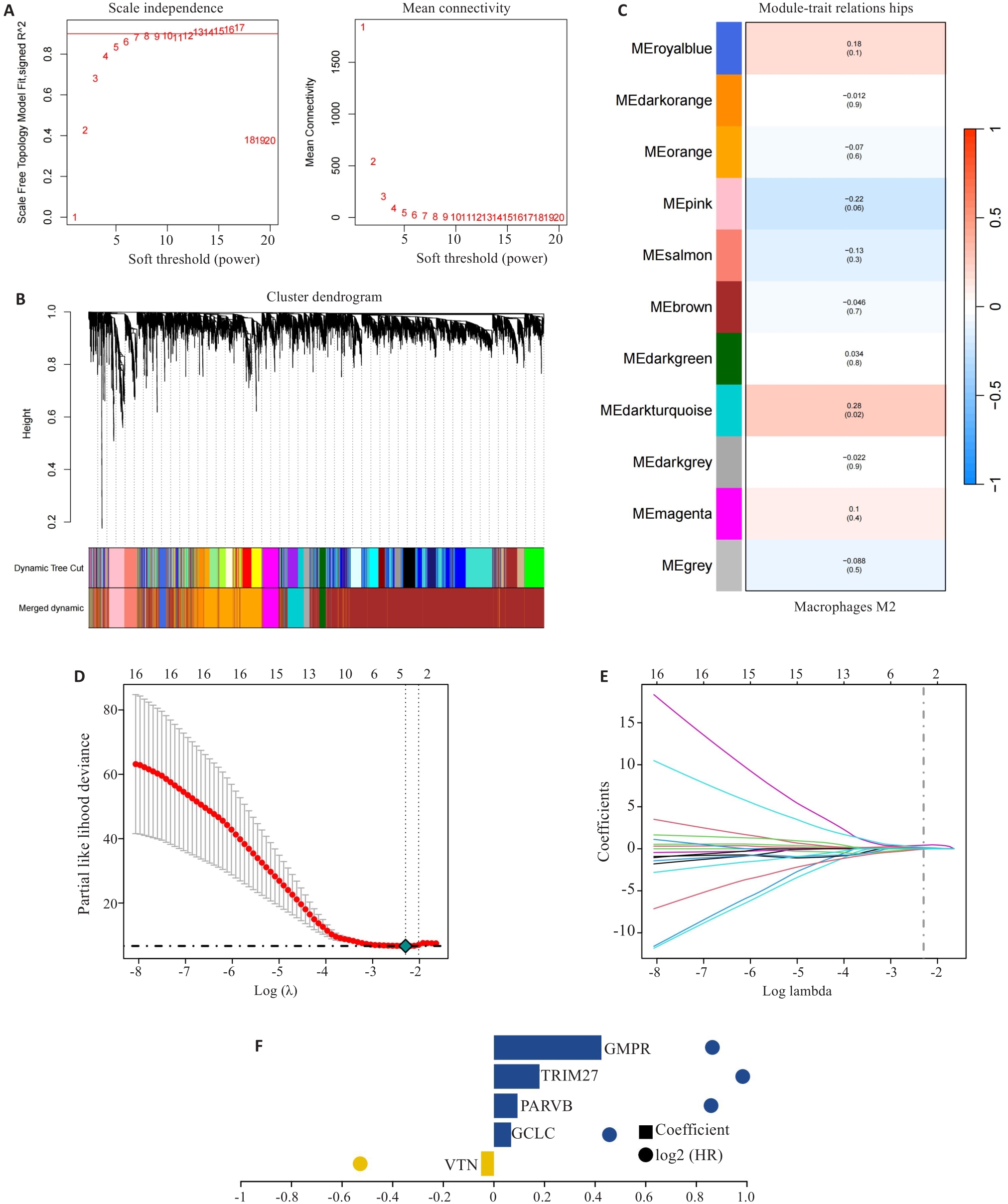
Fig.2 Construction of prognostic risk scoring model. A-C: Identification of M2 macrophage-related gene (MRG) modules by WGCNA. D-F: Five hub MRGs (VTN, GCLC, PARVB, TRIM27 and GMPR) identified by LASSO regression analysis for constructing the risk scoring model.
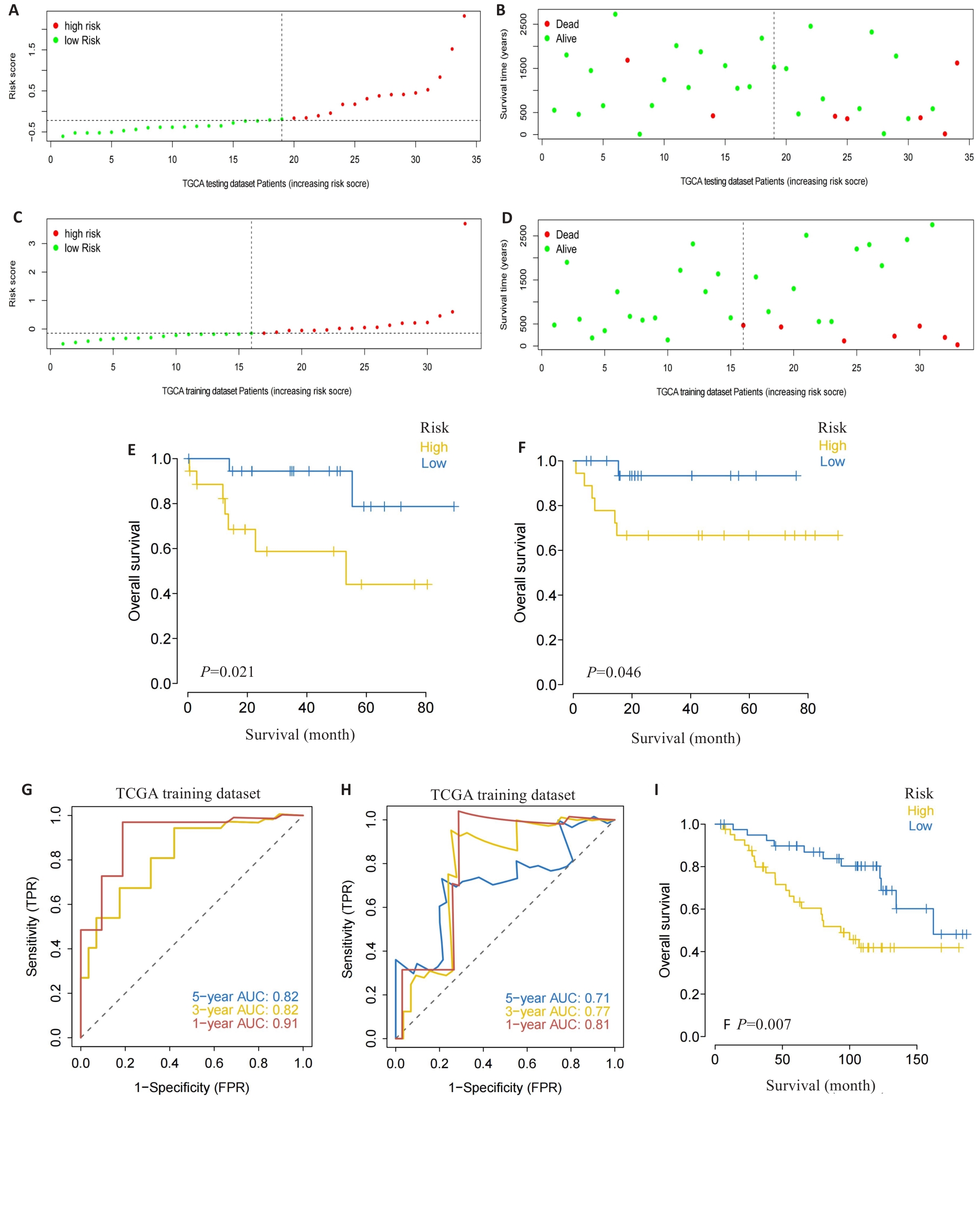
Fig.3 Validation of the risk scoring model. A-D: Different patterns of survival status and survival time between the high-risk group (A, B) and low-risk group (C, D). E, F: Validation of the MRG prognostic model in the training dataset (E) and testing dataset (F). G, H: ROC curves of the risk score for predicting overall survival (OS) in the training dataset (G) and testing dataset (H). I: Validation of the MRG prognostic model using the external dataset.
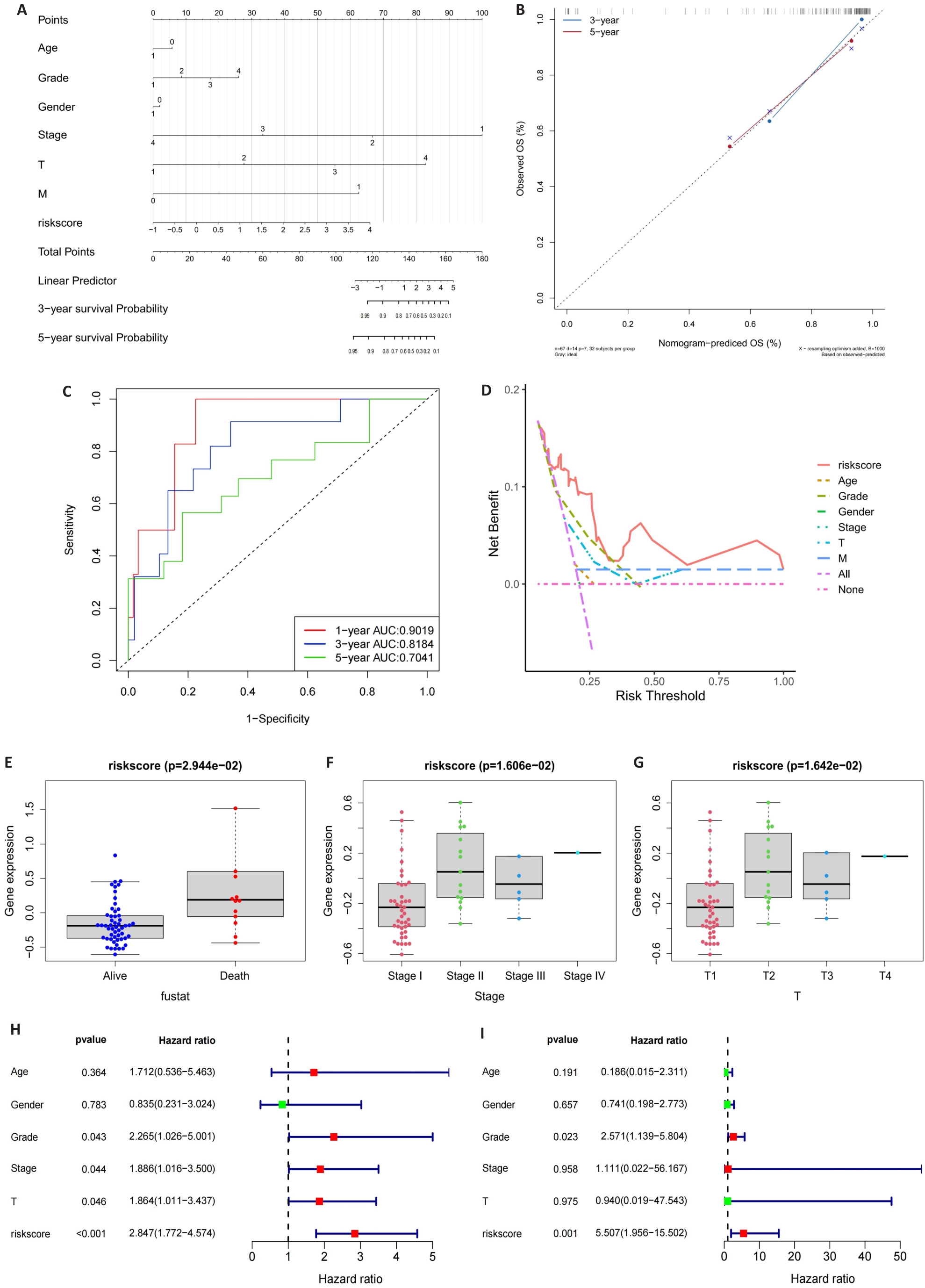
Fig.4 Construction of the nomogram. A: Nomogram of the risk scores and clinical characteristics. B: Calibration curves for evaluating OS predictions at 3 and 5 years. C, D: ROC and DCA curves for determining the accuracy of the nomogram for OS at 1, 3 and 5 years, respectively. E-G: Correlation of the risk scores with clinical stages. H, I: Univariate analysis and multivariate analysis for validating the independent prognostic value of the risk scores.
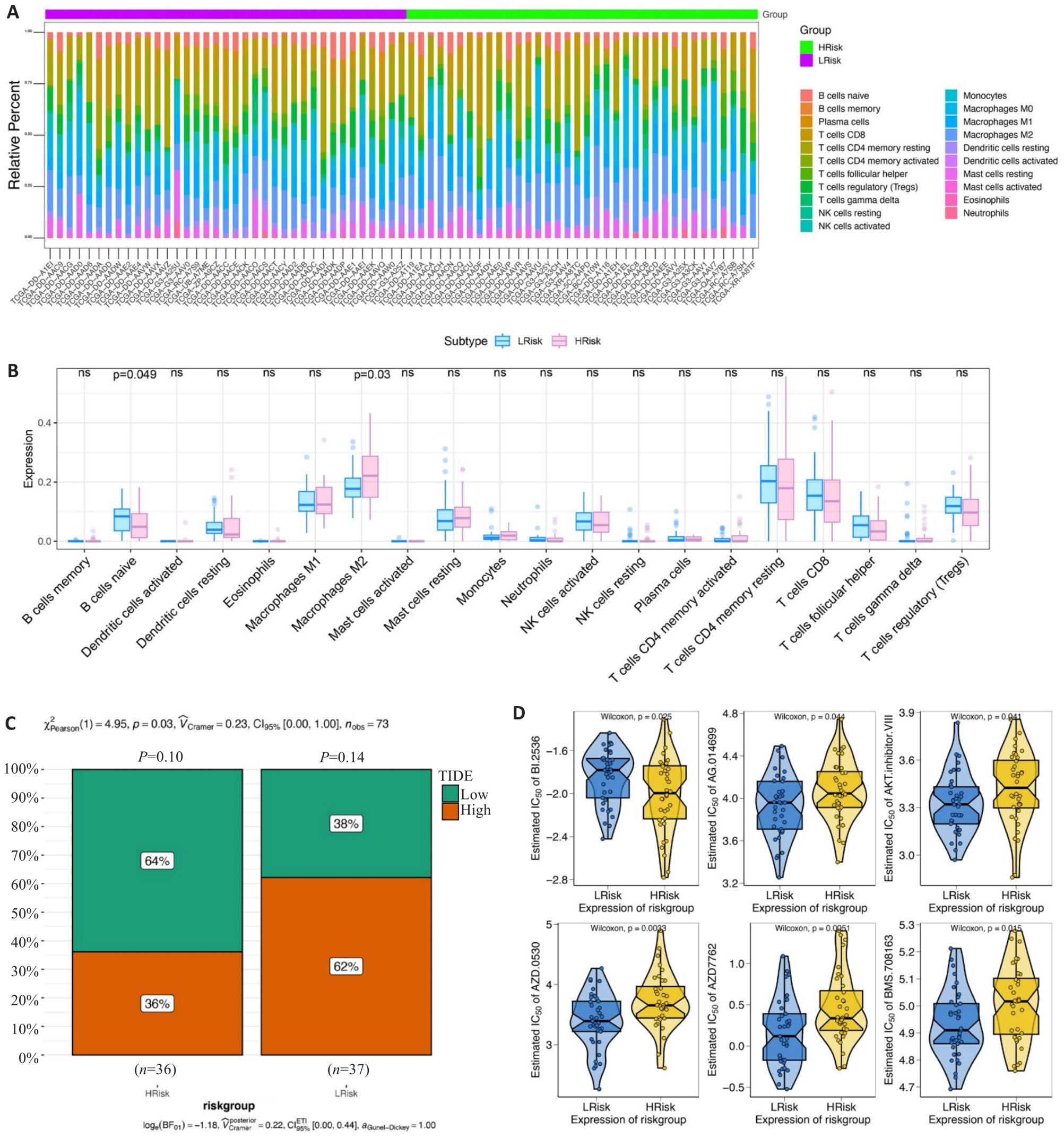
Fig.5 Clinical predictive value of the risk scoring model. A, B: Infiltrating level of immune cells in the high- and low-risk groups. C: Sensitivity of anti-tumor immunotherapy in the high- and low-risk groups. D: IC50 values of common chemotherapy drugs.
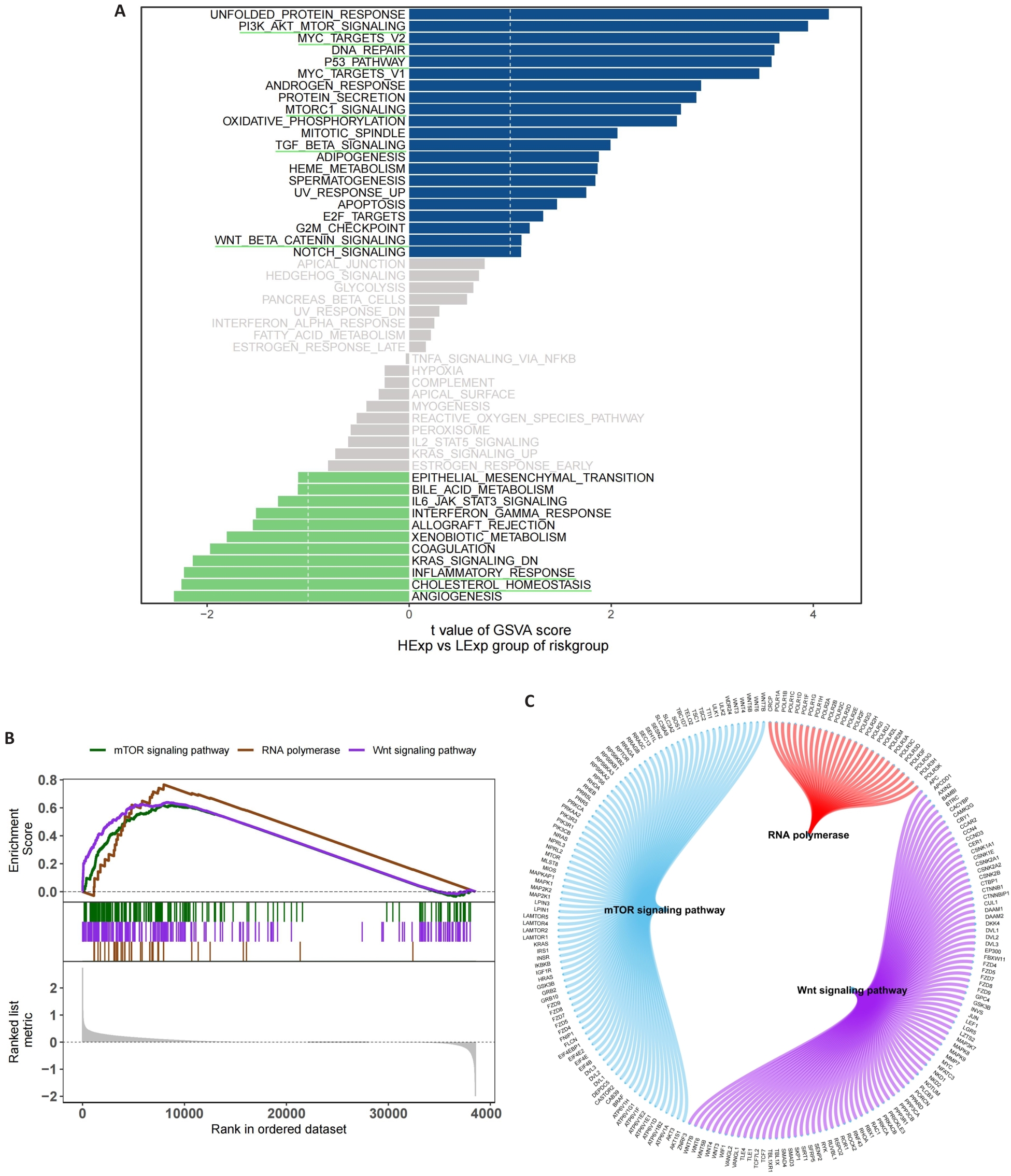
Fig.6 Functional enrichment analysis of signaling pathways related to the risk scoring model. A: GSVA in the high- and low-risk groups. B: GSEA in the high- and low-risk groups. C: Molecular interaction networks between the pathways.
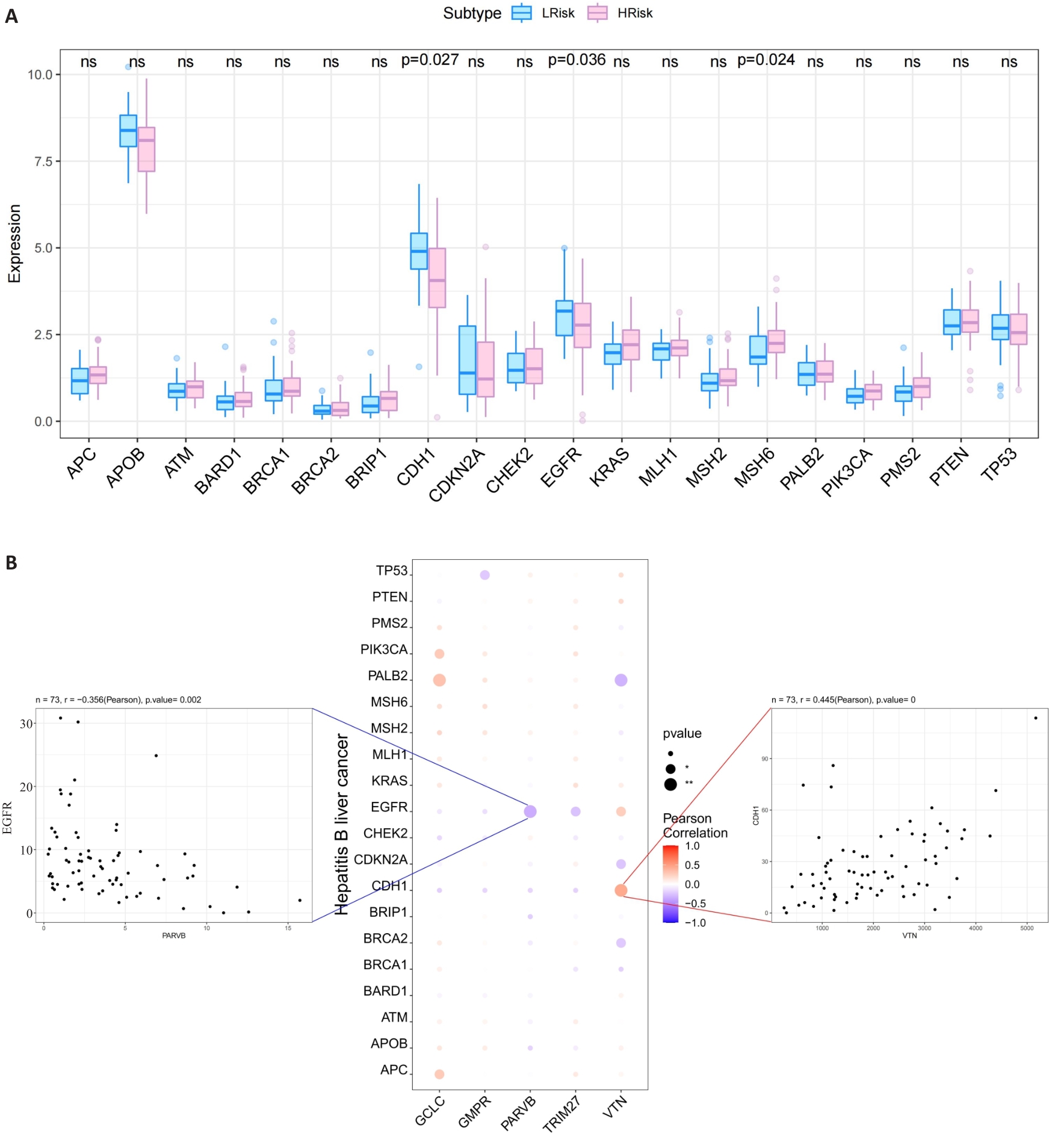
Fig.7 Relationship between the 5 hub MRGs and HCC pathogenic genes. A: Relationship between the hub genes and the top 20 HCC pathogenic genes. B: A bubble plot illustrating the Pearson correlation between 5 hub MRGs (VTN, GCLC, PARVB, TRIM27 and GMPR) and the top 20 HCC pathogenic genes.
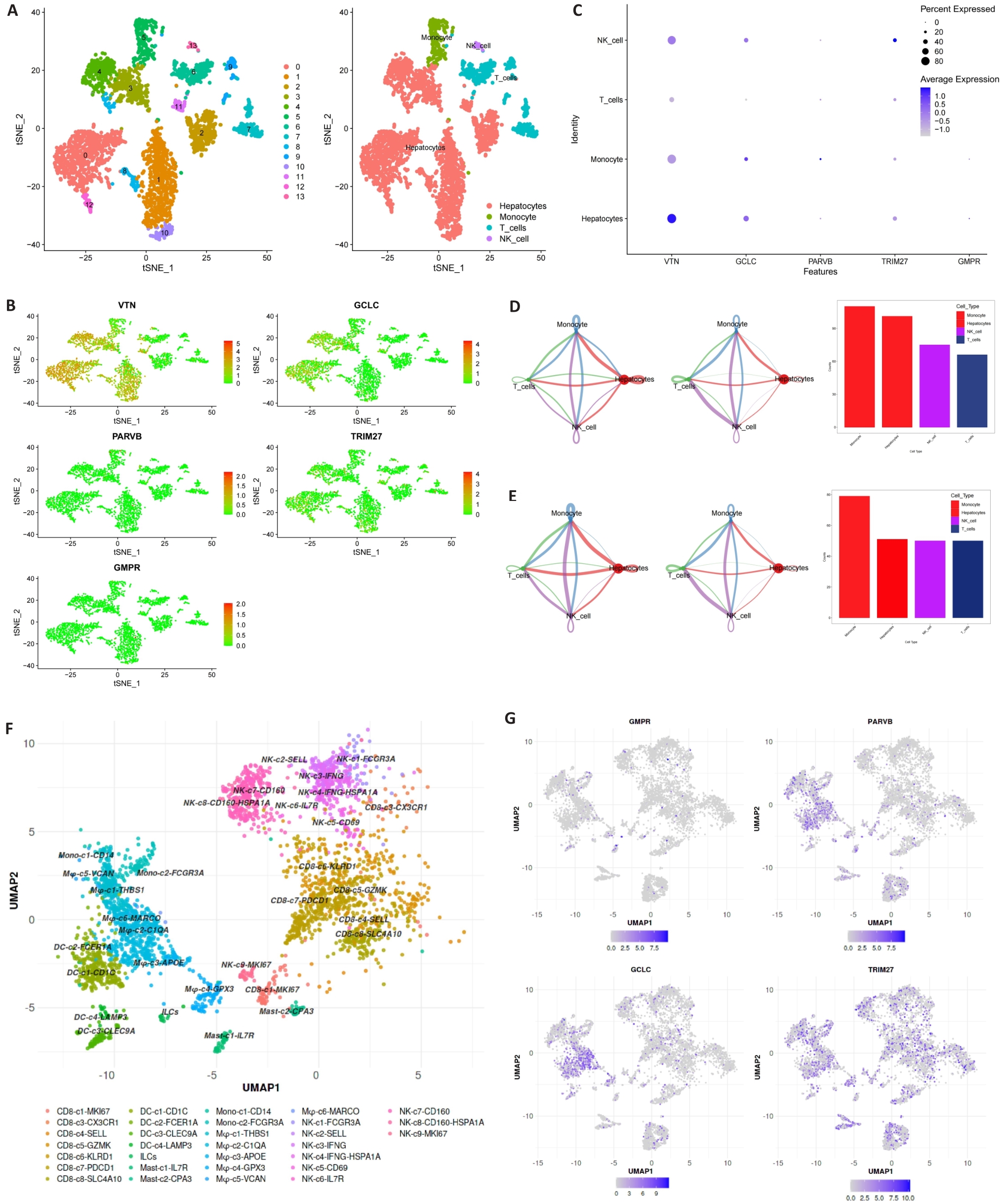
Fig.8 Expression of MRGs in different cell types. A-C: 5 hub MRGs are mainly expressed in hepatocytes, monocyte, T cells and NK cell. D, E: Cell chat intensity in high- and low-risk groups. F: UMAP projection showing the immune landscape of HCC, colored by cluster. G: Expressions of GCLC, PARVB, GMPR and TRIM27 in HCC immune microenvironment.
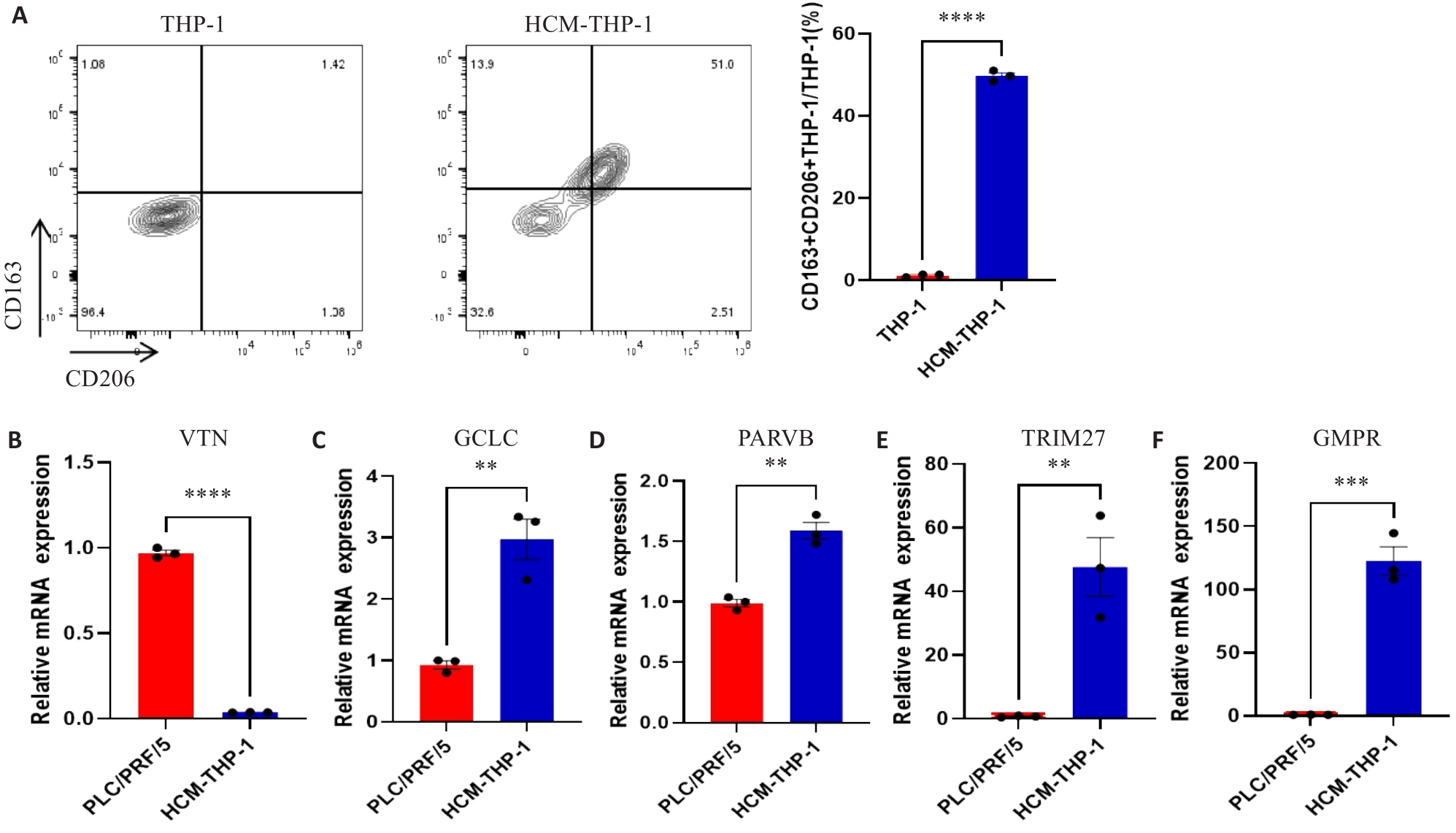
Fig.9 Expressions of MRGs in HCC cell lines and macrophages. A: Expression of CD163 and CD206 on THP-1 cells detected by flow cytometry. B-F: Expression of MRGs in PLC/PRF/5 and HCM-THP-1 cells. **P<0.01, ***P<0.001, ****P<0.0001.
| 1 | Craig AJ, von Felden J, Garcia-Lezana T, et al. Tumour evolution in hepatocellular carcinoma[J]. Nat Rev Gastroenterol Hepatol, 2020, 17(3): 139-52. DOI: 10.1038/s41575-019-0229-4 |
| 2 | Vogel A, Meyer T, Sapisochin G, et al. Hepatocellular carcinoma [J]. Lancet, 2022, 400(10360): 1345-62. DOI: 10.1016/s0140-6736(22)01200-4 |
| 3 | Koga H, Iwamoto H, Suzuki H, et al. Clinical practice guidelines and real-life practice in hepatocellular carcinoma: a Japanese perspective[J]. Clin Mol Hepatol, 2023, 29(2): 242-51. DOI: 10.3350/cmh.2023.0102 |
| 4 | Bruix J, Sherman M, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update[J]. Hepatology, 2011, 53(3): 1020-2. DOI: 10.1002/hep.24199 |
| 5 | Prospective validation of the CLIP score: a new prognostic system for patients with cirrhosis and hepatocellular carcinoma[J]. Hepatology, 2000, 31(4): 840-5. DOI: 10.1053/he.2000.5628 |
| 6 | Chevret S, Trinchet JC, Mathieu D, et al. A new prognostic classification for predicting survival in patients with hepatocellular carcinoma. Groupe d’Etude et de Traitement du Carcinome Hépatocellulaire[J]. J Hepatol, 1999, 31(1): 133-41. DOI: 10.1016/s0168-8278(99)80173-1 |
| 7 | Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification[J]. Semin Liver Dis, 1999, 19(3): 329-38. DOI: 10.1055/s-2007-1007122 |
| 8 | Yau T, Tang VYF, Yao TJ, et al. Development of Hong Kong Liver Cancer staging system with treatment stratification for patients with hepatocellular carcinoma[J]. Gastroenterology, 2014, 146(7): 1691-700.e3. DOI: 10.1053/j.gastro.2014.02.032 |
| 9 | Primack A, Vogel CL, Kyalwazi SK, et al. A staging system for hepatocellular carcinoma: prognostic factors in Ugandan patients[J]. Cancer, 1975, 35(5): 1357-64. DOI: 10.1002/1097-0142(197505)35:5<1357::aid-cncr2820350518>3.0.co;2-8 |
| 10 | Kamel HFM, Al-Amodi HSAB. Exploitation of gene expression and cancer biomarkers in paving the path to era of personalized medicine[J]. Genomics Proteomics Bioinformatics, 2017, 15(4): 220-35. DOI: 10.1016/j.gpb.2016.11.005 |
| 11 | Ahluwalia P, Kolhe R, Gahlay GK. The clinical relevance of gene expression based prognostic signatures in colorectal cancer[J]. Biochim Biophys Acta Rev Cancer, 2021, 1875(2): 188513. DOI: 10.1016/j.bbcan.2021.188513 |
| 12 | Ye ZM, Zou SM, Niu ZY, et al. A novel risk model based on lipid metabolism-associated genes predicts prognosis and indicates immune microenvironment in breast cancer[J]. Front Cell Dev Biol, 2021, 9: 691676. DOI: 10.3389/fcell.2021.691676 |
| 13 | Bussard KM, Mutkus L, Stumpf K, et al. Tumor-associated stromal cells as key contributors to the tumor microenvironment[J]. Breast Cancer Res, 2016, 18(1): 84. DOI: 10.1186/s13058-016-0740-2 |
| 14 | Jensen C, Nissen NI, Von Arenstorff CS, et al. Serological assessment of collagen fragments and tumor fibrosis may guide immune checkpoint inhibitor therapy[J]. J Exp Clin Cancer Res, 2021, 40(1): 326. DOI: 10.1186/s13046-021-02133-z |
| 15 | de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth[J]. Cancer Cell, 2023, 41(3): 374-403. DOI: 10.1016/j.ccell.2023.02.016 |
| 16 | Sherman MH, Beatty GL. Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance[J]. Annu Rev Pathol, 2023, 18: 123-48. DOI: 10.1146/annurev-pathmechdis-031621-024600 |
| 17 | Liu YD, Li CF, Lu YP, et al. Tumor microenvironment-mediated immune tolerance in development and treatment of gastric cancer[J]. Front Immunol, 2022, 13: 1016817. DOI: 10.3389/fimmu.2022.1016817 |
| 18 | Donne R, Lujambio A. The liver cancer immune microenvironment: therapeutic implications for hepatocellular carcinoma[J]. Hepatology, 2023, 77(5): 1773-96. DOI: 10.1002/hep.32740 |
| 19 | Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment[J]. Cancer Cell, 2012, 21(3): 309-22. DOI: 10.1016/j.ccr.2012.02.022 |
| 20 | Zhou DX, Luan JJ, Huang C, et al. Tumor-associated macrophages in hepatocellular carcinoma: friend or foe[J]? Gut Liver, 2021, 15(4): 500-16. DOI: 10.5009/gnl20223 |
| 21 | Pan YY, Yu YD, Wang XJ, et al. Tumor-associated macrophages in tumor immunity[J]. Front Immunol, 2020, 11: 583084. DOI: 10.3389/fimmu.2020.583084 |
| 22 | Zhang QM, He Y, Luo N, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma[J]. Cell, 2019, 179(4): 829-45.e20. DOI: 10.1016/j.cell.2019.10.003 |
| 23 | Yeung OWH, Lo CM, Ling CC, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma[J]. J Hepatol, 2015, 62(3): 607-16. DOI: 10.1016/j.jhep.2014.10.029 |
| 24 | Nosaka T, Murata Y, Takahashi K, et al. Hepatocellular carcinoma progression promoted by 5-lipoxygenase activity in CD163+ tumor-associated macrophages[J]. Biomed Pharmacother, 2023, 162: 114592. DOI: 10.1016/j.biopha.2023.114592 |
| 25 | Song HQ, Wang XX, Zhang C, et al. Construction of an M2 macrophage-related prognostic model in hepatocellular carcinoma[J]. Front Oncol, 2023, 13: 1170775. DOI: 10.3389/fonc.2023.1170775 |
| 26 | Chen BB, Khodadoust MS, Liu CL, et al. Profiling tumor infiltrating immune cells with CIBERSORT[J]. Methods Mol Biol, 2018, 1711: 243-59. DOI: 10.1007/978-1-4939-7493-1_12 |
| 27 | Shin SW, Ahn KS, Kim SW, et al. Liver resection versus local ablation therapies for hepatocellular carcinoma within the Milan criteria: a systematic review and meta-analysis[J]. Ann Surg, 2021, 273(4): 656-66. DOI: 10.1097/sla.0000000000004350 |
| 28 | Jiang P, Gu SQ, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response[J]. Nat Med, 2018, 24(10): 1550-8. DOI: 10.1038/s41591-018-0136-1 |
| 29 | Sun JL, Zhou CH, Ma QN, et al. High GCLC level in tumor tissues is associated with poor prognosis of hepatocellular carcinoma after curative resection[J]. J Cancer, 2019, 10(15): 3333-43. DOI: 10.7150/jca.29769 |
| 30 | Bianchi-Smiraglia A, Bagati A, Fink EE, et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools[J]. Oncogene, 2017, 36(1): 84-96. DOI: 10.1038/onc.2016.178 |
| 31 | Ng L, Tung-Ping Poon R, Yau S, et al. Suppression of actopaxin impairs hepatocellular carcinoma metastasis through modulation of cell migration and invasion[J]. Hepatology, 2013, 58(2): 667-79. DOI: 10.1002/hep.26396 |
| 32 | Wang HC, Zhang Y, Yan L, et al. Analysis of TRIM27 prognosis value and immune infiltrates in hepatocellular carcinoma[J]. Int J Immunopathol Pharmacol, 2022, 36: 3946320221132986. DOI: 10.1177/03946320221132986 |
| 33 | Casanova-Acebes M, Dalla E, Leader AM, et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells[J]. Nature, 2021, 595(7868): 578-84. DOI: 10.1038/s41586-021-03651-8 |
| 34 | Zhang L, Li ZY, Skrzypczynska KM, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer[J]. Cell, 2020, 181(2): 442-59.e29. DOI: 10.1016/j.cell.2020.03.048 |
| 35 | Zheng LT, Qin SS, Si W, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells[J]. Science, 2021, 374(6574): abe6474. DOI: 10.1126/science.abe6474 |
| 36 | Liu CL, Yang MX, Zhang DZ, et al. Clinical cancer immunotherapy: current progress and prospects[J]. Front Immunol, 2022, 13: 961805. DOI: 10.3389/fimmu.2022.961805 |
| 37 | Liu Y, Xun ZZ, Ma K, et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy[J]. J Hepatol, 2023, 78(4): 770-82. DOI: 10.1016/j.jhep.2023.01.011 |
| 38 | Wei CY, Zhu MX, Zhang PF, et al. PKCα/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti-PD1 resistance in hepatocellular carcinoma[J]. J Hepatol, 2022, 77(1): 163-76. DOI: 10.1016/j.jhep.2022.02.019 |
| 39 | Yang Y, Ye YC, Chen Y, et al. Crosstalk between hepatic tumor cells and macrophages via Wnt/β‑catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors[J]. Cell Death Dis, 2018, 9(8): 793. DOI: 10.1038/s41419-018-0818-0 |
| 40 | Lian GH, Chen SJ, Ouyang M, et al. Colon cancer cell secretes EGF to promote M2 polarization of TAM through EGFR/PI3K/AKT/mTOR pathway[J]. Technol Cancer Res Treat, 2019, 18: 1533033819849068. DOI: 10.1177/1533033819849068 |
| [1] | GAO Zhiqiang, LIN Jie, HONG Peng, HU Zaihong, DONG Junjun, SHI Qinlin, TIAN Xiaomao, LIU Feng, WEI Guanghui. Identification of key genes in Wilms tumor based on high- throughput RNA sequencing and their impacts on prognosis and immune responses [J]. Journal of Southern Medical University, 2024, 44(4): 727-738. |
| [2] | CHEN Hao, LI Zhenhan, WANG Mingting, LU Linming, TANG Qianli, LUO Liangping. High expression of UBE2S promotes progression of hepatocellular carcinoma by increasing cancer cell stemness [J]. Journal of Southern Medical University, 2024, 44(3): 455-464. |
| [3] | ZHONG Weixiong, LIANG Fangrong, YANG Ruimeng, ZHEN Xin. Prediction of microvascular invasion in hepatocellular carcinoma based on multi-phase dynamic enhanced CT radiomics feature and multi-classifier hierarchical fusion model [J]. Journal of Southern Medical University, 2024, 44(2): 260-269. |
| [4] | ZHU Quan, HUANG Baisheng, WEI Leiyan, LUO Qizhi. Overexpression of LncRNA MEG3 promotes ferroptosis and enhances chemotherapy sensitivity of hepatocellular carcinoma cells to cisplatin [J]. Journal of Southern Medical University, 2024, 44(1): 17-24. |
| [5] | CAO Danping, CAI Juan, LI Yanna, DONG Runyu, WANG Zhixiong, ZUO Xueliang. TMEM64 is highly expressed in hepatocellular carcinoma and promotes tumor cell proliferation and invasion [J]. Journal of Southern Medical University, 2023, 43(8): 1345-1355. |
| [6] | LAN Yu, WANG Kaifeng, LAN Zhixian, ZHOU Heqi, SUN Jian. Dealcoholized red wine inhibits occurrence and progression of hepatocellular carcinoma possibly by inducing cell cycle arrest and apoptosis [J]. Journal of Southern Medical University, 2023, 43(8): 1297-1305. |
| [7] | FAN Yifan, FENG Zhiwei, FAN Kuohai, YIN wei, SUN Na, SUN Panpan, SUN Yaogui, LI Hongquan. Procine recombinant NK-lysin inhibits hepatocellular carcinoma metastasis by downregulating FKBP3 and inhibiting oxidative phosphorylation and glycolysis: a proteomic analysis [J]. Journal of Southern Medical University, 2023, 43(7): 1116-1126. |
| [8] | GONG Gao, CAO Shi, XIAO Hui, FANG Weiyang, QUE Yuqing, LIU Ziwei, CHEN Chaomin. Prediction of microvascular invasion in hepatocellular carcinoma with magnetic resonance imaging using models combining deep attention mechanism with clinical features [J]. Journal of Southern Medical University, 2023, 43(5): 839-851. |
| [9] | XIE Siyu, LI Miaosheng, JIANG Fengle, YI Qian, YANG Wei. EHHADH is a key gene in fatty acid metabolism pathways in hepatocellular carcinoma: a transcriptomic analysis [J]. Journal of Southern Medical University, 2023, 43(5): 680-693. |
| [10] | SU Lili, LIANG Wanqing, LÜ Zhenyu, HAN Xiao. PLXNA1 is highly expressed in hepatocellular carcinoma and affects patients' survival and immune microenvironment [J]. Journal of Southern Medical University, 2023, 43(11): 1909-1918. |
| [11] | ZHANG Ling, ZHAO Chunyu, XU Yaoyao, CHEN Yansen, CAI Zhixiong, LIN Haowei, CAI Qiaoyan. Circular RNA hsa_circ_0006834 is a potential prognostic biomarker for hepatocellular carcinoma [J]. Journal of Southern Medical University, 2023, 43(11): 1850-1856. |
| [12] | LIN Jie, OU Huohui, WANG Weidong, MA Jing, ZHANG Weijie, LIU Qingbo. Overexpression of CLEC5A inhibits cell proliferation and metastasis and reverses epithelial-mesenchymal transition in hepatocellular carcinoma [J]. Journal of Southern Medical University, 2023, 43(1): 85-91. |
| [13] | ZHANG Huanhuan, CHEN Zhuo, ZHAO Xiangdi, HUO Qiang, CHENG Xiu. Shikonin induces hepatocellular carcinoma cell apoptosis by suppressing PKM2/PHD3/HIF-1α signaling pathway [J]. Journal of Southern Medical University, 2023, 43(1): 92-98. |
| [14] | NING Shiyu, HE Chunmei, GUO Zehao, ZHANG Hao, MO Zhijing. VIPR1 promoter methylation promotes transcription factor AP-2α binding to inhibit VIPR1 expression and promote hepatocellular carcinoma cell growth in vitro [J]. Journal of Southern Medical University, 2022, 42(7): 957-965. |
| [15] | YANG Xuejia, LI Yujie, WU Dengqiang, MA Yili, ZHOU Sufang. Screening and identification of key genes ATP1B3 and ENAH in the progression of hepatocellular carcinoma: based on data mining and clinical validation [J]. Journal of Southern Medical University, 2022, 42(6): 815-823. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||