Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (10): 1839-1849.doi: 10.12122/j.issn.1673-4254.2024.10.01
Wen ZHAO( ), Hejing RUAN(
), Hejing RUAN( ), Siyuan WANG, Yuzhe CHENG, Miao LEI, Jiufa ZHAO, Chuanmiao LIU(
), Siyuan WANG, Yuzhe CHENG, Miao LEI, Jiufa ZHAO, Chuanmiao LIU( )
)
Received:2024-04-12
Online:2024-10-20
Published:2024-10-31
Contact:
Chuanmiao LIU
E-mail:zhaowen6220@163.com;2930916898@qq.com;liuchuanmiao119@sina.com
Wen ZHAO, Hejing RUAN, Siyuan WANG, Yuzhe CHENG, Miao LEI, Jiufa ZHAO, Chuanmiao LIU. Inhibiting Yes-associated protein alleviates CCl4 liver fibrosis in mice by reducing epithelial mesenchymal transition[J]. Journal of Southern Medical University, 2024, 44(10): 1839-1849.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.10.01
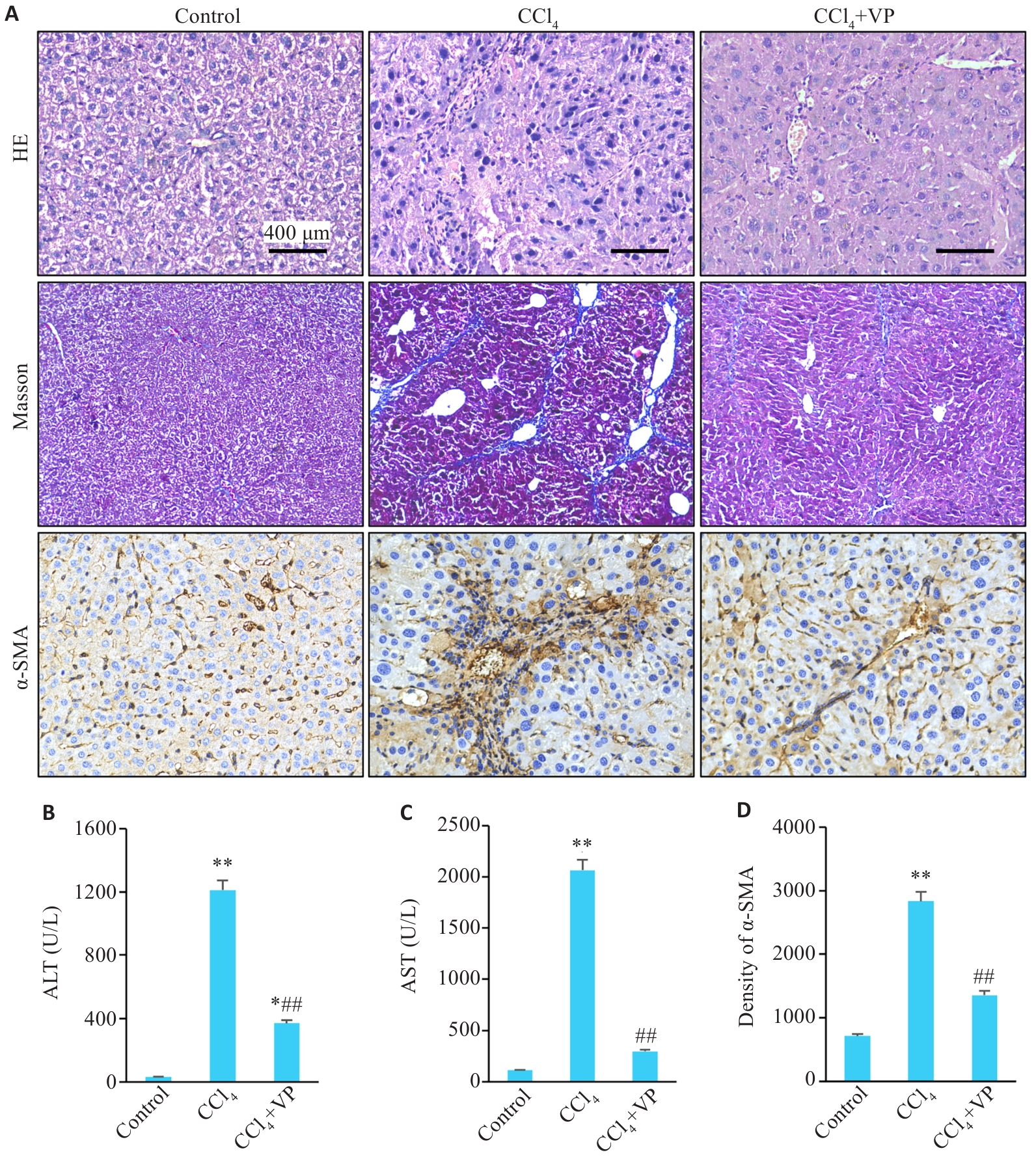
Fig.1 Liver histopathology and serum levels of ALT and AST in mice. A: HE staining (Original magnification: ×200), Masson's trichrome staining (×100), and immunohistochemical staining of α-SMA (×200). B, C: Serum levels of ALT and AST. D: Quantitative data of immunohistochemistry for α-SMA in liver tissues. *P<0.05, **P<0.01 vs control group; ##P<0.01 vs CCl4 group.
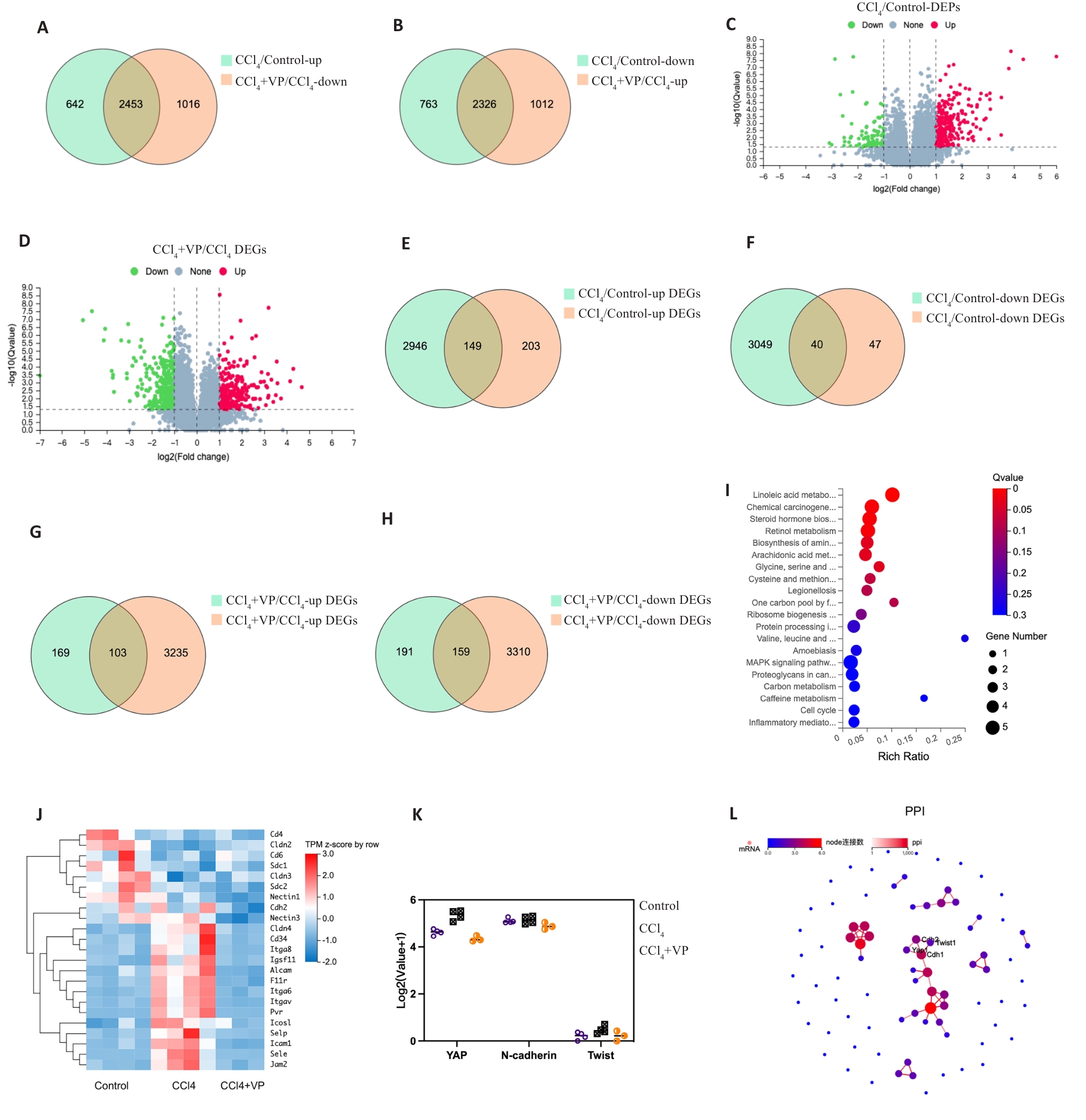
Fig.2 Transcriptomic analysis of and proteomic analysis of differentially expression mRNAs and proteins in the liver tissue of mice. A, B: Venn diagram showing differentially expressed genes (DEGs) between the groups. C, D: Volcano plot showing differentially expressed proteins (DEPs). The red and green dots indicate significantly upregulated and downregulated genes, respectively. E, F: Venn diagram showing differentially expressed mRNAs and proteins between control and CCl4 groups. G, H: Venn diagram showing differentially expressed mRNAs and proteins between the CCl4 and verteporfin (VP) treatment groups. I: Bubble chart of Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the DEGs. J: Heat map of the DEGs of cell adhesion molecules pathway. K: Expressions of YAP, N-cadherin and Twist mRNAs in the 3 groups. L: Protein-protein interaction (PPI) network of the DEGs.
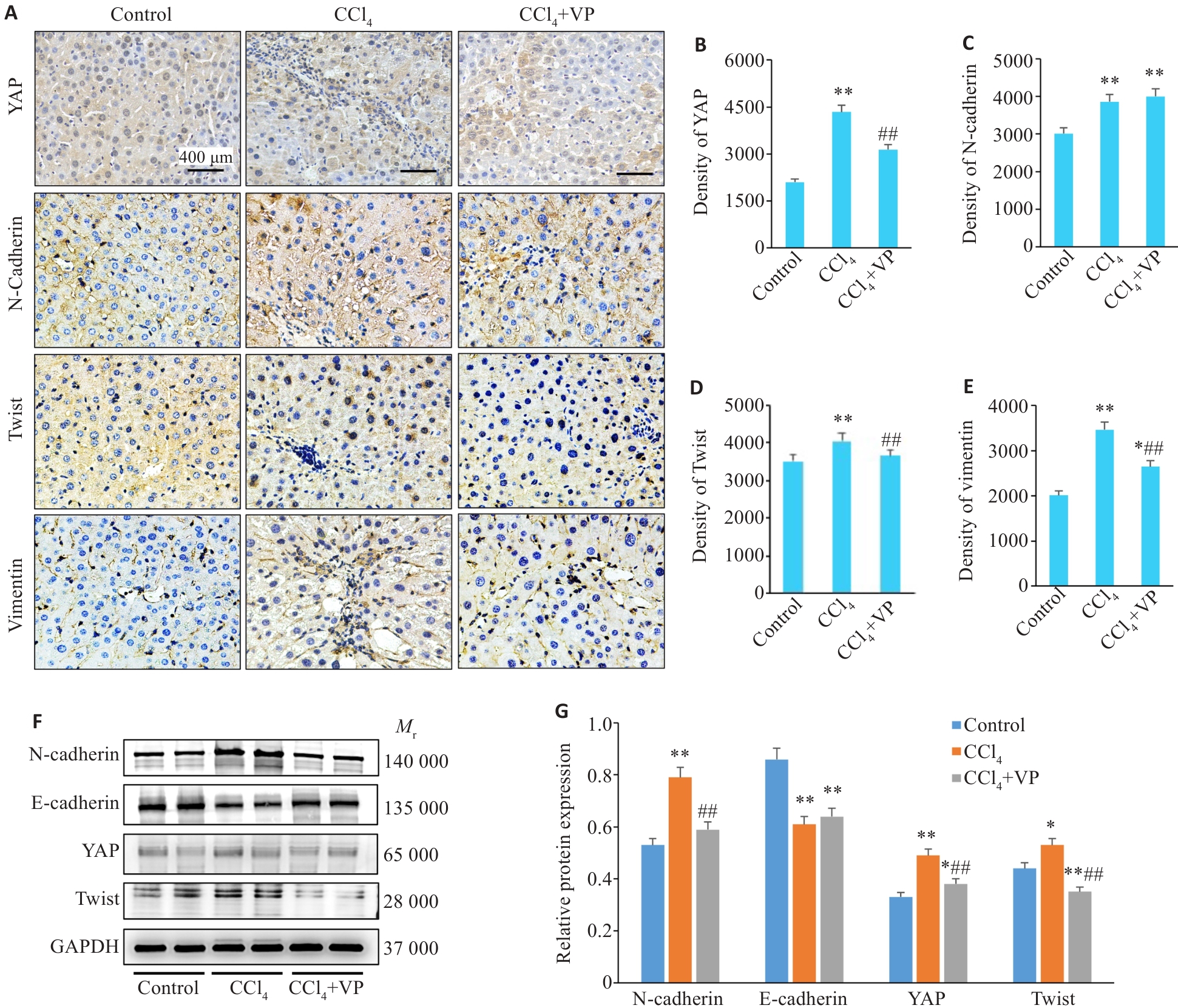
Fig.3 Expression of YAP and EMT-related proteins in the liver tissue of the mice. A: Immunohistochemical staining of YAP, N-cadherin, Twist and vimentin (×400). B-E: Quantitative data of immunohistochemical staining for YAP, N-cadherin, Twist and vimentin. F: Western blotting of hepatic YAP, N-cadherin, E-cadherin and Twist protein expressions. G: Quantification of the protein expression levels. *P<0.05, **P<0.01 vs control group; ##P<0.01 vs CCl4 group.
| Parameters | Healthy controls (n=60) | CHB (n=60) | Liver cirrhosis (n=60) | F/Z/χ2 | P |
|---|---|---|---|---|---|
| Male [n (%)] | 40 (66.7) | 30 (50.0) | 41 (68.3) | 5.217 | 0.074 |
| Age (year) | 50.1±0.5 | 46.6±1.1 | 51.0±1.3 | 5.377 | 0.095 |
| WBC (109/L) | 6.17 (5.38, 7.15) | 5.48 (4.54, 6.56) | 4.21 (2.72, 5.43) | 32.55 | <0.001 |
| PLT (109/L) | 232 (200, 275) | 187 (155, 227) | 89 (48, 134) | 54.95 | <0.001 |
| ALB (g/L) | 45.8 (42.8, 47.4) | 46.5 (44.6, 48.2) | 32.4 (28.0, 36.2) | 104.93 | <0.001 |
| ALT (U/L) | 22 (17, 32) | 18 (15, 25) | 32 (18, 56) | 19.42 | <0.001 |
| AST (U/L) | 21 (18, 25) | 22 (19, 26) | 47 (34., 76) | 72.43 | <0.001 |
| GGT (U/L) | 26 (18, 41.5) | 15 (12, 20) | 47 (28, 109) | 71.92 | <0.001 |
| APRI | 0.24 (0.17, 0.30) | 0.29 (0.23, 0.47) | 1.80 (0.90, 2.89) | 93.61 | <0.001 |
| FIB-4 | 0.96 (0.80, 1.19) | 1.29 (0.96, 1.69) | 5.06 (3.35, 10.02) | 97.86 | <0.001 |
Tab.1 General characteristics of healthy controls and patients with CHB and HBV-related liver cirrhosis
| Parameters | Healthy controls (n=60) | CHB (n=60) | Liver cirrhosis (n=60) | F/Z/χ2 | P |
|---|---|---|---|---|---|
| Male [n (%)] | 40 (66.7) | 30 (50.0) | 41 (68.3) | 5.217 | 0.074 |
| Age (year) | 50.1±0.5 | 46.6±1.1 | 51.0±1.3 | 5.377 | 0.095 |
| WBC (109/L) | 6.17 (5.38, 7.15) | 5.48 (4.54, 6.56) | 4.21 (2.72, 5.43) | 32.55 | <0.001 |
| PLT (109/L) | 232 (200, 275) | 187 (155, 227) | 89 (48, 134) | 54.95 | <0.001 |
| ALB (g/L) | 45.8 (42.8, 47.4) | 46.5 (44.6, 48.2) | 32.4 (28.0, 36.2) | 104.93 | <0.001 |
| ALT (U/L) | 22 (17, 32) | 18 (15, 25) | 32 (18, 56) | 19.42 | <0.001 |
| AST (U/L) | 21 (18, 25) | 22 (19, 26) | 47 (34., 76) | 72.43 | <0.001 |
| GGT (U/L) | 26 (18, 41.5) | 15 (12, 20) | 47 (28, 109) | 71.92 | <0.001 |
| APRI | 0.24 (0.17, 0.30) | 0.29 (0.23, 0.47) | 1.80 (0.90, 2.89) | 93.61 | <0.001 |
| FIB-4 | 0.96 (0.80, 1.19) | 1.29 (0.96, 1.69) | 5.06 (3.35, 10.02) | 97.86 | <0.001 |
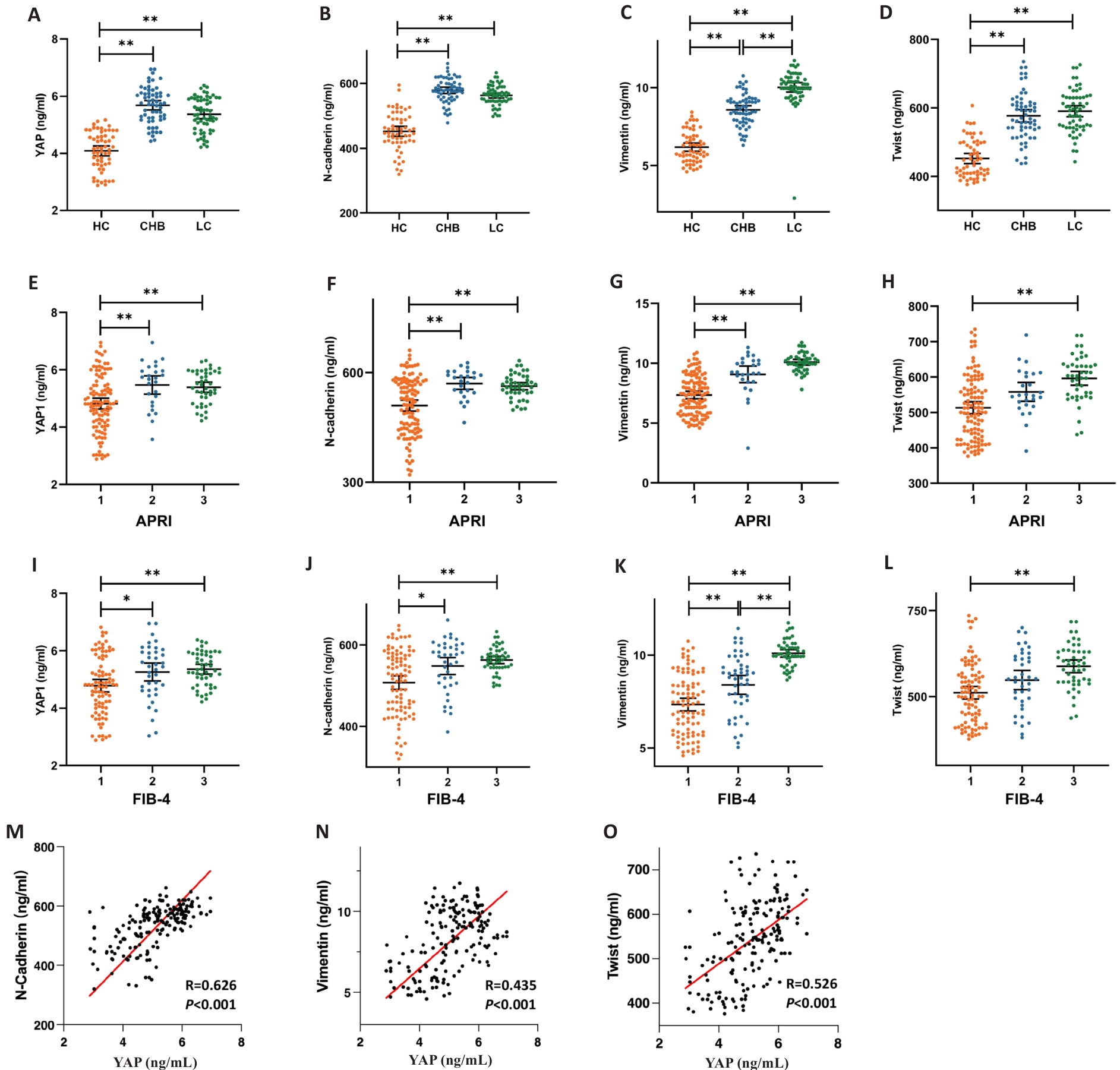
Fig.4 Plasma YAP, N-cadherin, vimentin and Twist levels in healthy controls and patients with CHB and HBV-related liver cirrhosis. A-D: Distribution of plasma YAP, N-cadherin, vimentin and Twist levels in the 3 groups. E-H: Distribution of plasma YAP, N-cadherin, vimentin and Twist levels in patients with APRI <0.5, =0.5-1.5, and >1.5. I-L: Distribution of plasma YAP, N-cadherin, vimentin and Twist levels in patients with FIB-4<1.45, 1.45-3.25, and >3.25. M-O: Correlation analysis of serum YAP level with serum N-cadherin, vimentin and Twist levels. *P<0.05, **P<0.01.
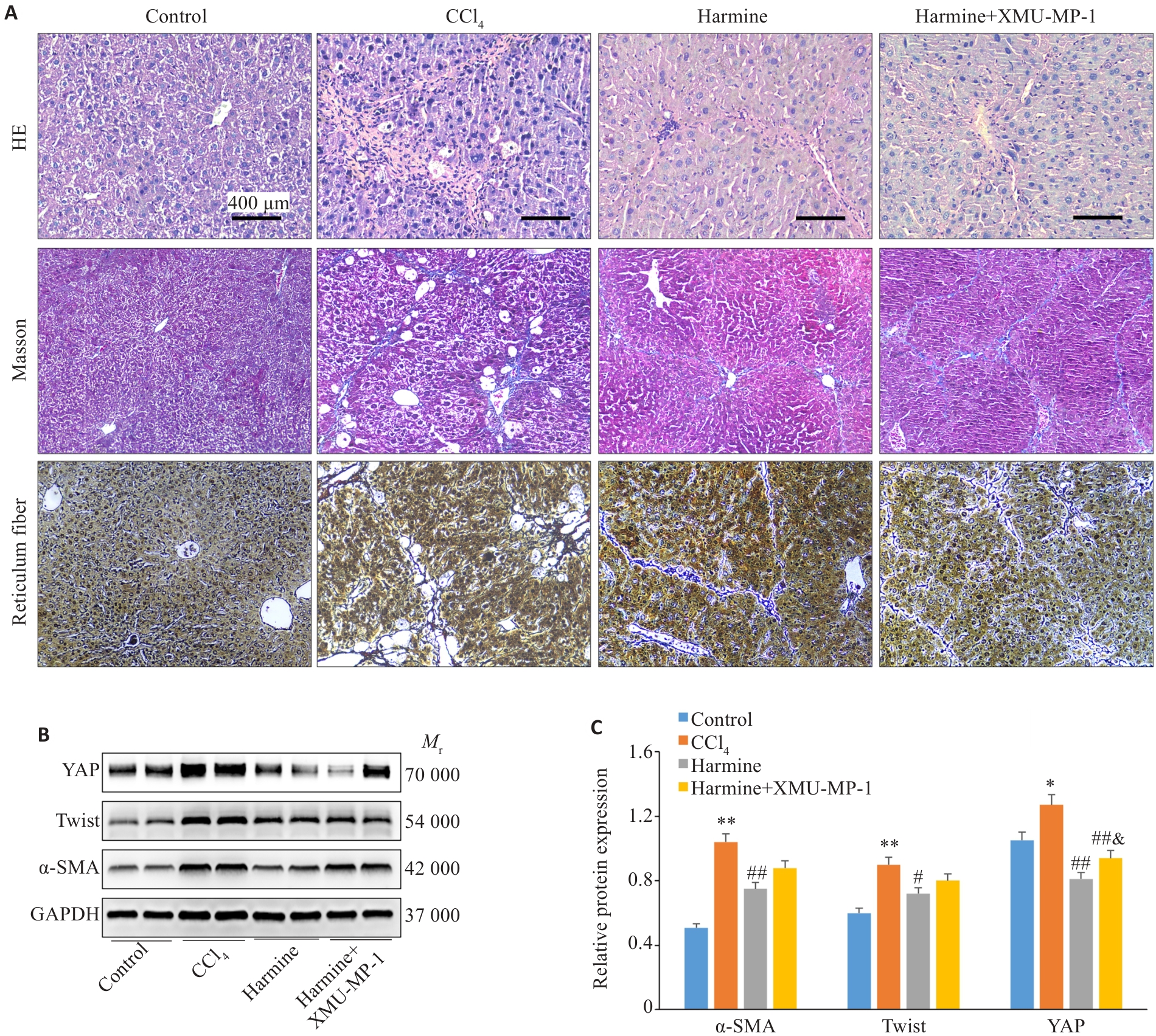
Fig.5 Activation of YAP reverses Twist inhibition-induced improvement of liver fibrosis in mice. A: HE staining (×200), Masson's trichrome staining (×100), and Reticulum fiber staining (×100). B: Western blotting of protein levels of hepatic YAP, Twist, and α-SMA in mice. C: Quantification of protein expression levels. *P<0.05, **P<0.01 vs control group; #P<0.05, ##P<0.01 vs CCl4 group; &P<0.05 vs Harmine group.
| 1 | Devarbhavi H, Asrani SK, Arab JP, et al. Global burden of liver disease: 2023 update[J]. J Hepatol, 2023, 79(2): 516-37. |
| 2 | Ginès P, Krag A, Abraldes JG, et al. Liver cirrhosis[J]. Lancet, 2021, 398(10308): 1359-76. |
| 3 | Younossi ZM, Wong G, Anstee QM, et al. The global burden of liver disease[J]. Clin Gastroenterol Hepatol, 2023, 21(8): 1978-91. |
| 4 | GBD Alcohol Collaborators. Alcohol use and burden for 195 countries and territories, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016[J]. Lancet, 2018, 392(10152): 1015-35. |
| 5 | Zhai MM, Long JH, Liu SS, et al. The burden of liver cirrhosis and underlying etiologies: results from the global burden of disease study 2017[J]. Aging, 2021, 13(1): 279-300. |
| 6 | Allen AM, Kim WR, Moriarty JP, et al. Time trends in the health care burden and mortality of acute on chronic liver failure in the United States[J]. Hepatology, 2016, 64(6): 2165-72. |
| 7 | Fabrellas N, Moreira R, Carol M, et al. Psychological burden of hepatic encephalopathy on patients and caregivers[J]. Clin Transl Gastroenterol, 2020, 11(4): e00159. |
| 8 | Arroyo V, Moreau R, Jalan R. Acute-on-chronic liver failure[J]. N Engl J Med, 2020, 382(22): 2137-45. |
| 9 | Zhang DY, Zhang YG, Sun B. The molecular mechanisms of liver fibrosis and its potential therapy in application[J]. Int J Mol Sci, 2022, 23(20): 12572. |
| 10 | Mannaerts I, Leite SB, Verhulst S, et al. The Hippo pathway effector YAP controls mouse hepatic stellate cell activation[J]. J Hepatol, 2015, 63(3): 679-88. |
| 11 | Martin K, Pritchett J, Llewellyn J, et al. PAK proteins and YAP-1 signalling downstream of integrin beta-1 in myofibroblasts promote liver fibrosis[J]. Nat Commun, 2016, 7: 12502. |
| 12 | Grijalva JL, Huizenga M, Mueller K, et al. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration[J]. Am J Physiol Gastrointest Liver Physiol, 2014, 307(2): G196-G204. |
| 13 | Machado MV, Michelotti GA, Pereira TA, et al. Accumulation of duct cells with activated YAP parallels fibrosis progression in non-alcoholic fatty liver disease[J]. J Hepatol, 2015, 63(4): 962-70. |
| 14 | Piersma B, Bank RA, Boersema M. Signaling in fibrosis: TGF-β, WNT, and YAP/TAZ converge[J]. Front Med, 2015, 2: 59. |
| 15 | Swiderska-Syn M, Xie GH, Michelotti GA, et al. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver[J]. Hepatology, 2016, 64(1): 232-44. |
| 16 | Grannas K, Arngården L, Lönn P, et al. Crosstalk between hippo and TGFβ: subcellular localization of YAP/TAZ/smad complexes[J]. J Mol Biol, 2015, 427(21): 3407-15. |
| 17 | Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications[J]. Mol Cancer, 2016, 15: 18. |
| 18 | Xie GH, Diehl AM. Evidence for and against epithelial-to-mesenchymal transition in the liver[J]. Am J Physiol Gastrointest Liver Physiol, 2013, 305(12): G881-90. |
| 19 | Zhao W, Zhang XX, Hou MM, et al. Traditional Chinese medicine Yiqi Huoxue recipe attenuates hepatic fibrosis via YAP/TAZ signaling[J]. Histol Histopathol, 2021, 36(9): 967-79. |
| 20 | Zhao W, Lei M, Li JF, et al. Yes-associated protein inhibition ameliorates liver fibrosis and acute and chronic liver failure by decreasing ferroptosis and necroptosis[J]. Heliyon, 2023, 9(4): e15075. |
| 21 | Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities[J]. Front Med, 2018, 12(4): 361-73. |
| 22 | Mizutani A, Koinuma D, Tsutsumi S, et al. Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4alpha in HepG2 cells[J]. J Biol Chem, 2011, 286(34): 29848-60. |
| 23 | Syn WK, Choi SS, Liaskou E, et al. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis[J]. Hepatology, 2011, 53(1): 106-15. |
| 24 | Xie GH, Karaca G, Swiderska-Syn M, et al. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice[J]. Hepatology, 2013, 58(5): 1801-13. |
| 25 | Xu XC, Zhang Y, Wang X, et al. Substrate stiffness drives epithelial to mesenchymal transition and proliferation through the NEAT1-wnt/β-catenin pathway in liver cancer[J]. Int J Mol Sci, 2021, 22(21): 12066. |
| 26 | Zhang K, Zhang MX, Yao QB, et al. The hepatocyte-specifically expressed lnc-HSER alleviates hepatic fibrosis by inhibiting hepatocyte apoptosis and epithelial-mesenchymal transition[J]. Theranostics, 2019, 9(25): 7566-82. |
| 27 | Ma SH, Meng ZP, Chen R, et al. The hippo pathway: biology and pathophysiology[J]. Annu Rev Biochem, 2019, 88: 577-604. |
| 28 | Zhan YT, Tao QQ, Meng QS, et al. LncRNA-MIAT activates hepatic stellate cells via regulating Hippo pathway and epithelial-to-mesenchymal transition[J]. Commun Biol, 2023, 6(1): 285. |
| 29 | Oh SH, Swiderska-Syn M, Jewell ML, et al. Liver regeneration requires Yap1-TGFβ-dependent epithelial-mesenchymal transition in hepatocytes[J]. J Hepatol, 2018, 69(2): 359-67. |
| 30 | Ge WS, Wang YJ, Wu JX, et al. β-catenin is overexpressed in hepatic fibrosis and blockage of Wnt/β‑catenin signaling inhibits hepatic stellate cell activation[J]. Mol Med Rep, 2014, 9(6): 2145-51. |
| 31 | Dong WH, Kong M, Zhu YW, et al. Activation of TWIST transcription by chromatin remodeling protein BRG1 contributes to liver fibrosis in mice[J]. Front Cell Dev Biol, 2020, 8: 340. |
| 32 | Yang XX, Ma LP, Wei R, et al. Twist1-induced miR-199a-3p promotes liver fibrosis by suppressing caveolin-2 and activating TGF-β pathway[J]. Signal Transduct Target Ther, 2020, 5(1): 75. |
| 33 | Luo XF, Zhang R, Schefczyk S, et al. Nuclear translocation of YAP drives BMI-associated hepatocarcinogenesis in hepatitis B virus infection[J]. Liver Int, 2023, 43(9): 2002-16. |
| 34 | Abdallah RA, Shaban MI, Taie DM, et al. Relation between immunohistochemical expression of hippo pathway effectors and chronic hepatitis induced fibrosis in Egyptian patients[J]. Turk Patoloji Derg, 2020, 36(1): 48-63. |
| 35 | Zhou ZX, Zhang RR, Li XM, et al. Circular RNA cVIM promotes hepatic stellate cell activation in liver fibrosis via miR-122-5p/miR-9-5p-mediated TGF-β signaling cascade[J]. Commun Biol, 2024, 7(1): 113. |
| [1] | Yuming ZHANG, Shicheng XIA, Linlin ZHANG, Mengxi CHEN, Xiaojing LIU, Qin GAO, Hongwei YE. Protective effect of Lonicerae japonicae flos extract against doxorubicin-induced liver injury in mice [J]. Journal of Southern Medical University, 2024, 44(8): 1571-1581. |
| [2] | Lili CHEN, Tianyu WU, Ming ZHANG, Zixia DING, Yan ZHANG, Yiqing YANG, Jiaqian ZHENG, Xiaonan ZHANG. Identification of potential biomarkers and immunoregulatory mechanisms of rheumatoid arthritis based on multichip co-analysis of GEO database [J]. Journal of Southern Medical University, 2024, 44(6): 1098-1108. |
| [3] | CAO Jiafan, SUN Yue, DING Xin, LI Shengwen, CHEN Bo, LAN Tian. Arbutin ameliorates liver fibrosis in mice by inhibiting macrophage recruitment and regulating the Akt/NF-κB and Smad signaling pathways [J]. Journal of Southern Medical University, 2024, 44(4): 652-659. |
| [4] | XU Xiaohui, FENG Jinmei, LUO Ying, HE Xinyu, ZANG Jinbao, HUANG Daochao. Adeno-associated virus-mediated hepatocyte-specific NDUFA13 overexpression protects against CCl4-induced liver fibrosis in mice by inhibiting hepatic NLRP3 activation [J]. Journal of Southern Medical University, 2024, 44(2): 201-209. |
| [5] | LIU Xuerou, YANG Yumei, CAI Hui, ZHANG Yaoshuai, FAN Fangtian, LI Xian, LI Shanshan. Aumolertinib inhibits proliferation, invasion and migration and promotes apoptosis of neuroblastoma cells by downregulating MMP2 and MMP9 expression [J]. Journal of Southern Medical University, 2023, 43(9): 1493-1499. |
| [6] | LIN Jiayi, LOU Anni, LI Xu. Lipopolysaccharide stimulates macrophages to secrete exosomes containing miR-155-5p to promote activation and migration of hepatic stellate cells [J]. Journal of Southern Medical University, 2023, 43(6): 994-1001. |
| [7] | CHENG Yang, HE Xuxu, WANG Lian, XU Yibo, SHEN Mengdi, ZHANG Wenjing, XIA Yongsheng, ZHANG Jie, ZHANG Min, WANG Yijun, HU Jianguo, ZHANG Jun. HSDL2 overexpression promotes rectal cancer progression by regulating cancer cell cycle and promoting cell proliferation [J]. Journal of Southern Medical University, 2023, 43(4): 544-551. |
| [8] | WANG Xuancheng, ZHU Yifan, ZHOU Hailin, HUANG Zongsheng, CHEN Hongwei, ZHANG Jiahao, YANG Shanyi, CHEN Guanghui, ZHANG Qisong. Integrated analysis of serum untargeted metabolomics and targeted bile acid metabolomics for identification of diagnostic biomarkers for colorectal cancer [J]. Journal of Southern Medical University, 2023, 43(3): 443-453. |
| [9] | YANG Xuejia, LI Yujie, WU Dengqiang, MA Yili, ZHOU Sufang. Screening and identification of key genes ATP1B3 and ENAH in the progression of hepatocellular carcinoma: based on data mining and clinical validation [J]. Journal of Southern Medical University, 2022, 42(6): 815-823. |
| [10] | HAN Yipeng, LU Xiaoxi, LAI Weinan, LIANG Renge, YANG Min, OUYANG Qingqing. Identification of serological biomarkers for diagnosis of rheumatoid arthritis using a protein array-based approach [J]. Journal of Southern Medical University, 2022, 42(5): 733-739. |
| [11] | ZHAN Weijie, YAN Tao, GAO Jiawen, SONG Minkai, WANG Ting, LIN Fei, ZHOU Haiyu, LI Li, ZHANG Chao. Role of circular RNAs in immune-related diseases [J]. Journal of Southern Medical University, 2022, 42(2): 163-170. |
| [12] | DENG Ya, WANG Chunyan, FU Yiming, LI Zhongbin, JI Dong. A high relapse risk of chronic drug-induced liver injury is correlated with a greater severity of liver fibrosis [J]. Journal of Southern Medical University, 2022, 42(11): 1655-1661. |
| [13] | ZHAO Chenling, DONG Ting, SUN Lunyan, HU Huibing, WANG Qiong, TIAN Liwei, JIANG Zhangsheng. Establishment and validation of a predictive nomogram for liver fibrosis in patients with Wilson disease and abnormal lipid metabolism [J]. Journal of Southern Medical University, 2022, 42(11): 1720-1725. |
| [14] | ZHAO Zhibin, DONG Hui, LI Binghang, SHEN Bo, GUO Yuecheng, GU Tianyi, QU Ying, CAI Xiaobo, LU Lungen. Hydroxynitone suppresses hepatic stellate cell activation by inhibiting TGF-β1 phosphorylation to alleviate CCl4-induced liver fibrosis in rats [J]. Journal of Southern Medical University, 2022, 42(10): 1511-1516. |
| [15] | ZHANG Chunyan, YAN Yuxin, GAO Xiaoyang, MA Yuehong. Therapeutic mechanism of the Mongolian medicine Qiwei Qinggan Powder against liver fibrosis based on UHPLC-TOF-MS combined with network pharmacological methods [J]. Journal of Southern Medical University, 2021, 41(8): 1131-1141. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||