Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (10): 2210-2222.doi: 10.12122/j.issn.1673-4254.2025.10.17
Jinguo WANG1( ), Yang MA2, Zhaoxin LI3, Lifei HE3, Yingze HUANG4(
), Yang MA2, Zhaoxin LI3, Lifei HE3, Yingze HUANG4( ), Xiaoming FAN5(
), Xiaoming FAN5( )
)
Received:2025-04-23
Online:2025-10-20
Published:2025-10-24
Contact:
Yingze HUANG, Xiaoming FAN
E-mail:wangjinguo401@alu.glmc.edu.cn;youshxicun@glmc.edu.cn;fanxiaom1987@ glmc.edu.cn
Jinguo WANG, Yang MA, Zhaoxin LI, Lifei HE, Yingze HUANG, Xiaoming FAN. PDZ-binding kinase as a prognostic biomarker for pancreatic cancer: a pan-cancer analysis and validation in pancreatic adenocarcinoma cells[J]. Journal of Southern Medical University, 2025, 45(10): 2210-2222.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.10.17
| Name | Detail | Tumor (TCGA) | Normal (source) |
|---|---|---|---|
| ACC | Adrenocortical carcinoma | 77 | 77 (adrenal gland, GTEx) |
| BLCA | Bladder urothelial carcinoma | 406 | 19 (TCGA) |
| BRCA | Breast invasive carcinoma | 1085 | 99 (TCGA) |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma | 306 | 13 (cervix uteri, GTEx) |
| CHOL | Cholangio carcinoma | 36 | 9 (TCGA) |
| COAD | Colon adenocarcinoma | 448 | 41 (TCGA) |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma | 47 | 47 (whole blood, GTEx) |
| ESCA | Esophageal carcinoma | 182 | 13 (TCGA) |
| GBM | Glioblastoma multiforme | 167 | 163 (brain cortex, GTEx) |
| HNSC | Head and neck squamous cell carcinoma | 519 | 44 (TCGA) |
| KICH | Kidney chromophobe | 66 | 25 (TCGA) |
| KIRC | Kidney renal clear cell carcinoma | 531 | 25 (TCGA) |
| KIRP | Kidney renal papillary cell carcinoma | 286 | 32 (TCGA) |
| LAML | Acute myeloid Leukemia | 173 | 173 (whole blood, GTEx) |
| LGG | Brain lower grade glioma | 524 | 255 (brain cortex, GTEx) |
| LIHC | Liver hepatocellular carcinoma | 369 | 50 (TCGA) |
| LUAD | Lung adenocarcinoma | 513 | 59 (TCGA) |
| LUSC | Lung squamous cell carcinoma | 486 | 50 (TCGA) |
| MESO | Mesothelioma | 87 | 87 (heart atrial appendage, GTEx) |
| OV | Ovarian serous cystadenocarcinoma | 426 | 180 (ovary, GTEx) |
| PAAD | Pancreatic adenocarcinoma | 178 | 4 (TCGA) |
| PCPG | Pheochromocytoma and paraganglioma | 183 | 182 (adrenal gland, GTEx) |
| PRAD | Prostate adenocarcinoma | 499 | 52 (TCGA) |
| READ | Rectum adenocarcinoma | 158 | 10 (TCGA) |
| SARC | Sarcoma | 262 | 262 (adipose subcutaneous, GTEx) |
| SKCM | Skin cutaneous melanoma | 461 | 461 (skin sun exposed lower, GTEx) |
| STAD | Stomach adenocarcinoma | 408 | 36 (TCGA) |
| TGCT | Testicular germ cell tumors | 139 | 137 (testis, GTEx) |
| THCA | Thyroid carcinoma | 512 | 59 (TCGA) |
| THYM | Thymoma | 118 | 118 (whole blood, GTEx) |
| UCEC | Uterine corpus endometrial carcinoma | 544 | 35 (TCGA) |
| UCS | Uterine carcinosarcoma | 57 | 57 (uterus, GTEx) |
| UVM | Uveal melanoma | 80 | 79 (EyeGEx retina, GTEx) |
| GSE15471 | 39 (GEO) | 39 (GEO) | |
| GSE16515 | 36 (GEO) | 16 (GEO) | |
| GSE62165 | 118 (GEO) | 13 (GEO) | |
Tab.1 Data of the samples from TCGA, GEO and GTEx
| Name | Detail | Tumor (TCGA) | Normal (source) |
|---|---|---|---|
| ACC | Adrenocortical carcinoma | 77 | 77 (adrenal gland, GTEx) |
| BLCA | Bladder urothelial carcinoma | 406 | 19 (TCGA) |
| BRCA | Breast invasive carcinoma | 1085 | 99 (TCGA) |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma | 306 | 13 (cervix uteri, GTEx) |
| CHOL | Cholangio carcinoma | 36 | 9 (TCGA) |
| COAD | Colon adenocarcinoma | 448 | 41 (TCGA) |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma | 47 | 47 (whole blood, GTEx) |
| ESCA | Esophageal carcinoma | 182 | 13 (TCGA) |
| GBM | Glioblastoma multiforme | 167 | 163 (brain cortex, GTEx) |
| HNSC | Head and neck squamous cell carcinoma | 519 | 44 (TCGA) |
| KICH | Kidney chromophobe | 66 | 25 (TCGA) |
| KIRC | Kidney renal clear cell carcinoma | 531 | 25 (TCGA) |
| KIRP | Kidney renal papillary cell carcinoma | 286 | 32 (TCGA) |
| LAML | Acute myeloid Leukemia | 173 | 173 (whole blood, GTEx) |
| LGG | Brain lower grade glioma | 524 | 255 (brain cortex, GTEx) |
| LIHC | Liver hepatocellular carcinoma | 369 | 50 (TCGA) |
| LUAD | Lung adenocarcinoma | 513 | 59 (TCGA) |
| LUSC | Lung squamous cell carcinoma | 486 | 50 (TCGA) |
| MESO | Mesothelioma | 87 | 87 (heart atrial appendage, GTEx) |
| OV | Ovarian serous cystadenocarcinoma | 426 | 180 (ovary, GTEx) |
| PAAD | Pancreatic adenocarcinoma | 178 | 4 (TCGA) |
| PCPG | Pheochromocytoma and paraganglioma | 183 | 182 (adrenal gland, GTEx) |
| PRAD | Prostate adenocarcinoma | 499 | 52 (TCGA) |
| READ | Rectum adenocarcinoma | 158 | 10 (TCGA) |
| SARC | Sarcoma | 262 | 262 (adipose subcutaneous, GTEx) |
| SKCM | Skin cutaneous melanoma | 461 | 461 (skin sun exposed lower, GTEx) |
| STAD | Stomach adenocarcinoma | 408 | 36 (TCGA) |
| TGCT | Testicular germ cell tumors | 139 | 137 (testis, GTEx) |
| THCA | Thyroid carcinoma | 512 | 59 (TCGA) |
| THYM | Thymoma | 118 | 118 (whole blood, GTEx) |
| UCEC | Uterine corpus endometrial carcinoma | 544 | 35 (TCGA) |
| UCS | Uterine carcinosarcoma | 57 | 57 (uterus, GTEx) |
| UVM | Uveal melanoma | 80 | 79 (EyeGEx retina, GTEx) |
| GSE15471 | 39 (GEO) | 39 (GEO) | |
| GSE16515 | 36 (GEO) | 16 (GEO) | |
| GSE62165 | 118 (GEO) | 13 (GEO) | |
| Gene name | Sequence | Tm | Amplicon length |
|---|---|---|---|
| PBK | F: TATGACTGCTCCTGCCTTCATAAC | 60 ℃ | 115 |
| R: CACAGCTTCTTTGGGTTTCCAT | |||
| GAPDH | F: TCTCTGCTCCTCCCTGTTC | 60 ℃ | 125 |
| R: ACACCGACCTTCACCATCT |
Tab.2 Primer sequences for RT-qPCR in this study
| Gene name | Sequence | Tm | Amplicon length |
|---|---|---|---|
| PBK | F: TATGACTGCTCCTGCCTTCATAAC | 60 ℃ | 115 |
| R: CACAGCTTCTTTGGGTTTCCAT | |||
| GAPDH | F: TCTCTGCTCCTCCCTGTTC | 60 ℃ | 125 |
| R: ACACCGACCTTCACCATCT |
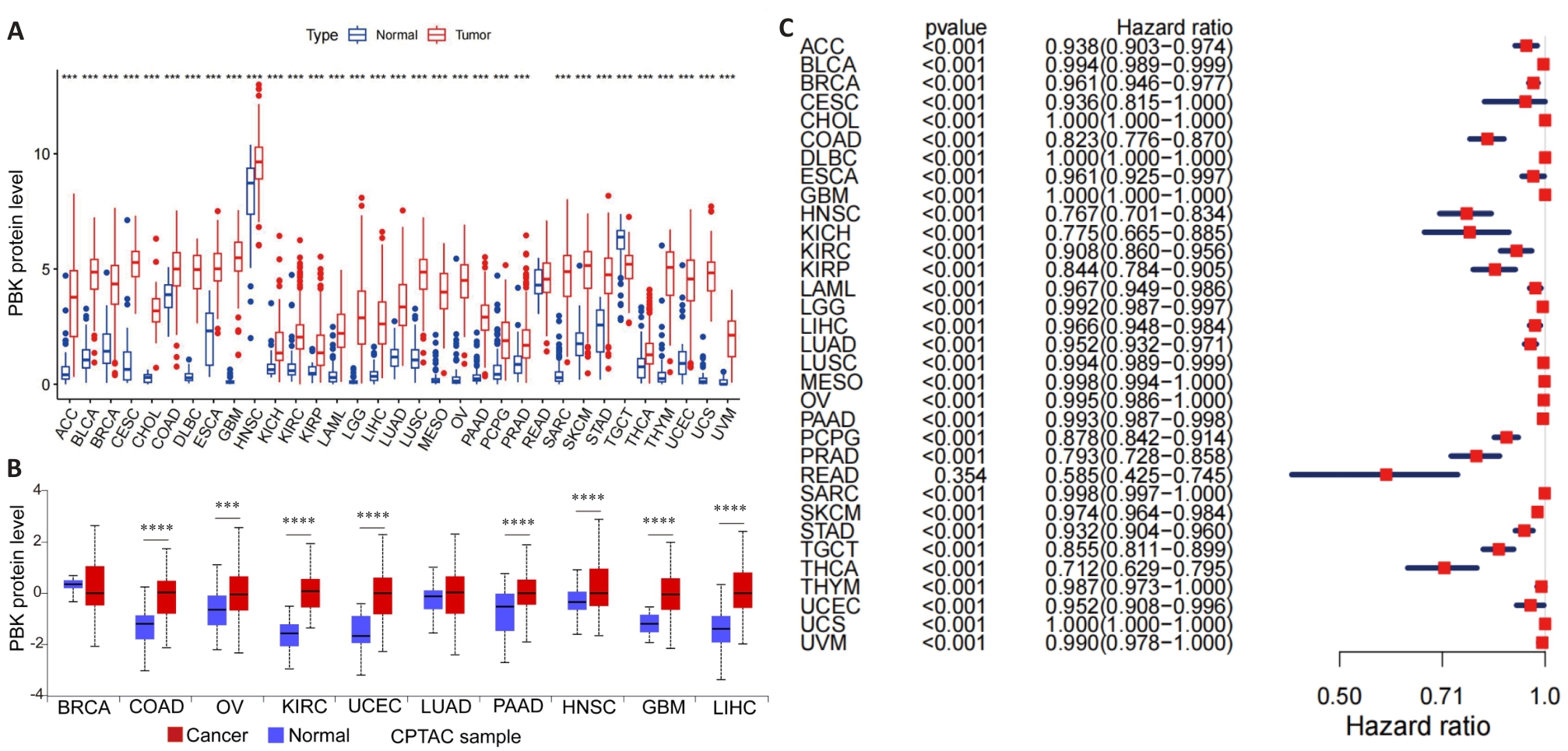
Fig.1 PBK expression in 33 cancer types. A: Distribution of PBK mRNA expression in 33 cancer types and their normal controls based on TCGA data. B: Protein expression levels of PBK in different cancers based on data from the CPTAC database. C: ROC curve analysis of PBK in 33 cancer types, demonstrating its diagnostic ability to differentiate cancer from normal tissues. ***P<0.001, ****P<0.0001.
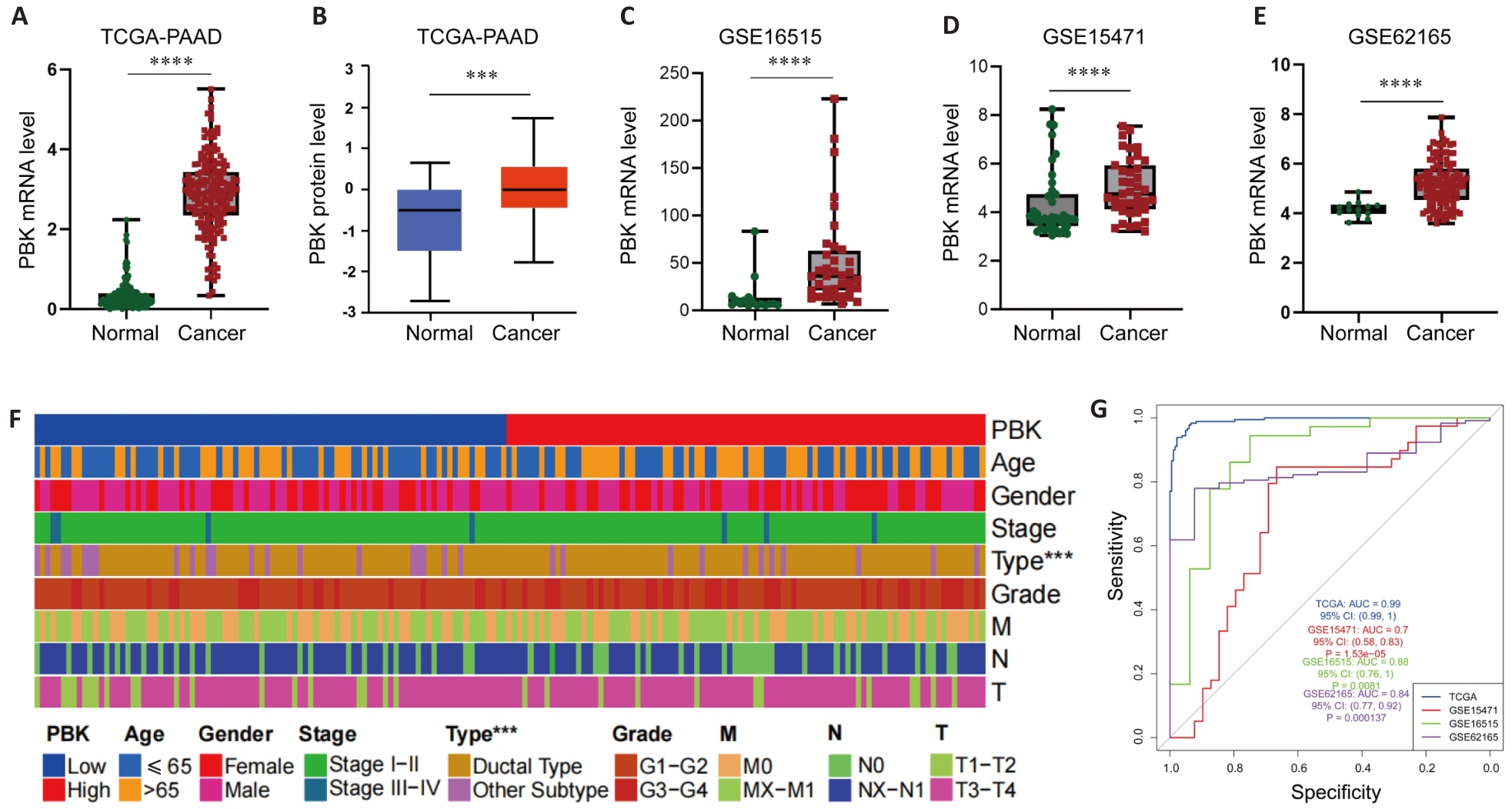
Fig.2 PBK expression in pancreatic cancer. A, B: TCGA and CPTAC data show significantly higher expression of PBK in pancreatic cancer tissues than in normal tissues. C-E: Differential expressions of PBK in pancreatic cancer and paired normal samples based on GSE16515, GSE15471 and GSE62165 datasets. F: Clinical feature heat map showing the association of PBK expression with pancreatic ductal adenocarcinoma and its correlation with tumor characteristics. G: ROC curves of the TCGA and GEO datasets further verify the reliability of PBK as a biomarker for diagnosis of pancreatic cancer. ***P<0.001, ****P<0.0001.
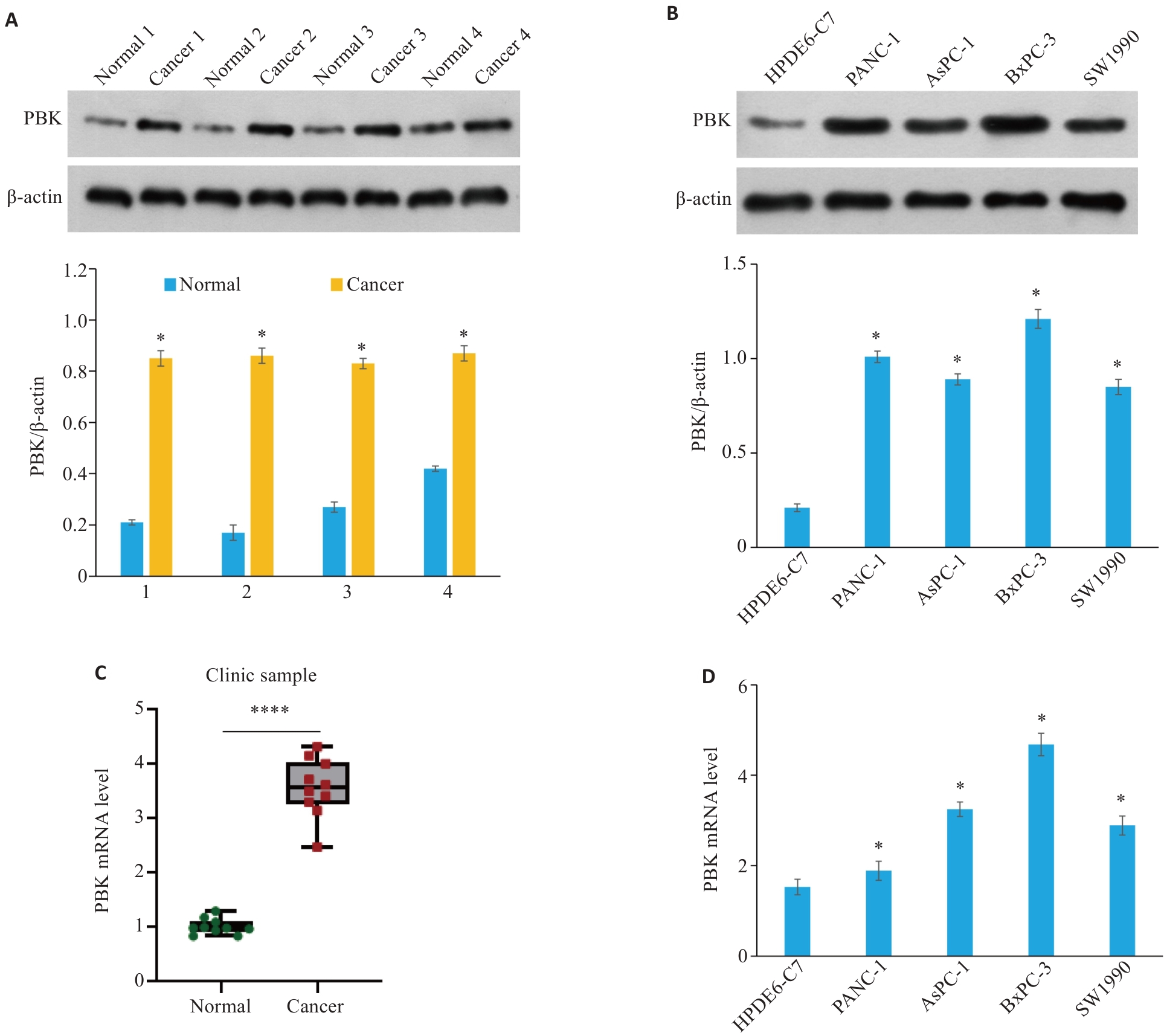
Fig.3 PBK is highly expressed in clinical samples and cell lines of pancreatic cancer. A: PBK protein expression levels in 4 clinical pancreatic cancer samples and their paired normal tissues. *P<0.05 vs Normal. B: PBK protein expression levels in normal pancreatic cell line HPDE6-C7 and 4 pancreatic cancer cell lines (PANC-1, AsPC-1, BxPC-3, and SW1990). *P<0.05 vs HPDE6-C7. C: PBK mRNA expression levels in 4 clinical pancreatic cancer samples and their paired normal tissues. ****P<0.0001. D: PBK mRNA expression levels in the normal pancreatic cell line HPDE6-C7 and 4 pancreatic cancer cell lines. *P<0.05 vs HPDE6-C7.
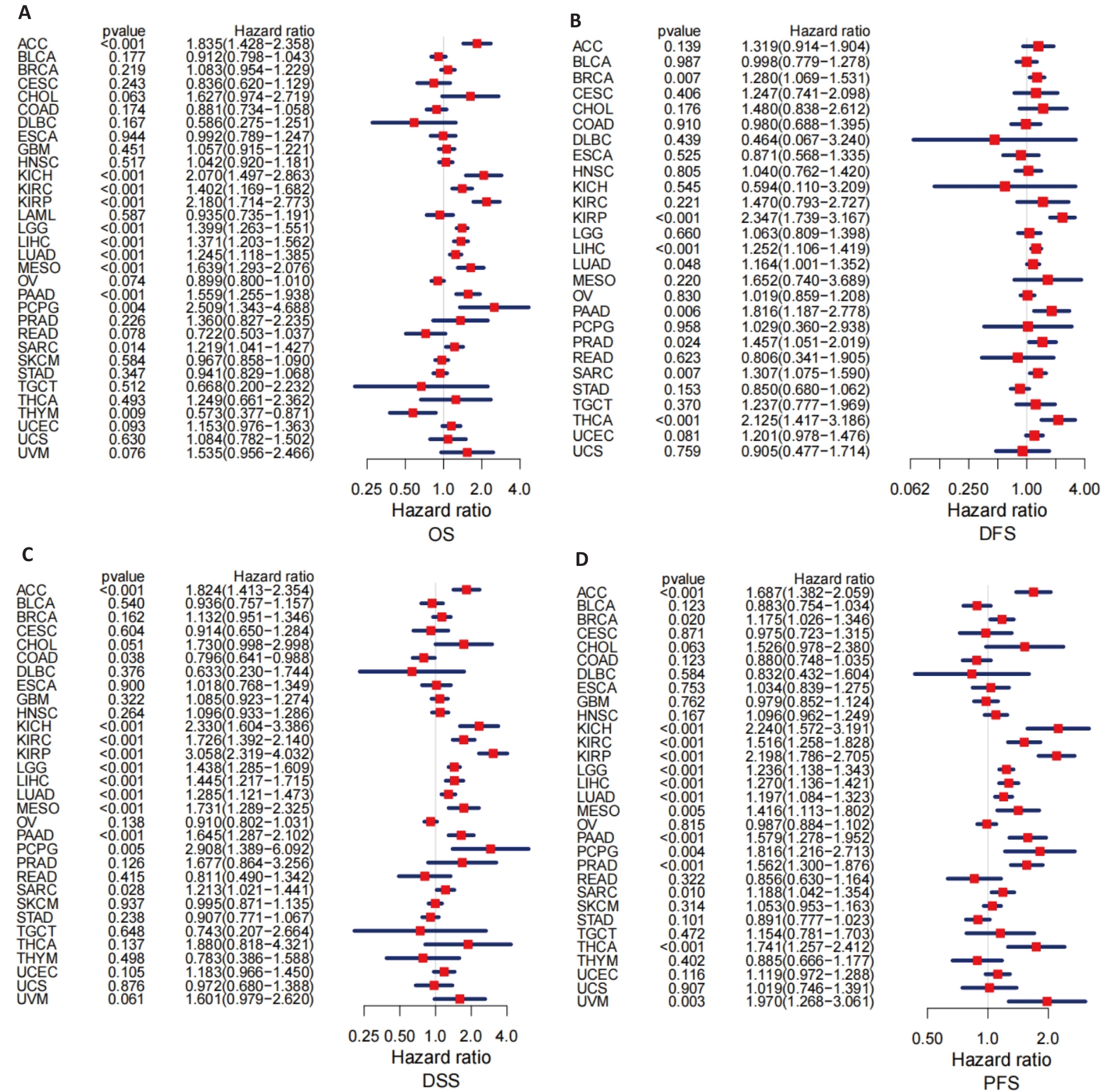
Fig.4 Prognostic value of PBK expression across 33 cancer types. A: Forest plot analysis of overall survival (OS) showing the prognostic value of PBK across various cancers. B: Forest plot analysis of disease-free survival (DFS) highlighting the potential impact of PBK in pancreatic cancer. C: Analysis of disease-specific survival (DSS) showing the ability of PBK for predicting disease-specific mortality. D: Progression-free survival (PFS) analysis further supports the role of PBK as a prognostic indicator.
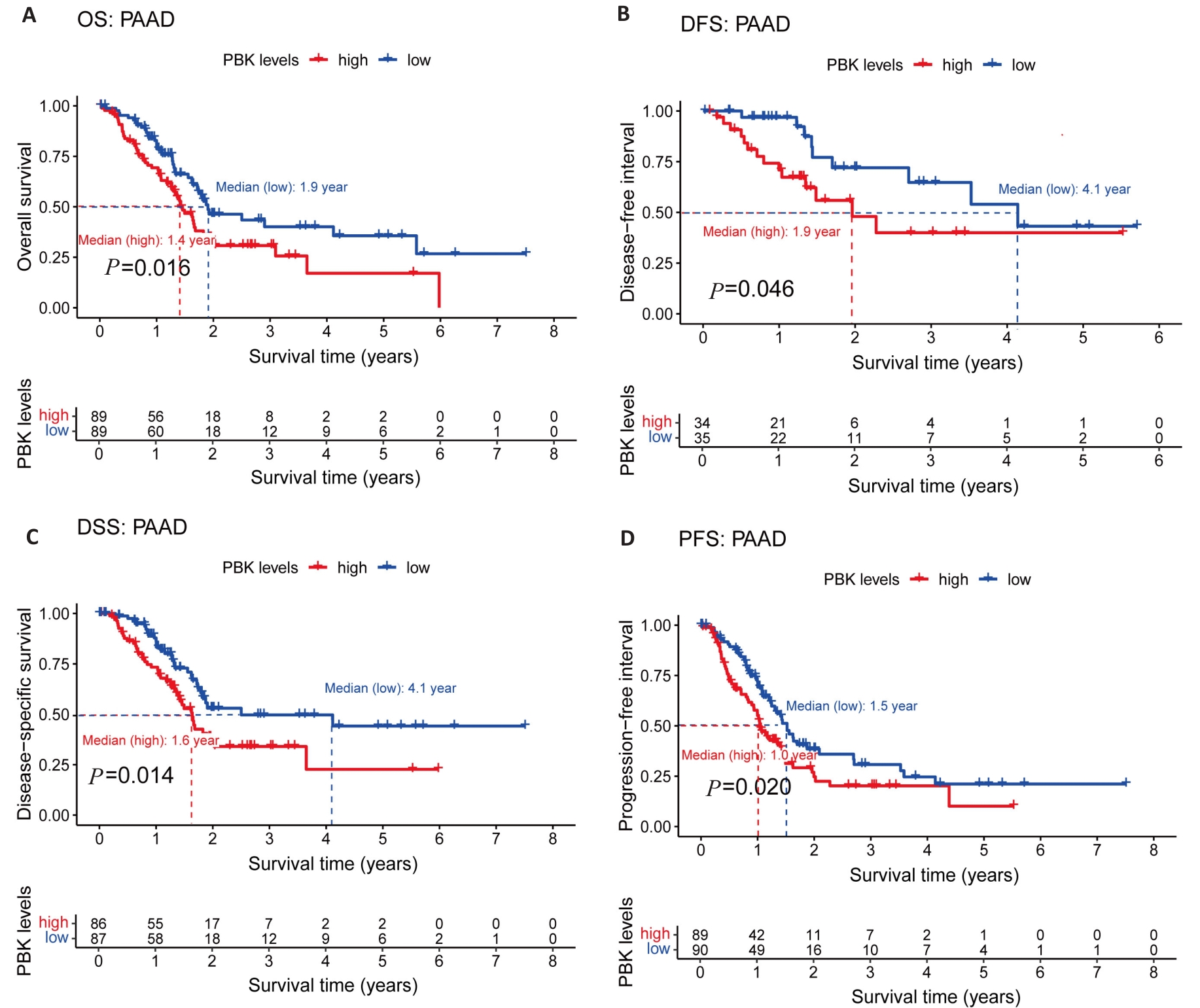
Fig.5 PBK expression and its association with prognosis in pancreatic cancer. A: Kaplan-Meier survival curves show a significant association between high PBK expression and decreased OS. B: DFS survival curve validates the influence of PBK on disease-free survival. C: DSS survival curve highlights the relationship between high PBK expression and lower disease-specific survival. D: PFS survival curve reveals the connection between PBK expression and progression-free survival.
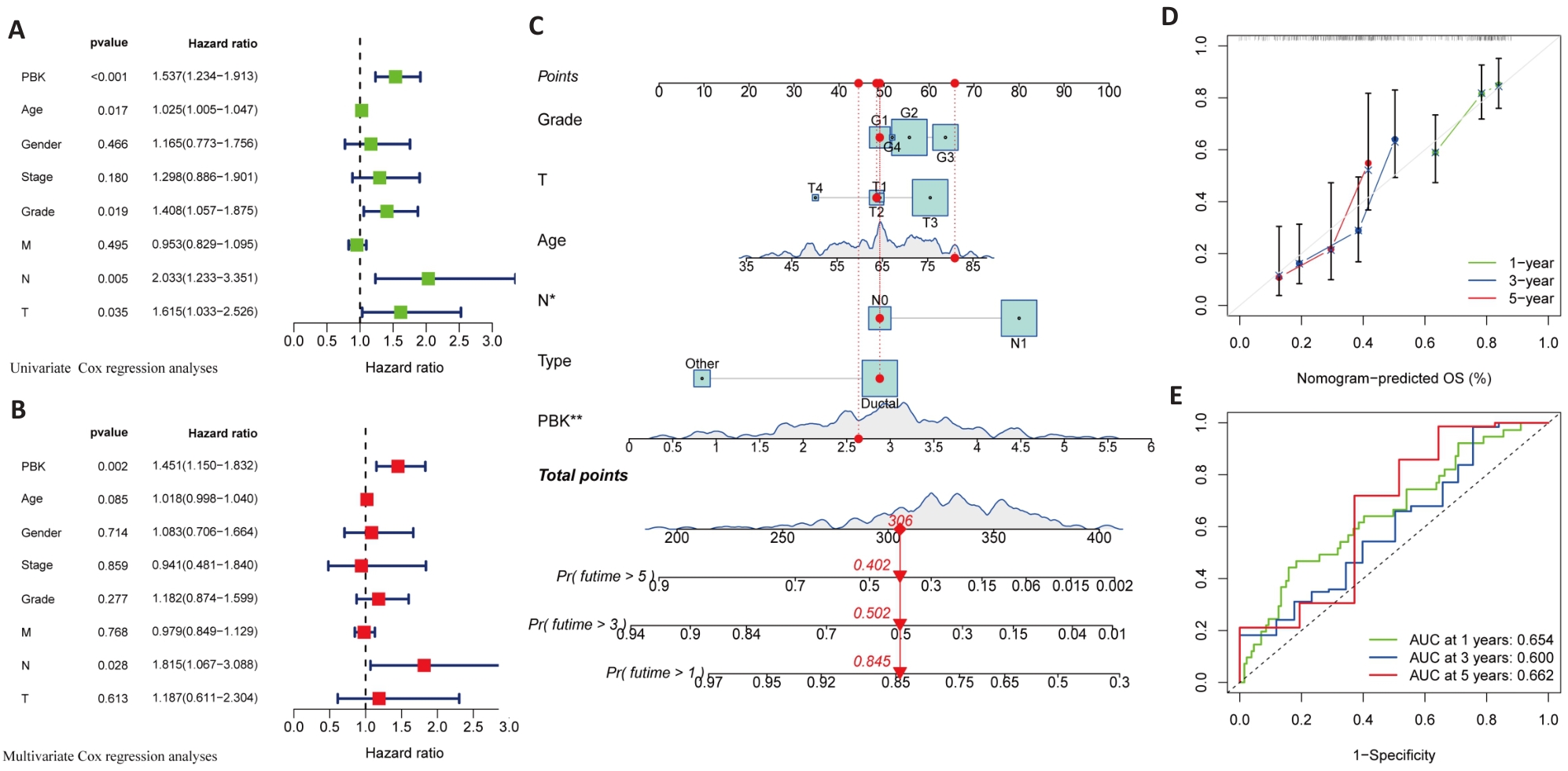
Fig.6 Univariate and multivariate analysis of the association of high PBK expression with prognosis of pancreatic cancer. A: Univariate analysis reveals significant association of PBK with clinicopathological factors of pancreatic cancer. B: Multivariate analysis validates PBK as an independent prognostic factor after adjusting for other clinical variables. C: A nomogram model for predicting survival probabilities of pancreatic cancer patients based on PBK expression. D: Calibration plot confirms the prediction accuracy of the nomogram model. E: ROC curves for 1-, 3-, and 5-year survival showing the predictive performance of PBK. *P<0.05, **P<0.01.
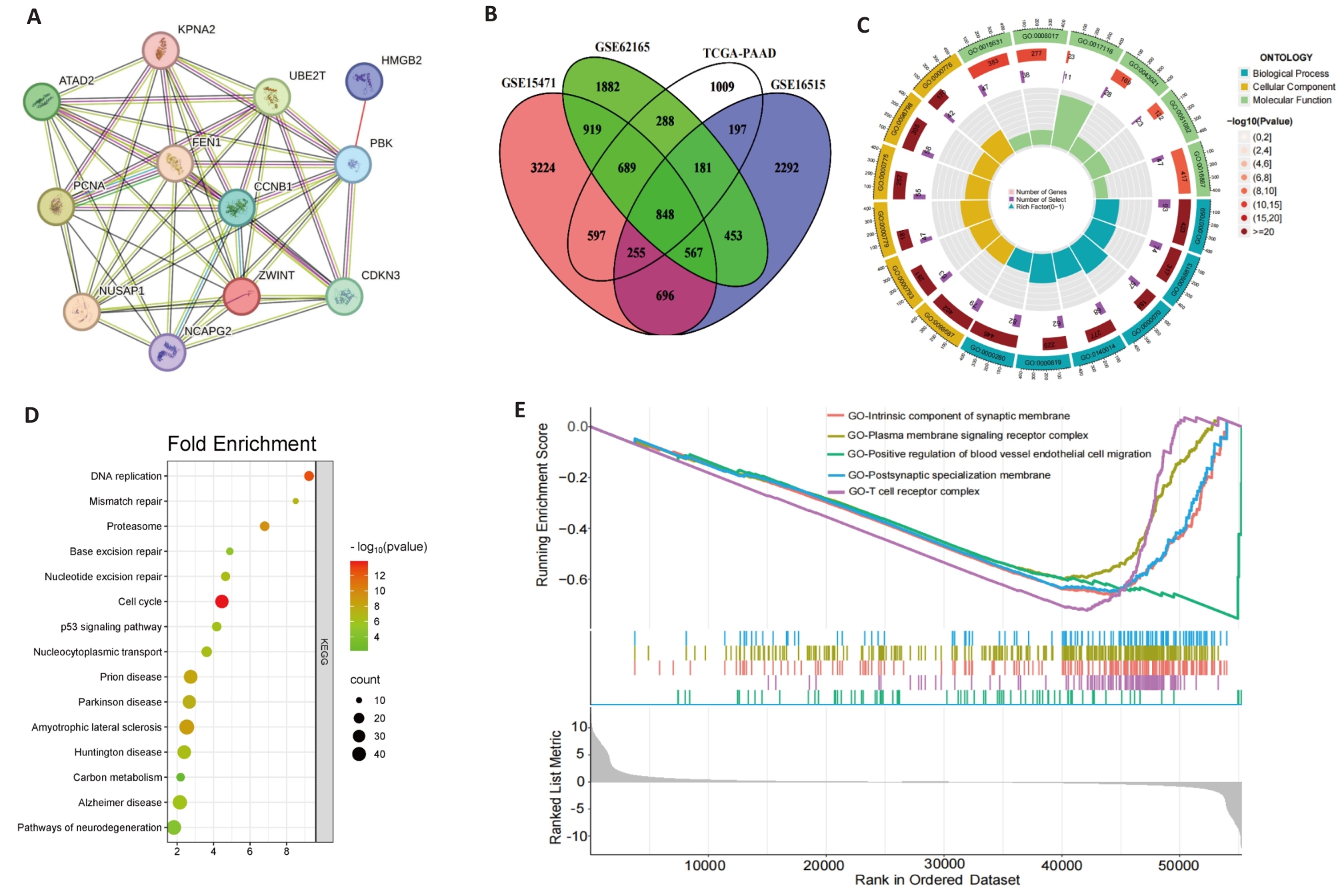
Fig.7 Gene correlation and enrichment analysis. A: Correlation analysis of PBK with 11 co-expressed genes across 33 cancer types. B: Intersection of PBK-related genes in pancreatic cancer, revealing key associated genes. C: GO analysis showing that PBK co-expressed genes are enriched in immune and cell cycle-related biological processes. D: KEGG pathway analysis shows that PBK is involved in cancer-related signaling pathways. E: GSEA analysis validates PBK-associated gene enrichment in immune regulation and tumor-related pathways.
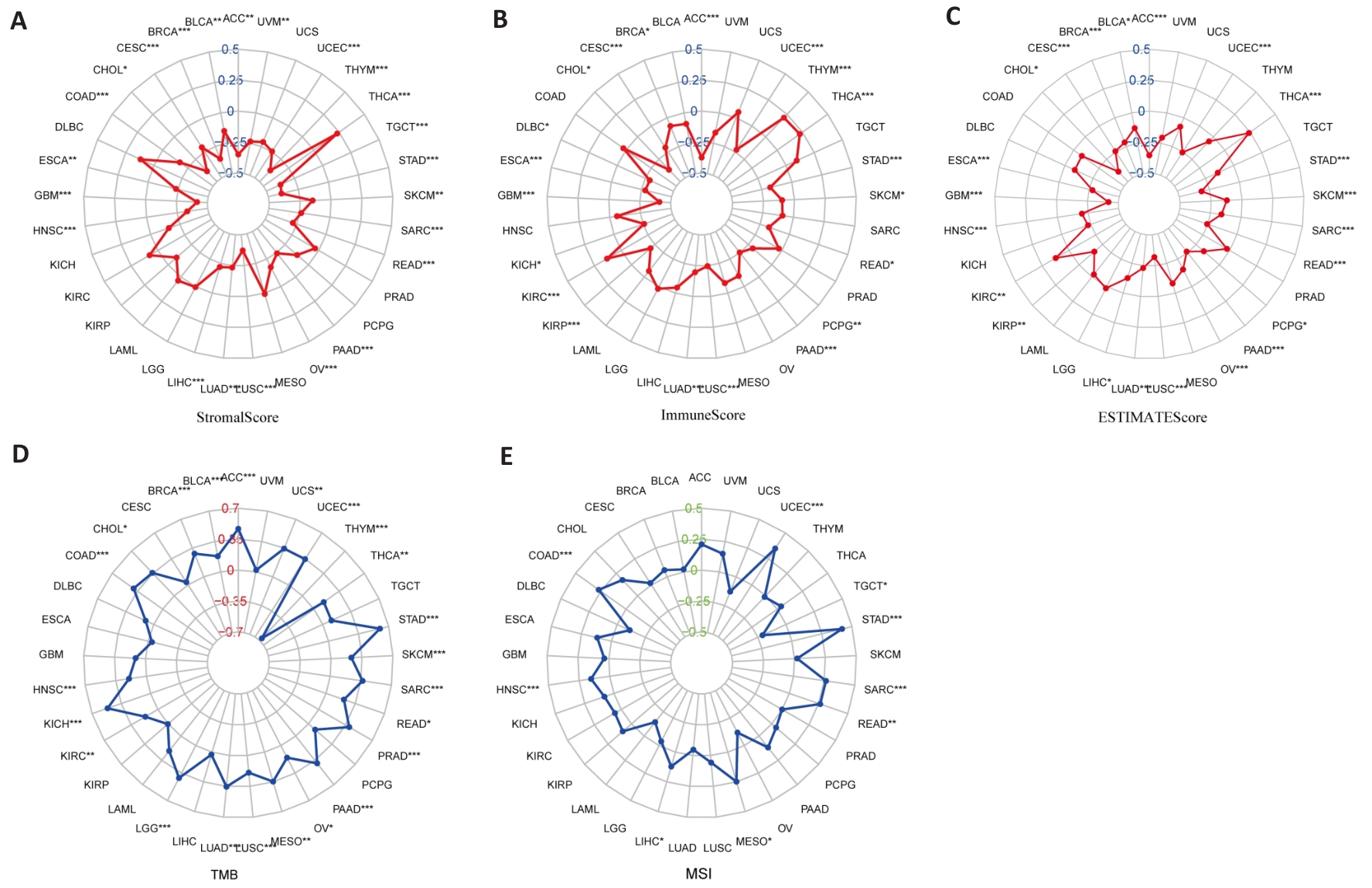
Fig.8 PBK expression and tumor microenvironment analysis. A: Stromal Score analysis shows a correlation between PBK expression and tumor stromal content. B: Immune Score analysis indicates that PBK expression is associated with tumor immune infiltration. C: ESTIMATE Score analysis validates the role of PBK in the tumor microenvironment. D: Correlation between PBK expression and tumor mutation burden (TMB). E: Correlation between PBK expression and microsatellite instability (MSI), suggesting its potential immune regulatory mechanisms. *P<0.05, **P<0.01, ***P<0.001.
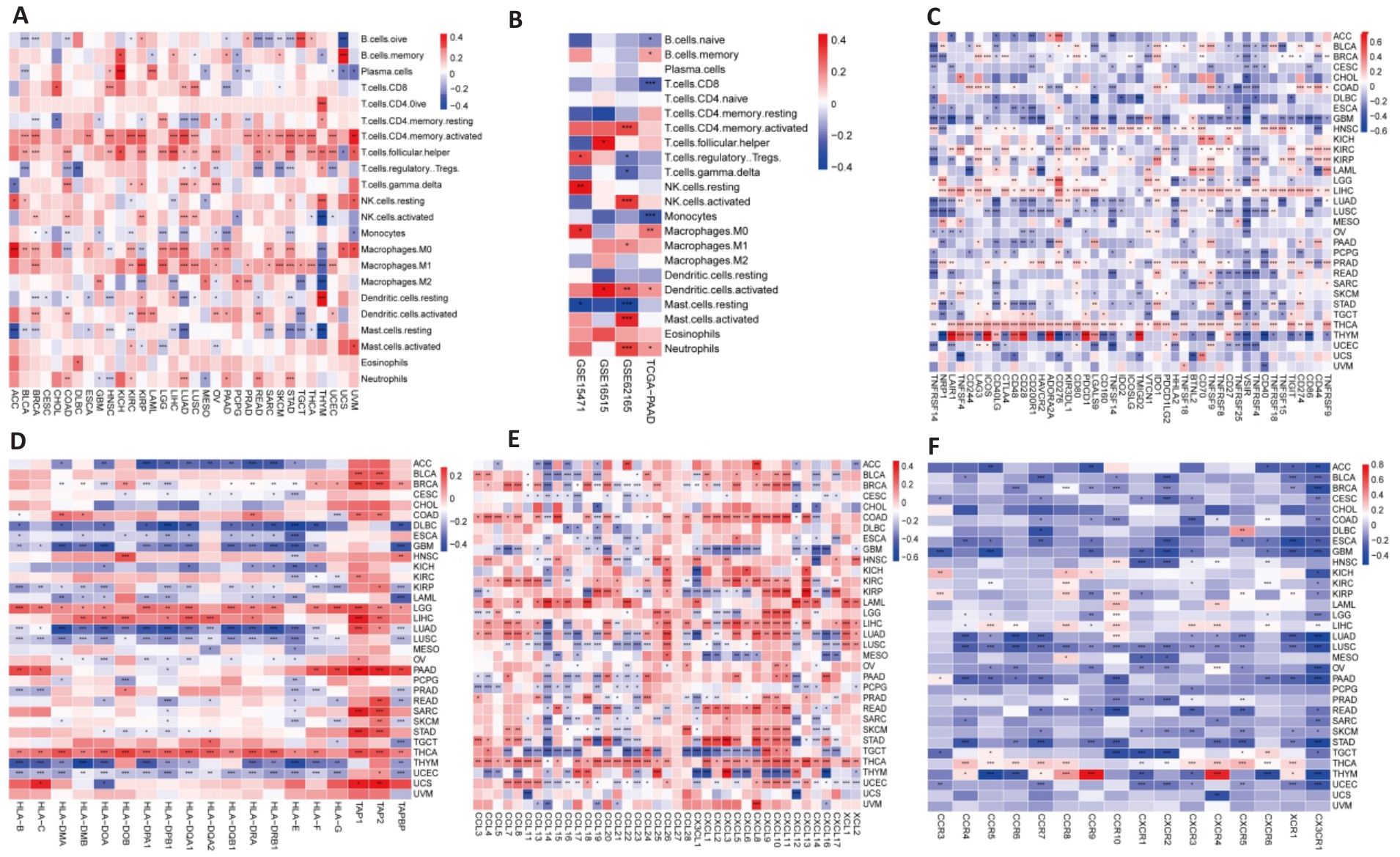
Fig.9 Relationship between PBK expression and immune system. A: Analysis of PBK expression and immune infiltration across 33 cancer types. B: Immune infiltration analysis of PBK in the pancreatic cancer immune microenvironment based on combined TCGA and GEO data. C: Correlation analysis of PBK expression with immune checkpoint molecules in 33 cancer types. D-F: Complex associations of PBK expression with MHC molecules (D), chemokines (E), and their receptors (F), suggesting its involvement in immune regulatory networks. *P<0.05, **P<0.01, ***P<0.001.
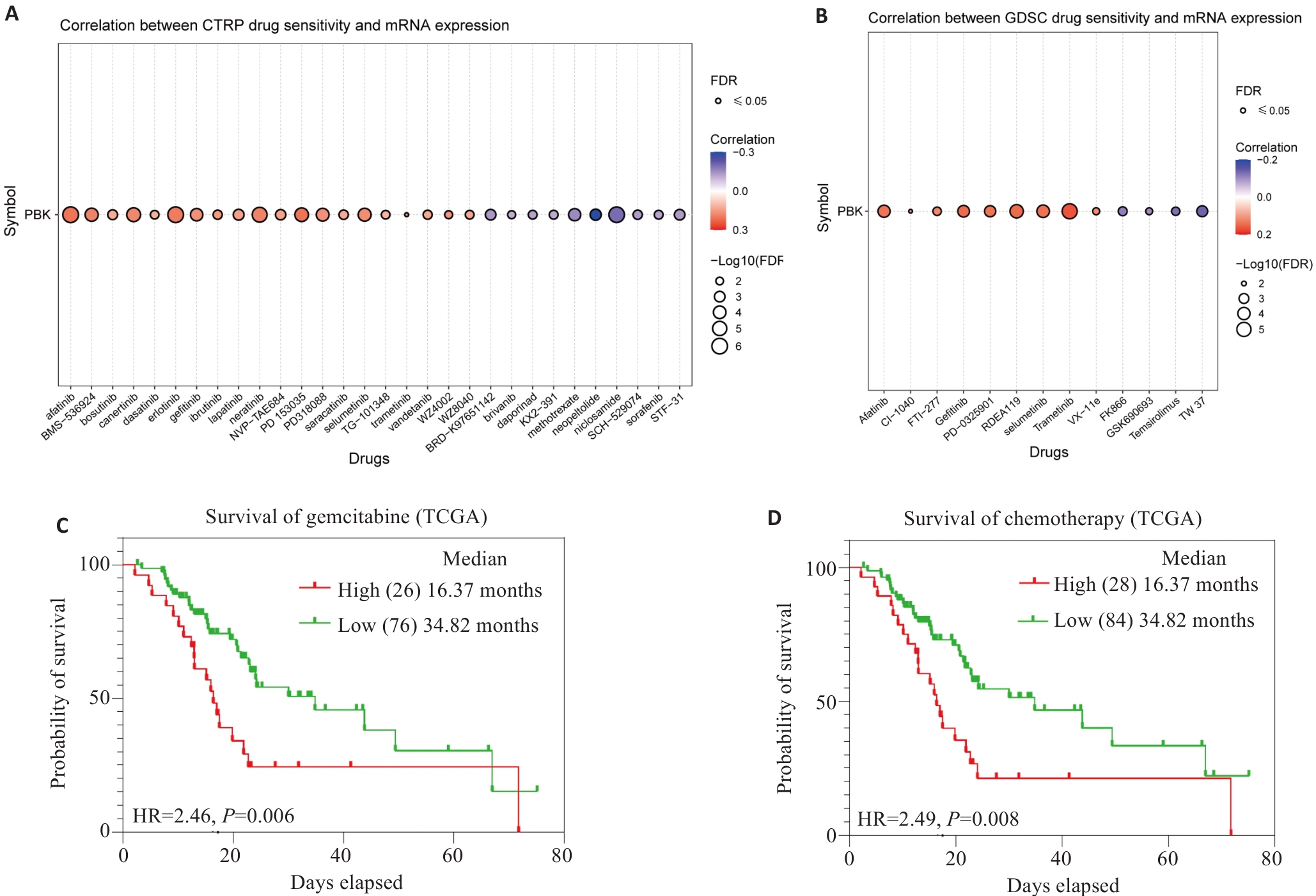
Fig.10 PBK expression and drug sensitivity analysis. A: Analysis from the CTRPC database shows that PBK mRNA expression affects drug sensitivity. B: Analysis from the GDSC database shows that PBK mRNA expression affects drug sensitivity. C: High PBK expression predicts poor response to gemcitabine treatment. D: High PBK expression is associated with chemotherapeutic drug resistance, indicating its potential as a drug target.
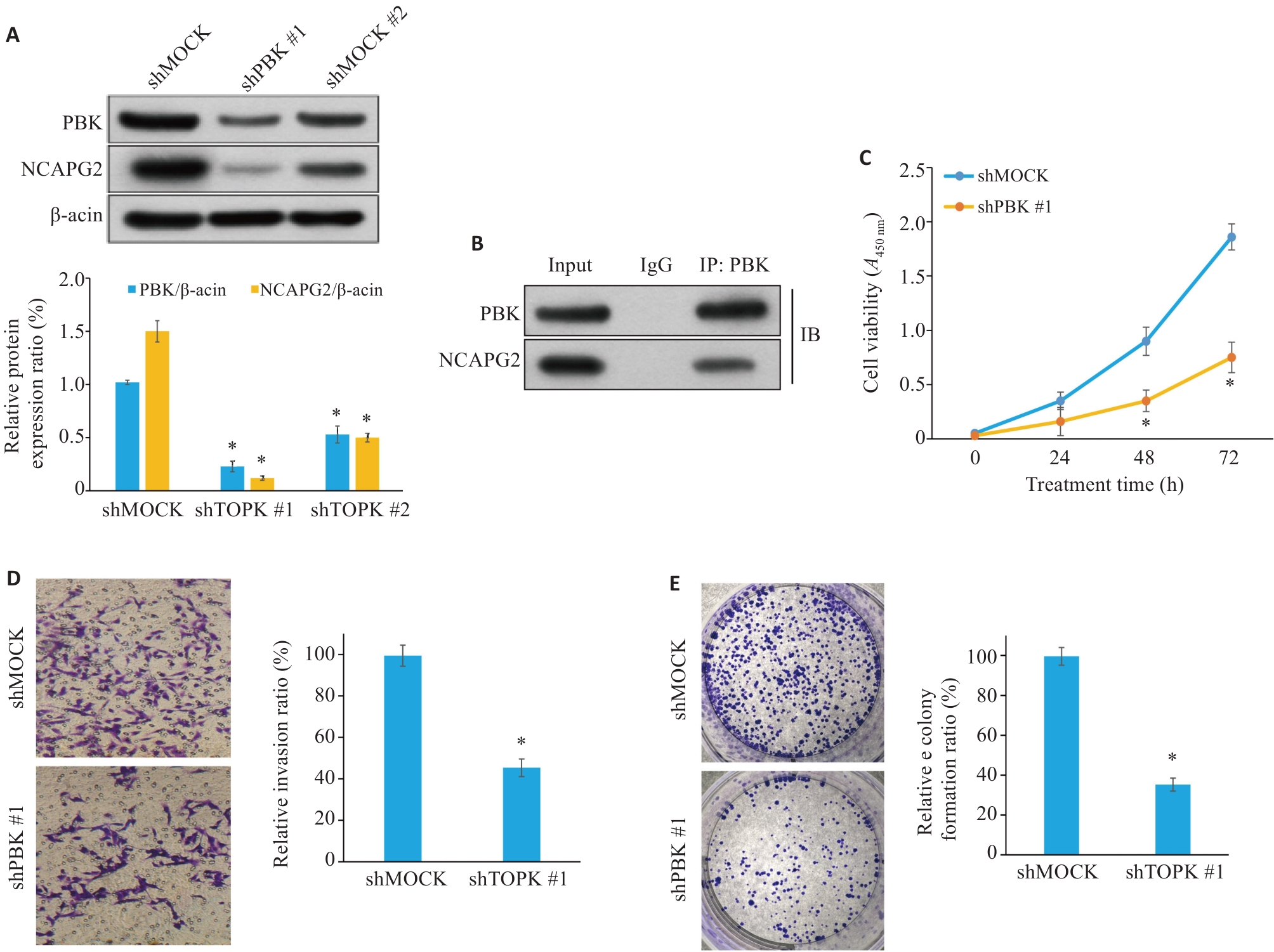
Fig.11 PBK knockdown inhibits proliferation, migration and invasion of pancreatic cancer cells. A: Western blotting showing efficient knockdown of PBK expression. B: Co-immunoprecipitation experiment shows that PBK directly interacts with NCAPG2, suggesting that the PBK-NCAPG2 axis plays a role in tumor progression. C: CCK-8 assay shows that PBK knockdown significantly inhibits cell proliferation. D: Transwell assay validating suppressed invasion ability of pancreatic cancer cells after PBK knockdown (Original magnification: ×200). E: Effect of shPBK on clone formation ability of pancreatic cancer cells. *P<0.05 vs shMock.
| [1] | Mei QJ, Li KX, Tang TY, et al. miR-203-3p promotes senescence of mouse bone marrow mesenchymal stem cells via downregulation of Pbk[J]. Aging Cell, 2024, 23(11): e14293. doi:10.1111/acel.14293 |
| [2] | Han ZP, Li LZ, Huang YY, et al. PBK/TOPK: a therapeutic target worthy of attention[J]. Cells, 2021, 10(2): 371. doi:10.3390/cells10020371 |
| [3] | Lee DH, Jeong YJ, Won JY, et al. PBK/TOPK is a favorable prognostic biomarker correlated with antitumor immunity in colon cancers[J]. Biomedicines, 2022, 10(2): 299. doi:10.3390/biomedicines10020299 |
| [4] | Herbert KJ, Ashton TM, Prevo R, et al. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics[J]. Cell Death Dis, 2018, 9: 1089. doi:10.1038/s41419-018-1131-7 |
| [5] | Deng MQ, Yang RY, Sun Q, et al. Small-molecule inhibitor HI-TOPK-032 improves NK-92MI cell infiltration into ovarian tumours[J]. Basic Clin Pharmacol Toxicol, 2024, 134(5): 629-42. doi:10.1111/bcpt.14002 |
| [6] | Huang H, Lee MH, Liu KD, et al. PBK/TOPK: an effective drug target with diverse therapeutic potential[J]. Cancers (Basel), 2021, 13(9): 2232. doi:10.3390/cancers13092232 |
| [7] | Niu NN, Shen XQ, Wang Z, et al. Tumor cell-intrinsic epigenetic dysregulation shapes cancer-associated fibroblasts heterogeneity to metabolically support pancreatic cancer[J]. Cancer Cell, 2024, 42(5): 869-84. e9. doi:10.1016/j.ccell.2024.03.005 |
| [8] | Li PY, Zhang HY, Gao XY, et al. Difference in fecal and oral microbiota between pancreatic cancer and benign/low-grade malignant tumor patients[J]. BMC Microbiol, 2024, 24(1): 527. doi:10.1186/s12866-024-03687-6 |
| [9] | De Santis MC, Bockorny B, Hirsch E, et al. Exploiting pancreatic cancer metabolism: challenges and opportunities[J]. Trends Mol Med, 2024, 30(6): 592-604. doi:10.1016/j.molmed.2024.03.008 |
| [10] | Feng TT, Zhang Y, Ling SB, et al. PDZ binding kinase/T-LAK cell-derived protein kinase plays an oncogenic role and promotes immune escape in human tumors[J]. J Oncol, 2021, 2021: 8892479. doi:10.1155/2021/8892479 |
| [11] | Fan XM, Tao JY, Cai Z, et al. Eupafolin suppresses esophagus cancer growth by targeting T-LAK cell-originated protein kinase[J]. Front Pharmacol, 2019, 10: 1248. doi:10.3389/fphar.2019.01248 |
| [12] | Lu H, Huang YZ, Ni XF, et al. TOPK promotes the development of psoriasis and worenine alleviates psoriasiform dermatitis by inhibiting TOPK activity[J]. J Eur Acad Dermatol Venereol, 2024, 38(5): 851-63. doi:10.1111/jdv.19724 |
| [13] | Brozos-Vázquez E, Toledano-Fonseca M, Costa-Fraga N, et al. Pancreatic cancer biomarkers: a pathway to advance in personalized treatment selection[J]. Cancer Treat Rev, 2024, 125: 102719. doi:10.1016/j.ctrv.2024.102719 |
| [14] | Pratticò F, Garajová I. Focus on pancreatic cancer microenvironment[J]. Curr Oncol, 2024, 31(8): 4241-60. doi:10.3390/curroncol31080316 |
| [15] | McGuigan A, Kelly P, Turkington RC, et al. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes[J]. World J Gastroenterol, 2018, 24(43): 4846-61. doi:10.3748/wjg.v24.i43.4846 |
| [16] | Cai J, Chen HD, Lu M, et al. Advances in the epidemiology of pancreatic cancer: Trends, risk factors, screening, and prognosis[J]. Cancer Lett, 2021, 520: 1-11. doi:10.1016/j.canlet.2021.06.027 |
| [17] | Nguyen N, Hoang TM, Huang T, et al. Macrophage-hitchhiked, effervescence-induced nanoemulsions for enhanced oral chemotherapy and immunotherapy: Impact on absorption route[J]. Biomaterials, 2025, 316: 123019. doi:10.1016/j.biomaterials.2024.123019 |
| [18] | Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and prevention of pancreatic cancer[J]. Trends Cancer, 2018, 4(6): 418-28. doi:10.1016/j.trecan.2018.04.001 |
| [19] | Feng TT, Jiang RB, Yin L, et al. PDZ-binding kinase aggravates pancreatic neuroendocrine neoplasm progression by activating the AKT/mTOR pathway[J]. Mol Carcinog, 2023, 62(5): 716-26. doi:10.1002/mc.23519 |
| [20] | Lv BB, Zhang FN, Zhang XY, et al. PBK as a novel biomarker performed excellent diagnostic and prognostic value in HCC associated with immune infiltration and methylation[J]. J Mol Histol, 2025, 56(2): 129. doi:10.1007/s10735-024-10324-z |
| [21] | Li BX, Yao TZ, Zhang M, et al. Correlation study of PBK/TOPK expression, prognosis, and immune infiltration in breast cancer[J]. Sci Rep, 2025, 15(1): 15052. doi:10.1038/s41598-025-96542-1 |
| [22] | Zhang QF, Zheng F, Chen YC, et al. The TOPK inhibitor HI-TOPK-032 enhances CAR T-cell therapy of hepatocellular carcinoma by upregulating memory T cells[J]. Cancer Immunol Res, 2024, 12(5): 631-43. doi:10.1158/2326-6066.cir-23-0587 |
| [23] | Song WH, Yu YB, Wang SQ, et al. Metabolic reprogramming shapes the immune microenvironment in pancreatic adenocarcinoma: prognostic implications and therapeutic targets[J]. Front Immunol, 2025, 16: 1555287. doi:10.3389/fimmu.2025.1555287 |
| [24] | Jing R, Wu N, Zhang Q, et al. DPP4 promotes an immunoenhancing tumor microenvironment through exhausted CD8+ T cells with activating IL13-IL13RA2 axis in papillary thyroid cancer[J]. Int Immunopharmacol, 2025, 145: 113760. doi:10.1016/j.intimp.2024.113760 |
| [25] | Pang SG, Zhang X, Li ZX, et al. TOPK inhibition enhances the sensitivity of colorectal cancer cells to radiotherapy by reducing the DNA damage response[J]. Curr Med Sci, 2024, 44(3): 545-53. doi:10.1007/s11596-024-2884-0 |
| [26] | Wang Q, Li ZZ, Zhou SJ, et al. NCAPG2 could be an immunological and prognostic biomarker: From pan-cancer analysis to pancreatic cancer validation[J]. Front Immunol, 2023, 14: 1097403. doi:10.3389/fimmu.2023.1097403 |
| [27] | Mi XH, Shan HF, Kang CB, et al. MYC and NCAPG2 as molecular targets of colorectal cancer and gastric cancer in nursing[J]. Medicine (Baltimore), 2024, 103(18): e38029. doi:10.1097/md.0000000000038029 |
| [28] | Zhang EC, Chen ZJ, Liu WM, et al. NCAPG2 promotes prostate cancer malignancy and stemness via STAT3/c-MYC signaling[J]. J Transl Med, 2024, 22(1): 12. doi:10.1186/s12967-023-04834-9 |
| [29] | Feng Z, Zhang LF, Liu YX, et al. NCAPG2 contributes to the progression of malignant melanoma through regulating proliferation and metastasis[J]. Biochem Cell Biol, 2022, 100(6): 473-84. doi:10.1139/bcb-2022-0048 |
| [30] | Ren WJ, Yang S, Chen X, et al. NCAPG2 is a novel prognostic biomarker and promotes cancer stem cell maintenance in low-grade glioma[J]. Front Oncol, 2022, 12: 918606. doi:10.3389/fonc.2022.918606 |
| [31] | Jiang SY, Huang JJ, He H, et al. NCAPG2 maintains cancer stemness and promotes erlotinib resistance in lung adenocarcinoma[J]. Cancers (Basel), 2022, 14(18): 4395. doi:10.3390/cancers14184395 |
| [32] | Meng FZ, Zhang SG, Song RP, et al. NCAPG2 overexpression promotes hepatocellular carcinoma proliferation and metastasis through activating the STAT3 and NF-κB/miR-188-3p pathways[J]. EBioMedicine, 2019, 44: 237-49. doi:10.1016/j.ebiom.2019.05.053 |
| [1] | Yu ZHANG, Haitao LI, Yuqing PAN, Jiexian CAO, Li ZHAI, Xi ZHANG. Pan-cancer analysis of MZB1 expression and its association with immune infiltration and clinical prognosis [J]. Journal of Southern Medical University, 2025, 45(9): 2006-2018. |
| [2] | Zhi GAO, Ao WU, Zhongxiang HU, Peiyang SUN. Bioinformatics analysis of oxidative stress and immune infiltration in rheumatoid arthritis [J]. Journal of Southern Medical University, 2025, 45(4): 862-870. |
| [3] | Xiangzhuo MIAO, Pengyu ZHU, Huohui OU, Qing ZHU, Linyuan YU, Baitang GUO, Wei LIAO, Yu HUANG, Leyang XIANG, Dinghua YANG. Overexpression of parathyroid hormone-like hormone facilitates hepatocellular carcinoma progression and correlates with adverse outcomes [J]. Journal of Southern Medical University, 2025, 45(10): 2135-2145. |
| [4] | Bihang SUN, Yujun GUO, Yulin QI, Dan YAO, Wenzhi CHEN, Nianzhi CHEN. Low-intensity pulsed ultrasound and oridonin synergistically induce ferroptosis of pancreatic cancer cells by activating PIEZO1 via the Nrf2/HO-1/GPX4 pathway [J]. Journal of Southern Medical University, 2025, 45(10): 2160-2170. |
| [5] | Yaobin WANG, Liuyan CHEN, Yiling LUO, Jiqing SHEN, Sufang ZHOU. Predictive value of NUF2 for prognosis and immunotherapy responses in pan-cancer [J]. Journal of Southern Medical University, 2025, 45(1): 137-149. |
| [6] | Yan HUANG, Lulu QIN, Shaoxing GUAN, Yanping GUANG, Yuru WEI, Ailing CAO, Dongmei LI, Guining WEI, Qibiao SU. Therapeutic mechanism of aqueous extract of Semiliquidambar cathayensis Chang root for pancreatic cancer: the active components, therapeutic targets and pathways [J]. Journal of Southern Medical University, 2024, 44(7): 1336-1344. |
| [7] | Lili CHEN, Tianyu WU, Ming ZHANG, Zixia DING, Yan ZHANG, Yiqing YANG, Jiaqian ZHENG, Xiaonan ZHANG. Identification of potential biomarkers and immunoregulatory mechanisms of rheumatoid arthritis based on multichip co-analysis of GEO database [J]. Journal of Southern Medical University, 2024, 44(6): 1098-1108. |
| [8] | Wen ZHAO, Hejing RUAN, Siyuan WANG, Yuzhe CHENG, Miao LEI, Jiufa ZHAO, Chuanmiao LIU. Inhibiting Yes-associated protein alleviates CCl4 liver fibrosis in mice by reducing epithelial mesenchymal transition [J]. Journal of Southern Medical University, 2024, 44(10): 1839-1849. |
| [9] | Jingting MIN, Shang PENG, Nana DU, Ran AN, Xiangcheng ZHEN, Jiawei CAO, Chenhang ZHOU, Zhenghong LI. Enhanced tumoricidal activity of PD-1 antibody-secreting c-Met CAR-T cells against pancreatic cancer cells [J]. Journal of Southern Medical University, 2024, 44(10): 1976-1984. |
| [10] | YAO Yina, LIU Jia, ZHOU Xiangjun, LIU Zeyu, QIU Shizhen, HE Yingzheng, ZHOU Xueqiong. A pan-cancer analysis of TTC9A expression level and its correlation with prognosis and immune microenvironment [J]. Journal of Southern Medical University, 2024, 44(1): 70-82. |
| [11] | SUN Jingjie, LU Peng, GUAN Shasha, LIU Songsong. Heterogeneity analysis of pancreatic cancer and identification of molecular subtypes of tumor cells based on CEACAM5, LGALS1 and CENPF gene expression [J]. Journal of Southern Medical University, 2023, 43(9): 1567-1576. |
| [12] | YU Zhengtao, LI Jiameng, JIANG Junwen, LI You, LIN Long, XIA Ying, WANG Lei. miRNA-128-3p inhibits malignant behavior of glioma cells by downregulating KLHDC8A expression [J]. Journal of Southern Medical University, 2023, 43(9): 1447-1459. |
| [13] | HUANG Zhuo, ZENG Zhenyu, LI Jia, CAI Rui, HE Wenxia, HU Shuting. High expression of Circ-PALLD in heart failure is transcriptionally regulated by the transcription factor GATA4 [J]. Journal of Southern Medical University, 2023, 43(8): 1371-1378. |
| [14] | GUAN Shenyuan, SHEN Zhiyong, LIN Mingdao, DENG Haijun, FANG Yuan. STIP1 correlates with tumor immune infiltration and prognosis as a potential immunotherapy target: a pan-cancer bioinformatics analysis [J]. Journal of Southern Medical University, 2023, 43(7): 1179-1193. |
| [15] | LIANG Lidu, ZHANG Haojie, LU Qian, ZHOU Chenjie, LI Shulong. Advanced Faster RCNN: a non-contrast CT-based algorithm for detecting pancreatic lesions in multiple disease stages [J]. Journal of Southern Medical University, 2023, 43(5): 755-763. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||