南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (12): 2756-2766.doi: 10.12122/j.issn.1673-4254.2025.12.23
• • 上一篇
喻珍妮1( ), 高竟哲1, 孙惠1, 冯芹2, 那效旗1, 张宁1(
), 高竟哲1, 孙惠1, 冯芹2, 那效旗1, 张宁1( ), 沈昆双1, 王媛媛1, 王喜军1(
), 沈昆双1, 王媛媛1, 王喜军1( )
)
收稿日期:2025-04-14
出版日期:2025-12-20
发布日期:2025-12-22
通讯作者:
张宁,王喜军
E-mail:yzhennni@163.com;zhangning0454@163.com;xijunw@sina.com
作者简介:喻珍妮,在读硕士研究生,E-mail: yzhennni@163.com
基金资助:
Zhenni YU1( ), Jingzhe GAO1, Hui SUN1, Qin Feng2, Xiaoqi NA1, Ning ZHANG1(
), Jingzhe GAO1, Hui SUN1, Qin Feng2, Xiaoqi NA1, Ning ZHANG1( ), Kungshuang SHEN1, Yuanyuan WANG1, Xijun WANG1(
), Kungshuang SHEN1, Yuanyuan WANG1, Xijun WANG1( )
)
Received:2025-04-14
Online:2025-12-20
Published:2025-12-22
Contact:
Ning ZHANG, Xijun WANG
E-mail:yzhennni@163.com;zhangning0454@163.com;xijunw@sina.com
摘要:
目的 采用孟德尔随机化(MR)方法探讨肠道菌群、T细胞功能与结直肠癌(CRC)风险之间的因果关系。 方法 从MiBioGen数据库收集肠道菌群,T细胞和结直肠癌数据从公开的GWAS数据中获得,对三者进行双样本MR分析。将逆方差加权法作为主分析方法,同时采用MR-Egger、加权中位数法(Weighted median)、简单模式法(Simple mode)、加权模式法(Weighted mode)作为补充,MR-PRESSO和MR-Egger回归方法来检测水平多效性,通过Cochran's Q检验来识别异质性,采用留一法进行敏感性分析。 结果 在肠道菌群与T细胞的正向MR分析中,有11种肠道菌群存在因果关系,其中有6种肠道菌与T细胞是正相关(Prevotella7属:P=0.003;Ruminococcaceae UCG011属:P=0.033;Ruminococcaceae UCG004属:P=0.010;Eubacterium brachy group属:P=0.005;Lachnospiraceae FCS020 group属:P=0.028;Coprobacter属:P=0.033),另外5种肠道菌呈负相关;在T细胞与结直肠癌的正向MR分析中,发现CD25++CD45RA-CD4非调节性T细胞与结直肠癌风险呈负相关(IVW: OR=0.935,95% CI:0.878~0.995,P=0.035);在肠道菌群与结直肠癌的正向MR分析中,有11种肠道菌群存在因果关系,其中有6种肠道菌与结直肠癌是正相关(Eubacterium xylanophilum group属:P=0.039;Selenomonadales目:P=0.014;Negativicutes纲:P=0.014;Bifidobacteriaceae科:P=0.048;Bifidobacteriales目:P=0.048;Coprococcus1属:P=0.033),另外5种肠道菌呈负相关。 结论 在肠道菌群、T细胞和结直肠癌三者关系中,Coprobacter属和Eubacterium xylanophilum group属是共有的菌,Eubacterium xylanophilum group属菌可通过促进CD25++ CD45RA- CD4非调节性T细胞的活性从而抑制结直肠癌的发展;而Coprobacter属菌可导致CD25++CD45RA-CD4非调节性T细胞失活从而使结直肠癌恶化。
喻珍妮, 高竟哲, 孙惠, 冯芹, 那效旗, 张宁, 沈昆双, 王媛媛, 王喜军. 肠道菌群、T细胞在结直肠癌发病中的因果关联:孟德尔随机化分析[J]. 南方医科大学学报, 2025, 45(12): 2756-2766.
Zhenni YU, Jingzhe GAO, Hui SUN, Qin Feng, Xiaoqi NA, Ning ZHANG, Kungshuang SHEN, Yuanyuan WANG, Xijun WANG. Causal relationship between gut microbiota and T cell subsets in the development of colorectal cancer: a Mendelian randomization analysis[J]. Journal of Southern Medical University, 2025, 45(12): 2756-2766.
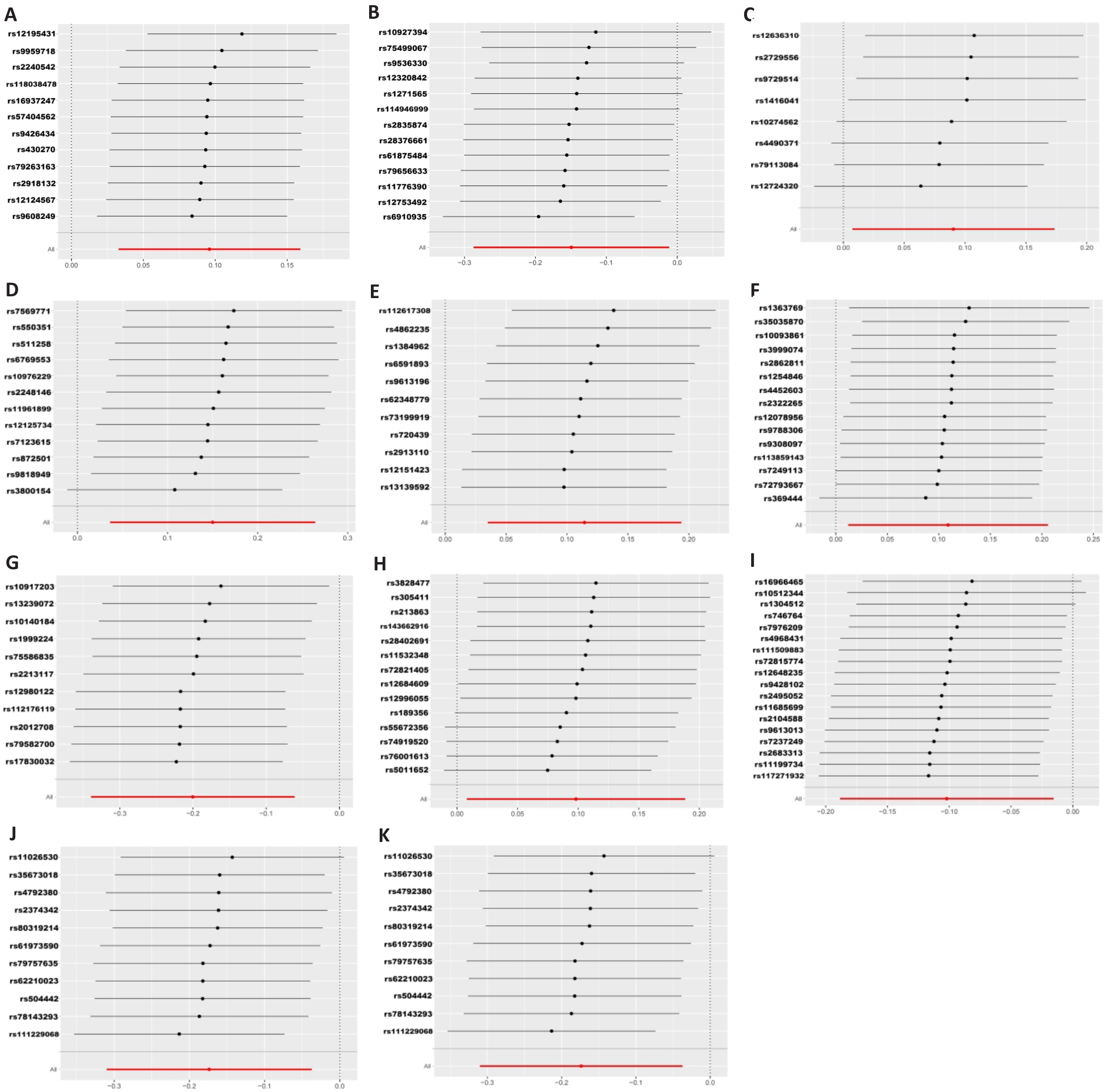
图2 孟德尔随机化分析下肠道菌群与结直肠癌之间的留一图
Fig.2 Leave-one-out plot of the association between gut microbiota and colorectal cancer under Mendelian randomization analysis. A: Prevotella7. B: Faecalibacterium. C: Ruminococcaceae UCG011. D: Ruminococcaceae UCG004. E: Eubacterium brachy group. F: Lachnospiraceae FCS020 group. G: Eubacterium xylanophilum group. H: Coprobacter. I: Prevotella9. J: Enterobacteriaceae. K: Enterobacteriales.
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Prevotella7 | 22 | 0.076 | 0.944 | 1.005 | 0.866-1.1167 |
| genus.Faecalibacterium | 23 | 0.040 | 0.216 | 1.051 | 0.972-1.136 |
| genus.Ruminococcaceae UCG011 | 22 | 0.057 | 0.347 | 0.948 | 0.848-1.06 |
| genus.Ruminococcaceae UCG004 | 23 | 0.036 | 0.406 | 0.970 | 0.904-1.042 |
| genus.Eubacterium brachy group | 22 | 0.054 | 0.734 | 1.019 | 0.916-1.132 |
| genus.Lachnospiraceae FCS020 group | 23 | 0.033 | 0.427 | 0.974 | 0.913-1.039 |
| genus.Eubacterium xylanophilum group | 23 | 0.035 | 0.774 | 0.990 | 0.924-1.061 |
| genus.Coprobacter | 23 | 0.052 | 0.468 | 1.038 | 0.938-1.149 |
| genus.Prevotella9 | 23 | 0.039 | 0.835 | 1.008 | 0.934-1.088 |
| family.Enterobacteriaceae | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| order.Enterobacteriales | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
表1 肠道菌群与结直肠癌的孟德尔随机化逆方差加权法分析结果
Tab.1 Results of inverse variance weighted method analysis in Mendelian randomization study on gut microbiota and colorectal cancer
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Prevotella7 | 22 | 0.076 | 0.944 | 1.005 | 0.866-1.1167 |
| genus.Faecalibacterium | 23 | 0.040 | 0.216 | 1.051 | 0.972-1.136 |
| genus.Ruminococcaceae UCG011 | 22 | 0.057 | 0.347 | 0.948 | 0.848-1.06 |
| genus.Ruminococcaceae UCG004 | 23 | 0.036 | 0.406 | 0.970 | 0.904-1.042 |
| genus.Eubacterium brachy group | 22 | 0.054 | 0.734 | 1.019 | 0.916-1.132 |
| genus.Lachnospiraceae FCS020 group | 23 | 0.033 | 0.427 | 0.974 | 0.913-1.039 |
| genus.Eubacterium xylanophilum group | 23 | 0.035 | 0.774 | 0.990 | 0.924-1.061 |
| genus.Coprobacter | 23 | 0.052 | 0.468 | 1.038 | 0.938-1.149 |
| genus.Prevotella9 | 23 | 0.039 | 0.835 | 1.008 | 0.934-1.088 |
| family.Enterobacteriaceae | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| order.Enterobacteriales | 23 | 0.032 | 0.987 | 1.001 | 0.940-1.065 |
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 3 | 0.032 | 0.035 | 0.935 | 0.878-0.995 |
| Weighted median | 3 | 0.031 | 0.006 | 0.919 | 0.866-0.976 |
| MR-Egger | 3 | 0.107 | 0.566 | 0.917 | 0.743-1.131 |
| Simple mode | 3 | 0.044 | 0.176 | 0.913 | 0.837-0.996 |
| Weighted mode | 3 | 0.041 | 0.144 | 0.909 | 0.839-0.985 |
表2 T细胞与结直肠癌的孟德尔随机化分析结果
Tab.2 Results of Mendelian randomization analysis for T Cells and colorectal cancer
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 3 | 0.032 | 0.035 | 0.935 | 0.878-0.995 |
| Weighted median | 3 | 0.031 | 0.006 | 0.919 | 0.866-0.976 |
| MR-Egger | 3 | 0.107 | 0.566 | 0.917 | 0.743-1.131 |
| Simple mode | 3 | 0.044 | 0.176 | 0.913 | 0.837-0.996 |
| Weighted mode | 3 | 0.041 | 0.144 | 0.909 | 0.839-0.985 |
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 25 | 0.056 | 0.195 | 1.076 | 0.963-1.201 |
| Weighted median | 25 | 0.079 | 0.142 | 1.123 | 0.962-1.312 |
| MR-Egger | 25 | 0.231 | 0.838 | 0.953 | 0.607-1.499 |
| Simple mode | 25 | 0.140 | 0.277 | 1.168 | 0.888-1.536 |
| Weighted mode | 25 | 0.124 | 0.222 | 1.168 | 0.916-1.489 |
表3 结直肠癌与T细胞的孟德尔随机化分析结果
Tab.3 Results of Mendelian randomization analysis for colorectal cancer and T cells
| MR method | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| IVW | 25 | 0.056 | 0.195 | 1.076 | 0.963-1.201 |
| Weighted median | 25 | 0.079 | 0.142 | 1.123 | 0.962-1.312 |
| MR-Egger | 25 | 0.231 | 0.838 | 0.953 | 0.607-1.499 |
| Simple mode | 25 | 0.140 | 0.277 | 1.168 | 0.888-1.536 |
| Weighted mode | 25 | 0.124 | 0.222 | 1.168 | 0.916-1.489 |
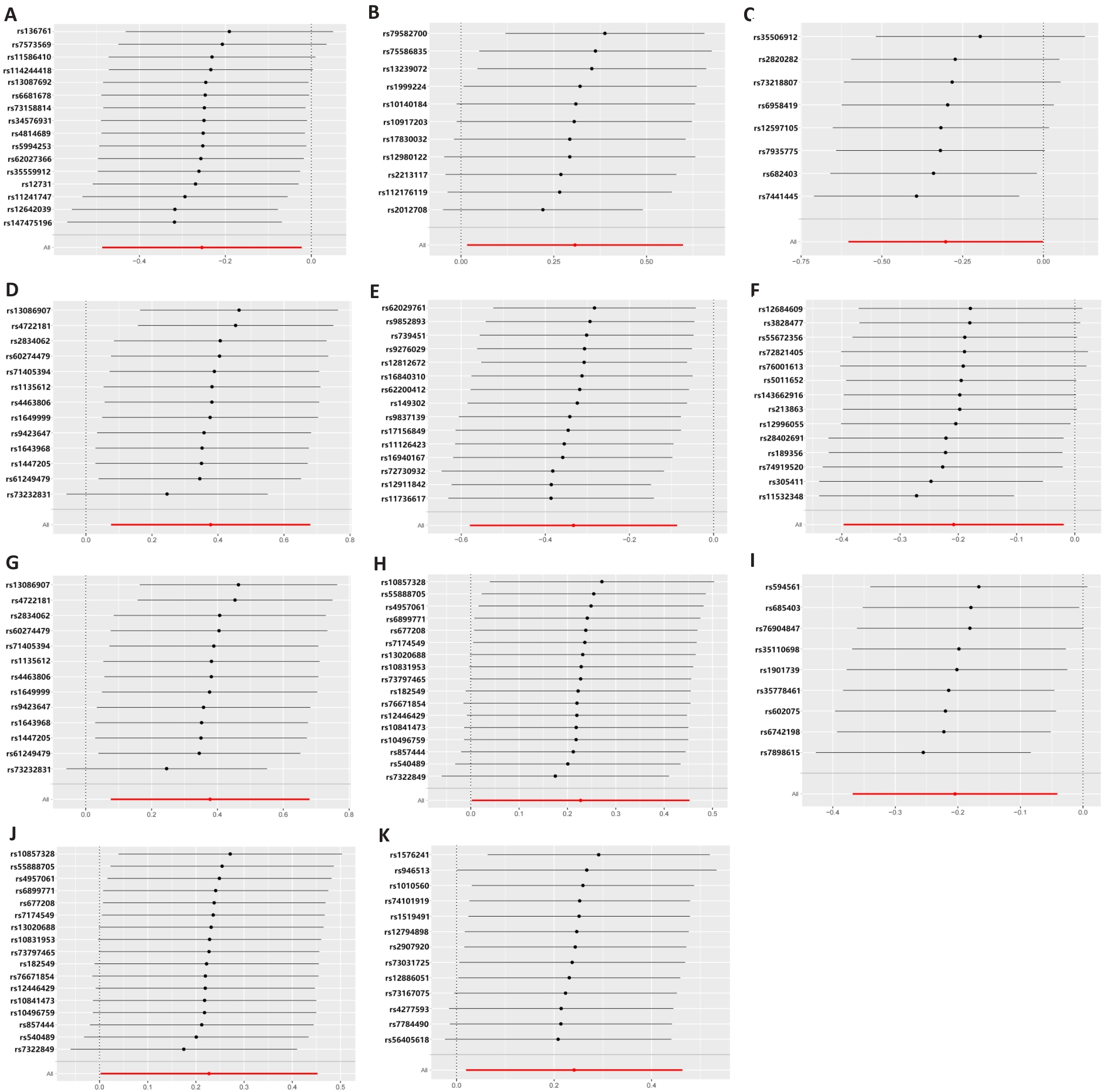
图5 孟德尔随机化分析下肠道菌群与T细胞之间的留一图
Fig.5 Leave-one-out plot of the association between gut microbiota and T cells under Mendelian randomization analysis.A: Ruminococcaceae NK4A214 group. B: Eubacterium xylanophilum group. C: Ruminococcaceae UCG010. D: Selenomonadales. E: XIII AD3011 group. F: Coprobacter. G: Negativicutes. H: Bifidobacteriaceae. I: Allisonella. J: Bifidobacteriales. K: Coprococcus1.
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Ruminococcaceae NK4A214 group | 5 | 0.028 | 0.947 | 0.998 | 0.945-1.054 |
| genus.Eubacterium xylanophilum group | 5 | 0.030 | 0.321 | 0.971 | 0.916-1.029 |
| genus.Ruminococcaceae UCG010 | 5 | 0.037 | 0.527 | 0.977 | 0.909-1.050 |
| order.Selenomonadales | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| genus.Family XIII AD3011 group | 5 | 0.029 | 0.762 | 1.009 | 0.953-1.067 |
| genus.Coprobacter | 5 | 0.069 | 0.843 | 1.014 | 0.885-1.162 |
| Class.Negativicutes | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| family.Bifidobacteriaceae | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Allisonella | 3 | 0.122 | 0.028 | 0.765 | 0.603-0.971 |
| order.Bifidobacteriales | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Coprococcus1 | 5 | 0.029 | 0.892 | 0.996 | 0.942-1.054 |
表4 T细胞与肠道菌群的孟德尔随机化逆方差加权法分析结果
Tab.4 Results of inverse variance weighted method analysis in Mendelian randomization study on colorectal cancer and T cells
| Outcome | nSNP | SE | P | OR | 95% CI |
|---|---|---|---|---|---|
| genus.Ruminococcaceae NK4A214 group | 5 | 0.028 | 0.947 | 0.998 | 0.945-1.054 |
| genus.Eubacterium xylanophilum group | 5 | 0.030 | 0.321 | 0.971 | 0.916-1.029 |
| genus.Ruminococcaceae UCG010 | 5 | 0.037 | 0.527 | 0.977 | 0.909-1.050 |
| order.Selenomonadales | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| genus.Family XIII AD3011 group | 5 | 0.029 | 0.762 | 1.009 | 0.953-1.067 |
| genus.Coprobacter | 5 | 0.069 | 0.843 | 1.014 | 0.885-1.162 |
| Class.Negativicutes | 5 | 0.037 | 0.483 | 0.974 | 0.906-1.048 |
| family.Bifidobacteriaceae | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Allisonella | 3 | 0.122 | 0.028 | 0.765 | 0.603-0.971 |
| order.Bifidobacteriales | 5 | 0.029 | 0.675 | 0.988 | 0.934-1.045 |
| genus.Coprococcus1 | 5 | 0.029 | 0.892 | 0.996 | 0.942-1.054 |
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| genus.Prevotella7 | CRC | MR-Rgger | 7.239 | 10 | 0.703 |
| genus.Prevotella7 | CRC | IVW | 9.043 | 11 | 0.618 |
| genus.Faecalibacterium | CRC | MR-Rgger | 13.261 | 11 | 0.277 |
| genus.Faecalibacterium | CRC | IVW | 13.262 | 12 | 0.350 |
| genus.Ruminococcaceae UCG011 | CRC | MR-Rgger | 6.409 | 6 | 0.379 |
| genus.Ruminococcaceae UCG011 | CRC | IVW | 6.820 | 7 | 0.448 |
| genus.Ruminococcaceae UCG004 | CRC | MR-Rgger | 10.434 | 10 | 0.403 |
| genus.Ruminococcaceae UCG004 | CRC | IVW | 11.115 | 11 | 0.434 |
| genus.Eubacterium brachy group | CRC | MR-Rgger | 9.150 | 9 | 0.424 |
| genus.Eubacterium brachy group | CRC | IVW | 9.264 | 10 | 0.507 |
| genus.Lachnospiraceae FCS020 group | CRC | MR-Rgger | 6.834 | 13 | 0.910 |
| genus.Lachnospiraceae FCS020 group | CRC | IVW | 6.848 | 14 | 0.940 |
| genus.Eubacterium xylanophilum group | CRC | MR-Rgger | 4.699 | 9 | 0.860 |
| genus.Eubacterium xylanophilum group | CRC | IVW | 7.284 | 10 | 0.698 |
| genus.Coprobacter | CRC | MR-Rgger | 15.480 | 12 | 0.216 |
| genus.Coprobacter | CRC | IVW | 15.643 | 13 | 0.269 |
| genus.Prevotella9 | CRC | MR-Rgger | 16.705 | 16 | 0.405 |
| genus.Prevotella9 | CRC | IVW | 16.715 | 17 | 0.474 |
| family.Enterobacteriaceae | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| family.Enterobacteriaceae | CRC | IVW | 9.399 | 10 | 0.495 |
| order.Enterobacteriales | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| order.Enterobacteriales | CRC | IVW | 9.399 | 10 | 0.495 |
| T Cells | CRC | MR-Rgger | 3.227 | 1 | 0.072 |
| T Cells | CRC | IVW | 3.356 | 2 | 0.187 |
| genus.Ruminococcaceae NK4A214 group | T Cells | MR-Rgger | 13.403 | 14 | 0.495 |
| genus.Ruminococcaceae NK4A214 group | T Cells | IVW | 14.064 | 15 | 0.521 |
| genus.Eubacterium xylanophilum group | T Cells | MR-Rgger | 12.167 | 9 | 0.204 |
| genus.Eubacterium xylanophilum group | T Cells | IVW | 12.770 | 10 | 0.237 |
| genus.Ruminococcaceae UCG010 | T Cells | MR-Rgger | 5.807 | 6 | 0.445 |
| genus.Ruminococcaceae UCG010 | T Cells | IVW | 6.325 | 7 | 0.502 |
| order.Selenomonadales | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| order.Selenomonadales | T Cells | IVW | 13.547 | 12 | 0.331 |
| genus.Family XIII AD3011 group | T Cells | MR-Rgger | 16.241 | 13 | 0.236 |
| genus.Family XIII AD3011 group | T Cells | IVW | 16.387 | 14 | 0.290 |
| genus.Coprobacter | T Cells | MR-Rgger | 17.482 | 12 | 0.132 |
| genus.Coprobacter | T Cells | IVW | 17.762 | 13 | 0.167 |
| Class.Negativicutes | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| Class.Negativicutes | T Cells | IVW | 13.547 | 12 | 0.331 |
| family.Bifidobacteriaceae | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| family.Bifidobacteriaceae | T Cells | IVW | 7.605 | 16 | 0.960 |
| genus.Allisonella | T Cells | MR-Rgger | 5.460 | 7 | 0.604 |
| genus.Allisonella | T Cells | IVW | 6.660 | 8 | 0.574 |
| order.Bifidobacteriales | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| order.Bifidobacteriales | T Cells | IVW | 7.605 | 16 | 0.960 |
表5 肠道菌群、T细胞与结直肠癌的异质性检验正向结果
Tab.5 Heterogeneity test for gut microbiota, T cells, and colorectal cancer
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| genus.Prevotella7 | CRC | MR-Rgger | 7.239 | 10 | 0.703 |
| genus.Prevotella7 | CRC | IVW | 9.043 | 11 | 0.618 |
| genus.Faecalibacterium | CRC | MR-Rgger | 13.261 | 11 | 0.277 |
| genus.Faecalibacterium | CRC | IVW | 13.262 | 12 | 0.350 |
| genus.Ruminococcaceae UCG011 | CRC | MR-Rgger | 6.409 | 6 | 0.379 |
| genus.Ruminococcaceae UCG011 | CRC | IVW | 6.820 | 7 | 0.448 |
| genus.Ruminococcaceae UCG004 | CRC | MR-Rgger | 10.434 | 10 | 0.403 |
| genus.Ruminococcaceae UCG004 | CRC | IVW | 11.115 | 11 | 0.434 |
| genus.Eubacterium brachy group | CRC | MR-Rgger | 9.150 | 9 | 0.424 |
| genus.Eubacterium brachy group | CRC | IVW | 9.264 | 10 | 0.507 |
| genus.Lachnospiraceae FCS020 group | CRC | MR-Rgger | 6.834 | 13 | 0.910 |
| genus.Lachnospiraceae FCS020 group | CRC | IVW | 6.848 | 14 | 0.940 |
| genus.Eubacterium xylanophilum group | CRC | MR-Rgger | 4.699 | 9 | 0.860 |
| genus.Eubacterium xylanophilum group | CRC | IVW | 7.284 | 10 | 0.698 |
| genus.Coprobacter | CRC | MR-Rgger | 15.480 | 12 | 0.216 |
| genus.Coprobacter | CRC | IVW | 15.643 | 13 | 0.269 |
| genus.Prevotella9 | CRC | MR-Rgger | 16.705 | 16 | 0.405 |
| genus.Prevotella9 | CRC | IVW | 16.715 | 17 | 0.474 |
| family.Enterobacteriaceae | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| family.Enterobacteriaceae | CRC | IVW | 9.399 | 10 | 0.495 |
| order.Enterobacteriales | CRC | MR-Rgger | 9.278 | 9 | 0.412 |
| order.Enterobacteriales | CRC | IVW | 9.399 | 10 | 0.495 |
| T Cells | CRC | MR-Rgger | 3.227 | 1 | 0.072 |
| T Cells | CRC | IVW | 3.356 | 2 | 0.187 |
| genus.Ruminococcaceae NK4A214 group | T Cells | MR-Rgger | 13.403 | 14 | 0.495 |
| genus.Ruminococcaceae NK4A214 group | T Cells | IVW | 14.064 | 15 | 0.521 |
| genus.Eubacterium xylanophilum group | T Cells | MR-Rgger | 12.167 | 9 | 0.204 |
| genus.Eubacterium xylanophilum group | T Cells | IVW | 12.770 | 10 | 0.237 |
| genus.Ruminococcaceae UCG010 | T Cells | MR-Rgger | 5.807 | 6 | 0.445 |
| genus.Ruminococcaceae UCG010 | T Cells | IVW | 6.325 | 7 | 0.502 |
| order.Selenomonadales | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| order.Selenomonadales | T Cells | IVW | 13.547 | 12 | 0.331 |
| genus.Family XIII AD3011 group | T Cells | MR-Rgger | 16.241 | 13 | 0.236 |
| genus.Family XIII AD3011 group | T Cells | IVW | 16.387 | 14 | 0.290 |
| genus.Coprobacter | T Cells | MR-Rgger | 17.482 | 12 | 0.132 |
| genus.Coprobacter | T Cells | IVW | 17.762 | 13 | 0.167 |
| Class.Negativicutes | T Cells | MR-Rgger | 9.926 | 11 | 0.537 |
| Class.Negativicutes | T Cells | IVW | 13.547 | 12 | 0.331 |
| family.Bifidobacteriaceae | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| family.Bifidobacteriaceae | T Cells | IVW | 7.605 | 16 | 0.960 |
| genus.Allisonella | T Cells | MR-Rgger | 5.460 | 7 | 0.604 |
| genus.Allisonella | T Cells | IVW | 6.660 | 8 | 0.574 |
| order.Bifidobacteriales | T Cells | MR-Rgger | 5.992 | 15 | 0.980 |
| order.Bifidobacteriales | T Cells | IVW | 7.605 | 16 | 0.960 |
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| CRC | genus.Prevotella7 | MR-Rgger | 33.667 | 20 | 0.028 |
| CRC | genus.Prevotella7 | IVW | 36.090 | 21 | 0.021 |
| CRC | genus.Faecalibacterium | MR-Rgger | 54.873 | 21 | <0.001 |
| CRC | genus.Faecalibacterium | IVW | 55.369 | 22 | <0.001 |
| CRC | genus.Ruminococcaceae UCG011 | MR-Rgger | 13.506 | 20 | 0.855 |
| CRC | genus.Ruminococcaceae UCG011 | IVW | 15.151 | 21 | 0.815 |
| CRC | genus.Ruminococcaceae UCG004 | MR-Rgger | 23.003 | 21 | 0.344 |
| CRC | genus.Ruminococcaceae UCG004 | IVW | 23.825 | 22 | 0.356 |
| CRC | genus.Eubacterium brachy group | MR-Rgger | 15.511 | 20 | 0.746 |
| CRC | genus.Eubacterium brachy group | IVW | 18.053 | 21 | 0.646 |
| CRC | genus.Lachnospiraceae FCS020 group | MR-Rgger | 29.009 | 21 | 0.114 |
| CRC | genus.Lachnospiraceae FCS020 group | IVW | 29.009 | 22 | 0.145 |
| CRC | genus.Eubacterium xylanophilum group | MR-Rgger | 30.851 | 21 | 0.076 |
| CRC | genus.Eubacterium xylanophilum group | IVW | 31.688 | 22 | 0.083 |
| CRC | genus.Coprobacter | MR-Rgger | 34.800 | 21 | 0.030 |
| CRC | genus.Coprobacter | IVW | 34.918 | 22 | 0.040 |
| CRC | genus.Prevotella9 | MR-Rgger | 19.765 | 21 | 0.536 |
| CRC | genus.Prevotella9 | IVW | 30.024 | 22 | 0.118 |
| CRC | family.Enterobacteriaceae | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | family.Enterobacteriaceae | IVW | 26.554 | 22 | 0.229 |
| CRC | order.Enterobacteriales | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | order.Enterobacteriales | IVW | 26.554 | 22 | 0.229 |
| CRC | T Cells | MR-Rgger | 17.594 | 23 | 0.779 |
| CRC | T Cells | IVW | 17.885 | 24 | 0.809 |
| T Cells | genus.Ruminococcaceae NK4A214 group | MR-Rgger | 1.310 | 3 | 0.727 |
| T Cells | genus.Ruminococcaceae NK4A214 group | IVW | 1.356 | 4 | 0.852 |
| T Cells | genus.Eubacterium xylanophilum group | MR-Rgger | 2.079 | 3 | 0.556 |
| T Cells | genus.Eubacterium xylanophilum group | IVW | 2.242 | 4 | 0.691 |
| T Cells | genus.Ruminococcaceae UCG010 | MR-Rgger | 0.939 | 3 | 0.816 |
| T Cells | genus.Ruminococcaceae UCG010 | IVW | 5.875 | 4 | 0.209 |
| T Cells | order.Selenomonadales | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | order.Selenomonadales | IVW | 8.155 | 4 | 0.086 |
| T Cells | genus.Family XIII AD3011 group | MR-Rgger | 1.403 | 3 | 0.705 |
| T Cells | genus.Family XIII AD3011 group | IVW | 1.482 | 4 | 0.830 |
| T Cells | genus.Coprobacter | MR-Rgger | 11.202 | 3 | 0.011 |
| T Cells | genus.Coprobacter | IVW | 11.278 | 4 | 0.024 |
| T Cells | Class.Negativicutes | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | Class.Negativicutes | IVW | 8.155 | 4 | 0.086 |
| T Cells | family.Bifidobacteriaceae | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | family.Bifidobacteriaceae | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Allisonella | MR-Rgger | 1.503 | 1 | 0.220 |
| T Cells | genus.Allisonella | IVW | 2.060 | 2 | 0.357 |
| T Cells | order.Bifidobacteriales | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | order.Bifidobacteriales | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Coprococcus1 | MR-Rgger | 4.329 | 3 | 0.228 |
| T Cells | genus.Coprococcus1 | IVW | 4.585 | 4 | 0.333 |
表6 肠道菌群、T细胞与结直肠癌的异质性检验反向结果
Tab.6 Heterogeneity test for gut microbiota, T cells, and colorectal cancer
| Exposure | Outcome | MR method | Q | Q_df | Q_pval |
|---|---|---|---|---|---|
| CRC | genus.Prevotella7 | MR-Rgger | 33.667 | 20 | 0.028 |
| CRC | genus.Prevotella7 | IVW | 36.090 | 21 | 0.021 |
| CRC | genus.Faecalibacterium | MR-Rgger | 54.873 | 21 | <0.001 |
| CRC | genus.Faecalibacterium | IVW | 55.369 | 22 | <0.001 |
| CRC | genus.Ruminococcaceae UCG011 | MR-Rgger | 13.506 | 20 | 0.855 |
| CRC | genus.Ruminococcaceae UCG011 | IVW | 15.151 | 21 | 0.815 |
| CRC | genus.Ruminococcaceae UCG004 | MR-Rgger | 23.003 | 21 | 0.344 |
| CRC | genus.Ruminococcaceae UCG004 | IVW | 23.825 | 22 | 0.356 |
| CRC | genus.Eubacterium brachy group | MR-Rgger | 15.511 | 20 | 0.746 |
| CRC | genus.Eubacterium brachy group | IVW | 18.053 | 21 | 0.646 |
| CRC | genus.Lachnospiraceae FCS020 group | MR-Rgger | 29.009 | 21 | 0.114 |
| CRC | genus.Lachnospiraceae FCS020 group | IVW | 29.009 | 22 | 0.145 |
| CRC | genus.Eubacterium xylanophilum group | MR-Rgger | 30.851 | 21 | 0.076 |
| CRC | genus.Eubacterium xylanophilum group | IVW | 31.688 | 22 | 0.083 |
| CRC | genus.Coprobacter | MR-Rgger | 34.800 | 21 | 0.030 |
| CRC | genus.Coprobacter | IVW | 34.918 | 22 | 0.040 |
| CRC | genus.Prevotella9 | MR-Rgger | 19.765 | 21 | 0.536 |
| CRC | genus.Prevotella9 | IVW | 30.024 | 22 | 0.118 |
| CRC | family.Enterobacteriaceae | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | family.Enterobacteriaceae | IVW | 26.554 | 22 | 0.229 |
| CRC | order.Enterobacteriales | MR-Rgger | 26.429 | 21 | 0.191 |
| CRC | order.Enterobacteriales | IVW | 26.554 | 22 | 0.229 |
| CRC | T Cells | MR-Rgger | 17.594 | 23 | 0.779 |
| CRC | T Cells | IVW | 17.885 | 24 | 0.809 |
| T Cells | genus.Ruminococcaceae NK4A214 group | MR-Rgger | 1.310 | 3 | 0.727 |
| T Cells | genus.Ruminococcaceae NK4A214 group | IVW | 1.356 | 4 | 0.852 |
| T Cells | genus.Eubacterium xylanophilum group | MR-Rgger | 2.079 | 3 | 0.556 |
| T Cells | genus.Eubacterium xylanophilum group | IVW | 2.242 | 4 | 0.691 |
| T Cells | genus.Ruminococcaceae UCG010 | MR-Rgger | 0.939 | 3 | 0.816 |
| T Cells | genus.Ruminococcaceae UCG010 | IVW | 5.875 | 4 | 0.209 |
| T Cells | order.Selenomonadales | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | order.Selenomonadales | IVW | 8.155 | 4 | 0.086 |
| T Cells | genus.Family XIII AD3011 group | MR-Rgger | 1.403 | 3 | 0.705 |
| T Cells | genus.Family XIII AD3011 group | IVW | 1.482 | 4 | 0.830 |
| T Cells | genus.Coprobacter | MR-Rgger | 11.202 | 3 | 0.011 |
| T Cells | genus.Coprobacter | IVW | 11.278 | 4 | 0.024 |
| T Cells | Class.Negativicutes | MR-Rgger | 6.388 | 3 | 0.094 |
| T Cells | Class.Negativicutes | IVW | 8.155 | 4 | 0.086 |
| T Cells | family.Bifidobacteriaceae | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | family.Bifidobacteriaceae | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Allisonella | MR-Rgger | 1.503 | 1 | 0.220 |
| T Cells | genus.Allisonella | IVW | 2.060 | 2 | 0.357 |
| T Cells | order.Bifidobacteriales | MR-Rgger | 2.909 | 3 | 0.406 |
| T Cells | order.Bifidobacteriales | IVW | 2.979 | 4 | 0.561 |
| T Cells | genus.Coprococcus1 | MR-Rgger | 4.329 | 3 | 0.228 |
| T Cells | genus.Coprococcus1 | IVW | 4.585 | 4 | 0.333 |
| [1] | Ionescu VA, Gheorghe G, Bacalbasa N, et al. Colorectal cancer: from risk factors to oncogenesis[J]. Medicina: Kaunas, 2023, 59(9): 1646. doi:10.3390/medicina59091646 |
| [2] | Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota[J]. Immunity, 2017, 46(4): 562-76. doi:10.1016/j.immuni.2017.04.008 |
| [3] | Hu ZJ, Zhu HR, Jin YJ, et al. Correlation between gut microbiota and tumor immune microenvironment: a bibliometric and visualized study[J]. World J Clin Oncol, 2025, 16(2): 101611. doi:10.5306/wjco.v16.i2.101611 |
| [4] | Schwabe RF, Jobin C. The microbiome and cancer[J]. Nat Rev Cancer, 2013, 13(11): 800-12. doi:10.1038/nrc3610 |
| [5] | Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis[J]. J Biomed Sci, 2018, 25(1): 79. doi:10.1186/s12929-018-0483-8 |
| [6] | Hou X, Zheng Z, Wei J, et al. Effects of gut microbiota on immune responses and immunotherapy in colorectal cancer[J]. Front Immunol, 2022, 13: 1030745. doi:10.3389/fimmu.2022.1030745 |
| [7] | Scarpellini E, Ianiro G, Attili F, et al. The human gut microbiota and virome: Potential therapeutic implications[J]. Dig Liver Dis, 2015, 47(12): 1007-12. doi:10.1016/j.dld.2015.07.008 |
| [8] | Quaglio AEV, Grillo TG, De Oliveira ECS, et al. Gut microbiota, inflammatory bowel disease and colorectal cancer[J]. World J Gastroenterol, 2022, 28(30): 4053-60. doi:10.3748/wjg.v28.i30.4053 |
| [9] | Shim JA, Ryu JH, Jo Y, et al. The role of gut microbiota in T cell immunity and immune mediated disorders[J]. Int J Biol Sci, 2023, 19(4): 1178-91. doi:10.7150/ijbs.79430 |
| [10] | Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life[J]. Immunity, 2018, 48(2): 202-13. doi:10.1016/j.immuni.2018.01.007 |
| [11] | Ni JJ, Li XS, Zhang H, et al. Mendelian randomization study of causal link from gut microbiota to colorectal cancer[J]. BMC Cancer, 2022, 22(1): 1371. doi:10.1186/s12885-022-10483-w |
| [12] | Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis[J]. Proc Natl Acad Sci USA, 2011, 108(37): 15354-9. doi:10.1073/pnas.1010203108 |
| [13] | Brennan CA, Clay SL, et al. Fusobacterium nucleatumdrives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression[J]. Gut Microbes, 2021, 13: 1987780. doi:10.1080/19490976.2021.1987780 |
| [14] | Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome[J]. Science, 2006, 313(5795): 1960-4. doi:10.1126/science.1129139 |
| [15] | Salama P, Phillips M, Grieu F, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer[J]. J Clin Oncol, 2009, 27(2): 186-92. doi:10.1200/jco.2008.18.7229 |
| [16] | Zheng Z, Wieder T, Mauerer B, et al. T cells in colorectal cancer: unravelling the function of different T cell subsets in the tumor microenvironment[J]. Int J Mol Sci, 2023, 24(14): 11673. doi:10.3390/ijms241411673 |
| [17] | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data[J]. Genet Epidemiol, 2013, 37(7): 658-65. doi:10.1002/gepi.21758 |
| [18] | Fong W, Li Q, Yu J. Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer[J]. Oncogene, 2020, 39(26): 4925-43. doi:10.1038/s41388-020-1341-1 |
| [19] | Yuan H, Gui R, Wang Z, et al. Gut microbiota: a novel and potential target for radioimmunotherapy in colorectal cancer[J]. Front Immunol, 2023, 14: 1128774. doi:10.3389/fimmu.2023.1128774 |
| [20] | Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental deter-minants of disease[J]? Int J Epidemiol, 2003, 32(1): 1-22. doi:10.1093/ije/dyg070 |
| [21] | 胡旭焘.肠道菌群,循环代谢物与胆石症:一项孟德尔随机化研究[D].吉林大学, 2024. 003850. |
| [22] | Shan J, Hu X, Chen T, et al. COVID-19 vaccination and the risk of autoimmune diseases: a Mendelian randomization study[J]. Front Public Health, 2024, 12: 1322140. doi:10.3389/fpubh.2024.1322140 |
| [23] | Burgess S, Bowden J, Fall T, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants[J]. Epidemiology, 2017, 28(1): 30-42. doi:10.1097/ede.0000000000000559 |
| [24] | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression[J]. Int J Epidemiol, 2015, 44(2): 512-25. doi:10.1093/ije/dyv080 |
| [25] | Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted Median estimator[J]. Genet Epidemiol, 2016, 40(4): 304-14. doi:10.1002/gepi.21965 |
| [26] | Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption[J]. Int J Epidemiol, 2017, 46(6): 1985-98. doi:10.1093/ije/dyx102 |
| [27] | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method[J]. Eur J Epidemiol, 2017, 32(5): 377-89. doi:10.1007/s10654-017-0255-x |
| [28] | Verbanck M, Chen CY, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases[J]. Nat Genet, 2018, 50(5): 693-8. doi:10.1038/s41588-018-0099-7 |
| [29] | Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies[J]. Hum Mol Genet, 2018, 27(r2): R195-208. doi:10.1093/hmg/ddy163 |
| [30] | Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data[J]. PLoS Genet, 2017, 13(11): e1007081. doi:10.1371/journal.pgen.1007081 |
| [31] | Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians[J]. BMJ, 2018, 362: k601. doi:10.1136/bmj.k601 |
| [32] | Teng NMY, Kiu R, Evans R, et al. Allocoprobacillus halotolerans gen. nov., sp. nov and Coprobacter tertius sp. nov., isolated from human gut microbiota[J]. Int J Syst Evol Microbiol, 2023, 73(7): 5950-8. doi:10.1099/ijsem.0.005950 |
| [33] | Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer[J]. Gut, 2017, 66(1): 70-8. doi:10.1136/gutjnl-2015-309800 |
| [34] | Silva M, Brunner V, Tschurtschenthaler M. Microbiota and colorectal cancer: from gut to bedside[J]. Front Pharmacol, 2021, 12: 760280. doi:10.3389/fphar.2021.760280 |
| [35] | Wang Y, Wan X, Wu X, et al. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis[J]. Gut Pathog, 2021, 13(1): 2. doi:10.1186/s13099-020-00396-z |
| [36] | Li HY, Wang Y, Shao SM, et al. Rabdosia serra alleviates dextran sulfate sodium salt-induced colitis in mice through anti-inflammation, regulating Th17/Treg balance, maintaining intestinal barrier integrity, and modulating gut microbiota[J]. J Pharm Anal, 2022, 12(6): 824-38. doi:10.1016/j.jpha.2022.08.001 |
| [37] | Park JS, Gazzaniga FS, Wu M, et al. Targeting PD-L2-RGMb overcomes microbiome-related immunotherapy resistance[J]. Nature, 2023, 617(7960): 377-85. doi:10.1038/s41586-023-06026-3 |
| [1] | 何榕茂, 方泽扬, 张芸芸, 吴友谅, 梁世秀, 计涛, 陈科全, 王斯琪. 铁死亡相关基因对溃疡性结肠炎具有诊断预测价值[J]. 南方医科大学学报, 2025, 45(9): 1927-1937. |
| [2] | 邓楚玉, 王雪莹, 甘立祥, 王大禹, 郑晓燕, 唐纯志. 电针足三里改善高脂血症小鼠的血脂紊乱:基于肠道菌群结构的改善[J]. 南方医科大学学报, 2025, 45(8): 1633-1642. |
| [3] | 张宏博, 闫梦宇, 张建东, 孙培旺, 汪蕊, 郭园园. 吡非尼酮抑制调节性T细胞延缓小鼠膀胱癌进展[J]. 南方医科大学学报, 2025, 45(7): 1513-1518. |
| [4] | 黄凯悦, 齐景馨, 罗文谦, 林怡萱, 陈梅妹, 甘慧娟. 温胆汤通过调控肠道菌群-胆汁酸轴改善代谢综合征痰证大鼠的代谢表型[J]. 南方医科大学学报, 2025, 45(6): 1174-1184. |
| [5] | 翁诺舟, 谭彬, 曾文涛, 古家宇, 翁炼基, 郑克鸿. 过表达RGL1通过激活CDC42/RAC1复合体上调运动型黏着斑组装促进结直肠癌转移[J]. 南方医科大学学报, 2025, 45(5): 1031-1038. |
| [6] | 马振南, 刘福全, 赵雪峰, 张晓微. DTX2促进奥沙利铂耐药的结直肠癌细胞增殖、侵袭和上皮间质转化[J]. 南方医科大学学报, 2025, 45(4): 829-836. |
| [7] | 李文婕, 洪耀南, 黄蕊, 李煜宸, 张莹, 张蕴, 吴迪炯. 自身免疫性疾病是再生障碍性贫血的危险因素:一项孟德尔随机化分析[J]. 南方医科大学学报, 2025, 45(4): 871-879. |
| [8] | 庆顺杰, 沈智勇. 过表达己糖激酶2通过激活JAK/STAT途径促进结直肠癌细胞的增殖、迁移和侵袭并调节肿瘤免疫微环境[J]. 南方医科大学学报, 2025, 45(3): 542-553. |
| [9] | 罗嘉纯, Sodnomjamts Batzaya, 高雪锋, 陈晶宇, 余政颖, 熊莎莎, 曹虹. Akkermansia muciniphila改善gp120转基因小鼠的肠-脑相互作用障碍[J]. 南方医科大学学报, 2025, 45(3): 554-565. |
| [10] | 殷丽霞, 牛民主, 张可妮, 耿志军, 胡建国, 李江艳, 李静. 升麻素抑制MAPK通路调节辅助性T细胞免疫平衡改善小鼠克罗恩病样结肠炎[J]. 南方医科大学学报, 2025, 45(3): 595-602. |
| [11] | 高俊杰, 叶开, 吴竞. 槲皮素通过调控TP53基因抑制肾透明细胞癌的增殖和迁移[J]. 南方医科大学学报, 2025, 45(2): 313-321. |
| [12] | 刘莹, 李柏睿, 李永财, 常禄博, 王娇, 杨琳, 颜永刚, 屈凯, 刘继平, 张岗, 沈霞. 加味逍遥丸通过神经递质调节、抗炎抗氧化及肠道菌群调控改善大鼠的抑郁样行为[J]. 南方医科大学学报, 2025, 45(2): 347-358. |
| [13] | 潘兴旭, 张秉祺, 张智华, 曹秋实. 戈登杆菌属丰度降低与肾结石风险增加相关:一项孟德尔随机化分析与动物实验研究[J]. 南方医科大学学报, 2025, 45(11): 2405-2415. |
| [14] | 马会华, 闫奎坡, 刘刚, 徐亚洲, 张磊, 李一卓. 1990~2021年心房颤动/扑动流行病学及其危险因素分析:基于2021年中国全球疾病负担研究与孟德尔随机化研究的系统分析[J]. 南方医科大学学报, 2025, 45(10): 2182-2190. |
| [15] | 姚辰, 李文佳, 庞瑞明, 周继红. 臀肌腱炎、原发性髋关节病可能导致髂胫束综合征—一项孟德尔随机化研究[J]. 南方医科大学学报, 2024, 44(9): 1821-1830. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||