南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (9): 1859-1866.doi: 10.12122/j.issn.1673-4254.2025.09.06
• • 上一篇
金家列1,2( ), 王菲1, 朱丽雅1, 赵晓静3,4, 王金鑫5, 朱超6(
), 王菲1, 朱丽雅1, 赵晓静3,4, 王金鑫5, 朱超6( ), 杨蓉西1(
), 杨蓉西1( )
)
收稿日期:2025-03-21
出版日期:2025-09-20
发布日期:2025-09-28
通讯作者:
朱超,杨蓉西
E-mail:jljin@cdc.zj.cn;zhuchaodoctor@163.com;rongxiyang@njmu.edu.cn
作者简介:金家列,硕士,E-mail: jljin@cdc.zj.cn
基金资助:
Jialie JIN1,2( ), Fei WANG1, Liya ZHU1, Xiaojing ZHAO3,4, Jinxin WANG5, Chao ZHU6(
), Fei WANG1, Liya ZHU1, Xiaojing ZHAO3,4, Jinxin WANG5, Chao ZHU6( ), Rongxi YANG1(
), Rongxi YANG1( )
)
Received:2025-03-21
Online:2025-09-20
Published:2025-09-28
Contact:
Chao ZHU, Rongxi YANG
E-mail:jljin@cdc.zj.cn;zhuchaodoctor@163.com;rongxiyang@njmu.edu.cn
Supported by:摘要:
目的 探讨中国人群外周血肿瘤抑制可转移候选基因1(TSSC1)和黑素亲和素(MLPH)基因甲基化水平与冠心病(CHD)之间的关系。 方法 收集86例CHD患者和95例健康对照,使用定量质谱法检测TSSC1和MLPH基因的CpG位点甲基化水平。使用Mann-Whitney秩和检验比较不同临床特征组间的甲基化水平差异。通过Spearman相关系数和列联系数分别评估甲基化水平与年龄、性别之间的相关性。 结果 与对照组相比,MLPH基因高甲基化与心肌梗死亚型相关(MLPH_CpG_2.7:P=0.045;MLPH_CpG_3/cg06639874:P=0.049;MLPH_CpG_5:P=0.019)。MLPH基因高甲基化与CHD的相关性在年龄<65岁组(MLPH_CpG_2.7:P=0.014;MLPH_CpG_4:P=0.001)和男性(MLPH_CpG_2.7:P=0.004;MLPH_CpG_3/cg06639874:P=0.044)中更加明显。未观察到外周血TSSC1基因甲基化改变与CHD及其亚型的相关性。 结论 中国人群外周血中MLPH基因高甲基化与CHD及心肌梗死具有相关性,尤其是65岁以下群体和男性群体。
金家列, 王菲, 朱丽雅, 赵晓静, 王金鑫, 朱超, 杨蓉西. 中国人群外周血MLPH基因高甲基化与冠心病相关[J]. 南方医科大学学报, 2025, 45(9): 1859-1866.
Jialie JIN, Fei WANG, Liya ZHU, Xiaojing ZHAO, Jinxin WANG, Chao ZHU, Rongxi YANG. Association between MLPH gene hypermethylation in peripheral blood and coronary heart disease[J]. Journal of Southern Medical University, 2025, 45(9): 1859-1866.
| Variables | CHD group (n=86) | Control group (n=95) | Z/χ2 | P |
|---|---|---|---|---|
| Gender [n (%)] | 3.113 | 0.078a | ||
| Male | 52 (60.5) | 45 (47.4) | ||
| Female | 34 (39.5) | 50 (52.6) | ||
| Smoking [n (%)] | 7.983 | 0.005a | ||
| Yes | 35 (40.7) | 20 (21.1) | ||
| No | 51 (59.3) | 74 (77.9) | ||
| Drinking [n (%)] | 0.034 | 0.853a | ||
| Yes | 23 (26.7) | 24 (25.3) | ||
| No | 63 (73.3) | 70 (73.7) | ||
| Hypertension [n (%)] | 6.004 | 0.014a | ||
| Yes | 55 (64.0) | 42 (44.2) | ||
| No | 31 (36.0) | 50 (52.6) | ||
| Diabetes [n (%)] | 4.314 | 0.038a | ||
| Yes | 34 (39.5) | 23 (24.2) | ||
| No | 52 (60.5) | 69 (72.6) | ||
| Age (years) | 65 (57, 73) | 63 (61,68) | -0.364 | 0.716b |
| TC (mmol/L) | 3.79 (3.26, 4.41) | 4.21 (3.59, 5.02) | -2.608 | 0.009b |
| TG (mmol/L) | 1.33 (0.97, 1.86) | 1.38 (1.06, 1.92) | -0.562 | 0.574b |
| HDL-C (mmol/L) | 1.03 (0.86, 1.29) | 1.18 (0.91, 1.41) | -1.829 | 0.067b |
| LDL-C (mmol/L) | 2.27 (1.85, 2.76) | 2.64 (2.07, 3.35) | -2.794 | 0.005b |
表1 CHD患者与正常对照的基线资料
Tab.1 Baseline data from patients with CHD and normal controls
| Variables | CHD group (n=86) | Control group (n=95) | Z/χ2 | P |
|---|---|---|---|---|
| Gender [n (%)] | 3.113 | 0.078a | ||
| Male | 52 (60.5) | 45 (47.4) | ||
| Female | 34 (39.5) | 50 (52.6) | ||
| Smoking [n (%)] | 7.983 | 0.005a | ||
| Yes | 35 (40.7) | 20 (21.1) | ||
| No | 51 (59.3) | 74 (77.9) | ||
| Drinking [n (%)] | 0.034 | 0.853a | ||
| Yes | 23 (26.7) | 24 (25.3) | ||
| No | 63 (73.3) | 70 (73.7) | ||
| Hypertension [n (%)] | 6.004 | 0.014a | ||
| Yes | 55 (64.0) | 42 (44.2) | ||
| No | 31 (36.0) | 50 (52.6) | ||
| Diabetes [n (%)] | 4.314 | 0.038a | ||
| Yes | 34 (39.5) | 23 (24.2) | ||
| No | 52 (60.5) | 69 (72.6) | ||
| Age (years) | 65 (57, 73) | 63 (61,68) | -0.364 | 0.716b |
| TC (mmol/L) | 3.79 (3.26, 4.41) | 4.21 (3.59, 5.02) | -2.608 | 0.009b |
| TG (mmol/L) | 1.33 (0.97, 1.86) | 1.38 (1.06, 1.92) | -0.562 | 0.574b |
| HDL-C (mmol/L) | 1.03 (0.86, 1.29) | 1.18 (0.91, 1.41) | -1.829 | 0.067b |
| LDL-C (mmol/L) | 2.27 (1.85, 2.76) | 2.64 (2.07, 3.35) | -2.794 | 0.005b |
| CpG sites | CHD group (n=86) | Control group (n=95) | Z | P* |
|---|---|---|---|---|
| TSSC1_CpG_1 | 0.89 (0.82, 0.95) | 0.85 (0.77, 0.93) | -1.258 | 0.208 |
| TSSC1_CpG_6/cg23245316 | 0.43 (0.33, 0.53) | 0.41 (0.32, 0.55) | -0.007 | 0.994 |
| TSSC1_CpG_7 | 1.00 (0.98, 1.00) | 1.00 (0.97, 1.00) | -0.389 | 0.697 |
| TSSC1_CpG_8 | 0.68 (0.60, 0.77) | 0.67 (0.59, 0.73) | -1.474 | 0.140 |
| TSSC1_CpG_12 | 1.00 (0.89, 1.00) | 1.00 (0.91, 1.00) | -0.315 | 0.753 |
| TSSC1_CpG_14 | 0.33 (0.26, 0.39) | 0.31 (0.24, 0.39) | -0.942 | 0.346 |
| TSSC1_CpG_15 | 0.38 (0.33, 0.50) | 0.42 (0.32, 0.49) | -0.329 | 0.742 |
| TSSC1_CpG_16 | 0.96 (0.92, 0.97) | 0.96 (0.93, 0.97) | -0.593 | 0.553 |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | -1.629 | 0.103 |
| TSSC1_CpG_18 | 0.98 (0.95, 0.99) | 0.99 (0.96, 0.99) | -0.468 | 0.640 |
| MLPH_CpG_2.7 | 0.85 (0.81, 0.93) | 0.84 (0.80, 0.90) | -1.715 | 0.086 |
| MLPH_CpG_3/cg06639874 | 0.92 (0.84, 0.97) | 0.90 (0.87, 0.94) | -0.657 | 0.511 |
| MLPH_CpG_4 | 0.94 (0.89, 0.99) | 0.94 (0.88, 0.96) | -1.410 | 0.159 |
| MLPH_CpG_5 | 0.87 (0.82, 0.95) | 0.86 (0.82, 0.90) | -1.763 | 0.078 |
| MLPH_CpG_6 | 0.59 (0.37, 0.69) | 0.56 (0.36, 0.67) | -1.217 | 0.224 |
表2 CHD组和对照组中TSSC1和MLPH基因甲基化差异
Tab.2 Differences in methylation levels of TSSC1 and MLPH genes between CHD and control groups [M(P25, P75)]
| CpG sites | CHD group (n=86) | Control group (n=95) | Z | P* |
|---|---|---|---|---|
| TSSC1_CpG_1 | 0.89 (0.82, 0.95) | 0.85 (0.77, 0.93) | -1.258 | 0.208 |
| TSSC1_CpG_6/cg23245316 | 0.43 (0.33, 0.53) | 0.41 (0.32, 0.55) | -0.007 | 0.994 |
| TSSC1_CpG_7 | 1.00 (0.98, 1.00) | 1.00 (0.97, 1.00) | -0.389 | 0.697 |
| TSSC1_CpG_8 | 0.68 (0.60, 0.77) | 0.67 (0.59, 0.73) | -1.474 | 0.140 |
| TSSC1_CpG_12 | 1.00 (0.89, 1.00) | 1.00 (0.91, 1.00) | -0.315 | 0.753 |
| TSSC1_CpG_14 | 0.33 (0.26, 0.39) | 0.31 (0.24, 0.39) | -0.942 | 0.346 |
| TSSC1_CpG_15 | 0.38 (0.33, 0.50) | 0.42 (0.32, 0.49) | -0.329 | 0.742 |
| TSSC1_CpG_16 | 0.96 (0.92, 0.97) | 0.96 (0.93, 0.97) | -0.593 | 0.553 |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | -1.629 | 0.103 |
| TSSC1_CpG_18 | 0.98 (0.95, 0.99) | 0.99 (0.96, 0.99) | -0.468 | 0.640 |
| MLPH_CpG_2.7 | 0.85 (0.81, 0.93) | 0.84 (0.80, 0.90) | -1.715 | 0.086 |
| MLPH_CpG_3/cg06639874 | 0.92 (0.84, 0.97) | 0.90 (0.87, 0.94) | -0.657 | 0.511 |
| MLPH_CpG_4 | 0.94 (0.89, 0.99) | 0.94 (0.88, 0.96) | -1.410 | 0.159 |
| MLPH_CpG_5 | 0.87 (0.82, 0.95) | 0.86 (0.82, 0.90) | -1.763 | 0.078 |
| MLPH_CpG_6 | 0.59 (0.37, 0.69) | 0.56 (0.36, 0.67) | -1.217 | 0.224 |
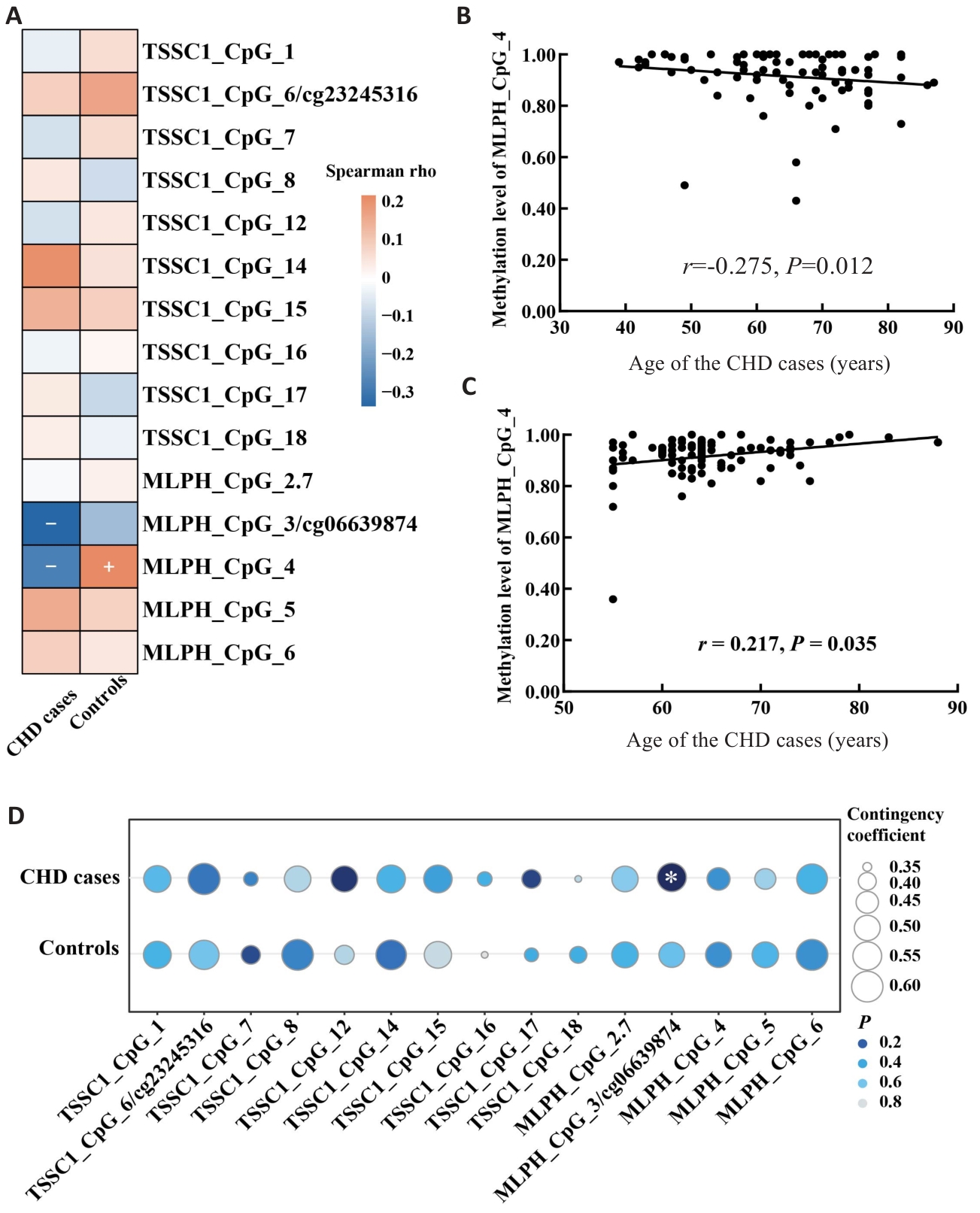
图1 TSSC1和MLPH基因甲基化水平与年龄、男性的相关性
Fig.1 Correlation of methylation levels of TSSC1 and MLPH genes with age and male. A: Heat map of the correlation between age and methylation levels. B: Scatter plot of the correlation between MLPH_CpG_4 methylation level and age in CHD cases. C: Scatter plot of the correlation between MLPH_CpG_4 methylation level and age in control group. D: Bubble chart of the correlation between male gender and methylation levels.
| CpG sites | Age<65 years (n=101) | Age≥65 year (n=80) | |||||
|---|---|---|---|---|---|---|---|
| CHD (n=41) | Control (n=0) | P* | CHD (n=45) | Control (n=35) | P * | ||
| TSSC1_CpG_1 | 0.92 (0.82, 0.95) | 0.85 (0.76, 0.93) | 0.151 | 0.89 (0.79, 0.95) | 0.86 (0.76, 0.95) | 0.725 | |
| TSSC1_CpG_6/cg23245316 | 0.42 (0.29, 0.53) | 0.40 (0.29, 0.55) | 0.730 | 0.44 (0.34, 0.54) | 0.45 (0.36, 0.58) | 0.470 | |
| TSSC1_CpG_7 | 1.00 (0.97, 1.00) | 1.00 (0.97, 1.00) | 0.888 | 1.00 (0.98, 1.00) | 1.00 (0.96, 1.00) | 0.419 | |
| TSSC1_CpG_8 | 0.68 (0.56, 0.76) | 0.68 (0.59, 0.73) | 0.921 | 0.69 (0.62, 0.77) | 0.66 (0.59, 0.73) | 0.233 | |
| TSSC1_CpG_12 | 1.00 (0.90, 1.00) | 1.00 (0.89, 1.00) | 0.687 | 1.00 (0.89, 1.00) | 1.00 (0.93, 1.00) | 0.352 | |
| TSSC1_CpG_14 | 0.33 (0.25, 0.39) | 0.31 (0.25, 0.38) | 0.888 | 0.34 (0.29, 0.42) | 0.32 (0.21, 0.43) | 0.384 | |
| TSSC1_CpG_15 | 0.37 (0.31, 0.51) | 0.42 (0.31, 0.47) | 0.969 | 0.40 (0.33, 0.50) | 0.42 (0.35, 0.50) | 0.507 | |
| TSSC1_CpG_16 | 0.96 (0.94, 0.97) | 0.96 (0.92, 0.97) | 1.000 | 0.96 (0.90, 0.97) | 0.96 (0.93, 0.97) | 0.405 | |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) | 0.701 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | 0.051 | |
| TSSC1_CpG_18 | 0.98 (0.94, 0.99) | 0.99 (0.95, 0.99) | 0.320 | 0.99 (0.96, 0.99) | 0.98 (0.96, 0.99) | 0.744 | |
| MLPH_CpG_2.7 | 0.86 (0.83, 0.94) | 0.83 (0.80, 0.89) | 0.014 | 0.85 (0.80, 0.91) | 0.85 (0.79, 0.93) | 0.941 | |
| MLPH_CpG_3/cg06639874 | 0.95 (0.87, 0.98) | 0.91 (0.88, 0.96) | 0.147 | 0.89 (0.82, 0.94) | 0.89 (0.83, 0.93) | 0.930 | |
| MLPH_CpG_4 | 0.97 (0.93, 1.00) | 0.93 (0.87, 0.95) | 0.001 | 0.92 (0.86, 0.98) | 0.94 (0.89, 0.97) | 0.187 | |
| MLPH_CpG_5 | 0.86 (0.81, 0.95) | 0.86 (0.82, 0.89) | 0.652 | 0.90 (0.84, 0.95) | 0.87 (0.84, 0.91) | 0.104 | |
| MLPH_CpG_6 | 0.58 (0.35, 0.71) | 0.55 (0.35, 0.67) | 0.568 | 0.62 (0.41, 0.69) | 0.57 (0.39, 0.66) | 0.284 | |
| *The P values were calculated by the Mann-Whitney U test. | |||||||
表4 CHD组和对照组中TSSC1和MLPH基因甲基化差异 (按年龄分层)
Tab.4 Differences in TSSC1 and MLPH gene methylation between CHD and control groups stratified by age [M (P25,P75)]
| CpG sites | Age<65 years (n=101) | Age≥65 year (n=80) | |||||
|---|---|---|---|---|---|---|---|
| CHD (n=41) | Control (n=0) | P* | CHD (n=45) | Control (n=35) | P * | ||
| TSSC1_CpG_1 | 0.92 (0.82, 0.95) | 0.85 (0.76, 0.93) | 0.151 | 0.89 (0.79, 0.95) | 0.86 (0.76, 0.95) | 0.725 | |
| TSSC1_CpG_6/cg23245316 | 0.42 (0.29, 0.53) | 0.40 (0.29, 0.55) | 0.730 | 0.44 (0.34, 0.54) | 0.45 (0.36, 0.58) | 0.470 | |
| TSSC1_CpG_7 | 1.00 (0.97, 1.00) | 1.00 (0.97, 1.00) | 0.888 | 1.00 (0.98, 1.00) | 1.00 (0.96, 1.00) | 0.419 | |
| TSSC1_CpG_8 | 0.68 (0.56, 0.76) | 0.68 (0.59, 0.73) | 0.921 | 0.69 (0.62, 0.77) | 0.66 (0.59, 0.73) | 0.233 | |
| TSSC1_CpG_12 | 1.00 (0.90, 1.00) | 1.00 (0.89, 1.00) | 0.687 | 1.00 (0.89, 1.00) | 1.00 (0.93, 1.00) | 0.352 | |
| TSSC1_CpG_14 | 0.33 (0.25, 0.39) | 0.31 (0.25, 0.38) | 0.888 | 0.34 (0.29, 0.42) | 0.32 (0.21, 0.43) | 0.384 | |
| TSSC1_CpG_15 | 0.37 (0.31, 0.51) | 0.42 (0.31, 0.47) | 0.969 | 0.40 (0.33, 0.50) | 0.42 (0.35, 0.50) | 0.507 | |
| TSSC1_CpG_16 | 0.96 (0.94, 0.97) | 0.96 (0.92, 0.97) | 1.000 | 0.96 (0.90, 0.97) | 0.96 (0.93, 0.97) | 0.405 | |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) | 0.701 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | 0.051 | |
| TSSC1_CpG_18 | 0.98 (0.94, 0.99) | 0.99 (0.95, 0.99) | 0.320 | 0.99 (0.96, 0.99) | 0.98 (0.96, 0.99) | 0.744 | |
| MLPH_CpG_2.7 | 0.86 (0.83, 0.94) | 0.83 (0.80, 0.89) | 0.014 | 0.85 (0.80, 0.91) | 0.85 (0.79, 0.93) | 0.941 | |
| MLPH_CpG_3/cg06639874 | 0.95 (0.87, 0.98) | 0.91 (0.88, 0.96) | 0.147 | 0.89 (0.82, 0.94) | 0.89 (0.83, 0.93) | 0.930 | |
| MLPH_CpG_4 | 0.97 (0.93, 1.00) | 0.93 (0.87, 0.95) | 0.001 | 0.92 (0.86, 0.98) | 0.94 (0.89, 0.97) | 0.187 | |
| MLPH_CpG_5 | 0.86 (0.81, 0.95) | 0.86 (0.82, 0.89) | 0.652 | 0.90 (0.84, 0.95) | 0.87 (0.84, 0.91) | 0.104 | |
| MLPH_CpG_6 | 0.58 (0.35, 0.71) | 0.55 (0.35, 0.67) | 0.568 | 0.62 (0.41, 0.69) | 0.57 (0.39, 0.66) | 0.284 | |
| *The P values were calculated by the Mann-Whitney U test. | |||||||
| CpG sites | Female (n=84) | Male (n=97) | |||||
|---|---|---|---|---|---|---|---|
| CHD (n=34) | Control (n=50) | P* | CHD (n = 52) | Control (n = 45) | P* | ||
| TSSC1_CpG_1 | 0.90 (0.83, 0.96) | 0.85 (0.75, 0.95) | 0.135 | 0.89 (0.76, 0.95) | 0.86 (0.79, 0.93) | 0.566 | |
| TSSC1_CpG_6/cg23245316 | 0.44 (0.33, 0.55) | 0.38 (0.31, 0.48) | 0.160 | 0.43 (0.32, 0.52) | 0.45 (0.35, 0.58) | 0.114 | |
| TSSC1_CpG_7 | 1.00 (0.97, 1.00) | 1.00 (0.97, 1.00) | 0.545 | 1.00 (0.98, 1.00) | 1.00 (0.96, 1.00) | 0.298 | |
| TSSC1_CpG_8 | 0.74 (0.68, 0.79) | 0.68 (0.59, 0.73) | 0.014 | 0.65 (0.52, 0.72) | 0.66 (0.58, 0.72) | 0.420 | |
| TSSC1_CpG_12 | 1.00 (0.91, 1.00) | 1.00 (0.92, 1.00) | 0.467 | 1.00 (0.88, 1.00) | 0.98 (0.90, 1.00) | 0.687 | |
| TSSC1_CpG_14 | 0.35 (0.27, 0.41) | 0.33 (0.24, 0.43) | 0.461 | 0.33 (0.25, 0.39) | 0.30 (0.24, 0.36) | 0.349 | |
| TSSC1_CpG_15 | 0.43 (0.34, 0.53) | 0.41 (0.31, 0.46) | 0.131 | 0.38 (0.30, 0.46) | 0.44 (0.34, 0.52) | 0.077 | |
| TSSC1_CpG_16 | 0.96 (0.90, 0.97) | 0.96 (0.92, 0.97) | 0.447 | 0.96 (0.93, 0.97) | 0.96 (0.93, 0.97) | 0.870 | |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) | 0.134 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | 0.290 | |
| TSSC1_CpG_18 | 0.98 (0.96, 0.99) | 0.99 (0.96, 0.99) | 0.677 | 0.99 (0.93, 0.99) | 0.99 (0.95, 0.99) | 0.756 | |
| MLPH_CpG_2.7 | 0.83 (0.77, 0.89) | 0.84 (0.80, 0.91) | 0.323 | 0.87 (0.83, 0.94) | 0.83 (0.78, 0.88) | 0.004 | |
| MLPH_CpG_3/cg06639874 | 0.90 (0.81, 0.95) | 0.91 (0.86, 0.96) | 0.215 | 0.94 (0.86, 0.97) | 0.90 (0.87, 0.92) | 0.044 | |
| MLPH_CpG_4 | 0.93 (0.86, 1.00) | 0.93 (0.89, 0.96) | 0.966 | 0.95 (0.90, 0.99) | 0.94 (0.88, 0.96) | 0.090 | |
| MLPH_CpG_5 | 0.86 (0.82, 0.93) | 0.85 (0.82, 0.89) | 0.294 | 0.90 (0.83, 0.95) | 0.87 (0.84, 0.91) | 0.228 | |
| MLPH_CpG_6 | 0.62 (0.44, 0.69) | 0.49 (0.31, 0.64) | 0.099 | 0.58 (0.34, 0.74) | 0.58 (0.41, 0.68) | 0.981 | |
表5 CHD组和对照组中TSSC1和MLPH基因甲基化差异(按性别分层)
Tab.5 Differences in TSSC1 and MLPH gene methylation between CHD and control groups stratified by sex [M (P25,P75)]
| CpG sites | Female (n=84) | Male (n=97) | |||||
|---|---|---|---|---|---|---|---|
| CHD (n=34) | Control (n=50) | P* | CHD (n = 52) | Control (n = 45) | P* | ||
| TSSC1_CpG_1 | 0.90 (0.83, 0.96) | 0.85 (0.75, 0.95) | 0.135 | 0.89 (0.76, 0.95) | 0.86 (0.79, 0.93) | 0.566 | |
| TSSC1_CpG_6/cg23245316 | 0.44 (0.33, 0.55) | 0.38 (0.31, 0.48) | 0.160 | 0.43 (0.32, 0.52) | 0.45 (0.35, 0.58) | 0.114 | |
| TSSC1_CpG_7 | 1.00 (0.97, 1.00) | 1.00 (0.97, 1.00) | 0.545 | 1.00 (0.98, 1.00) | 1.00 (0.96, 1.00) | 0.298 | |
| TSSC1_CpG_8 | 0.74 (0.68, 0.79) | 0.68 (0.59, 0.73) | 0.014 | 0.65 (0.52, 0.72) | 0.66 (0.58, 0.72) | 0.420 | |
| TSSC1_CpG_12 | 1.00 (0.91, 1.00) | 1.00 (0.92, 1.00) | 0.467 | 1.00 (0.88, 1.00) | 0.98 (0.90, 1.00) | 0.687 | |
| TSSC1_CpG_14 | 0.35 (0.27, 0.41) | 0.33 (0.24, 0.43) | 0.461 | 0.33 (0.25, 0.39) | 0.30 (0.24, 0.36) | 0.349 | |
| TSSC1_CpG_15 | 0.43 (0.34, 0.53) | 0.41 (0.31, 0.46) | 0.131 | 0.38 (0.30, 0.46) | 0.44 (0.34, 0.52) | 0.077 | |
| TSSC1_CpG_16 | 0.96 (0.90, 0.97) | 0.96 (0.92, 0.97) | 0.447 | 0.96 (0.93, 0.97) | 0.96 (0.93, 0.97) | 0.870 | |
| TSSC1_CpG_17 | 1.00 (1.00, 1.00) | 1.00 (0.99, 1.00) | 0.134 | 1.00 (1.00, 1.00) | 1.00 (0.98, 1.00) | 0.290 | |
| TSSC1_CpG_18 | 0.98 (0.96, 0.99) | 0.99 (0.96, 0.99) | 0.677 | 0.99 (0.93, 0.99) | 0.99 (0.95, 0.99) | 0.756 | |
| MLPH_CpG_2.7 | 0.83 (0.77, 0.89) | 0.84 (0.80, 0.91) | 0.323 | 0.87 (0.83, 0.94) | 0.83 (0.78, 0.88) | 0.004 | |
| MLPH_CpG_3/cg06639874 | 0.90 (0.81, 0.95) | 0.91 (0.86, 0.96) | 0.215 | 0.94 (0.86, 0.97) | 0.90 (0.87, 0.92) | 0.044 | |
| MLPH_CpG_4 | 0.93 (0.86, 1.00) | 0.93 (0.89, 0.96) | 0.966 | 0.95 (0.90, 0.99) | 0.94 (0.88, 0.96) | 0.090 | |
| MLPH_CpG_5 | 0.86 (0.82, 0.93) | 0.85 (0.82, 0.89) | 0.294 | 0.90 (0.83, 0.95) | 0.87 (0.84, 0.91) | 0.228 | |
| MLPH_CpG_6 | 0.62 (0.44, 0.69) | 0.49 (0.31, 0.64) | 0.099 | 0.58 (0.34, 0.74) | 0.58 (0.41, 0.68) | 0.981 | |
| [1] | Virani SS, Alonso A, Aparicio HJ, et al. Heart disease and stroke statistics-2021 update: a report from the American heart association[J]. Circulation, 2021, 143(8): e254-743. doi:10.1161/cir.0000000000000950 |
| [2] | 中国心血管健康与疾病报告2021概要[J]. 中国循环杂志, 2022, 37 (6): 553-78. doi:10.3969/j.issn.1004-8812.2022.07.001 |
| [3] | Desiderio A, Pastorino M, Campitelli M, et al. DNA methylation in cardiovascular disease and heart failure: novel prediction models?[J]. Clin Epigenetics, 2024, 16(1): 115. doi:10.1186/s13148-024-01722-x |
| [4] | Dai Y, Chen D, Xu T. DNA methylation aberrant in atherosclerosis[J]. Front Pharmacol, 2022, 13: 815977. doi:10.3389/fphar.2022.815977 |
| [5] | Zhu L, Zhu C, Wang J, et al. The association between DNA methylation of 6p21.33 and AHRR in blood and coronary heart disease in Chinese population[J]. BMC Cardiovasc Disord, 2022, 22(1): 370. doi:10.1186/s12872-022-02766-8 |
| [6] | Agha G, Mendelson MM, Ward-Caviness CK, et al. Blood leukocyte DNA methylation predicts risk of future myocardial infarction and coronary heart disease[J]. Circulation, 2019, 140(8): 645-57. doi:10.1161/circulationaha.118.039357 |
| [7] | Nam AR, Lee KH, Hwang HJ, et al. Alternative methylation of intron motifs is associated with cancer-related gene expression in both canine mammary tumor and human breast cancer[J]. Clin Epigenetics, 2020, 12(1): 110. doi:10.1186/s13148-020-00888-4 |
| [8] | Bi L, Jin J, Fan Y, et al. Blood-based HYAL2 methylation as a potential marker for the preclinical detection of coronary heart disease and stroke[J]. Clin Epigenetics, 2024, 16(1): 130. doi:10.1186/s13148-024-01742-7 |
| [9] | Yin Q, Yang X, Li L, et al. The association between breast cancer and blood-based methylation of S100P and HYAL2 in the Chinese population[J]. Front Genet, 2020, 11: 977. doi:10.3389/fgene.2020.00977 |
| [10] | Long P, Si J, Zhu Z, et al. Genome-wide DNA methylation profiling in blood reveals epigenetic signature of incident acute coronary syndrome[J]. Nat Commun, 2024, 15(1): 7431. doi:10.1038/s41467-024-51751-6 |
| [11] | Infante T, Franzese M, Ruocco A, et al. ABCA1, TCF7, NFATC1, PRKCZ, and PDGFA DNA methylation as potential epigenetic-sensitive targets in acute coronary syndrome via network analysis[J]. Epigenetics, 2022, 17(5): 547-63. doi:10.1080/15592294.2021.1939481 |
| [12] | Salah HT, Yang RK, Roy-Chowdhuri S, et al. Spitz melanocytic neoplasms with MLPH: : ALK fusions: report of two cases with previously unreported features and literature review[J]. J Cutan Pathol, 2024, 51(6): 407-14. doi:10.1111/cup.14605 |
| [13] | Krushkal J, Silvers T, Reinhold WC, et al. Epigenome-wide DNA methylation analysis of small cell lung cancer cell lines suggests potential chemotherapy targets[J]. Clin Epigenetics, 2020, 12(1): 93. doi:10.1186/s13148-020-00876-8 |
| [14] | Zhang T, Sun Y, Zheng T, et al. MLPH accelerates the epithelial-mesenchymal transition in prostate cancer[J]. Onco Targets Ther, 2020, 13: 701-8. doi:10.2147/ott.s225023 |
| [15] | Wang DC, Wang HF, Yuan ZN. Runx2 induces bone osteolysis by transcriptional suppression of TSSC1[J]. Biochem Biophys Res Commun, 2013, 438(4): 635-9. doi:10.1016/j.bbrc.2013.07.131 |
| [16] | Ramakrishnan S, Cortes-Gomez E, Athans SR, et al. Race-specific coregulatory and transcriptomic profiles associated with DNA methylation and androgen receptor in prostate cancer[J]. Genome Med, 2024, 16(1): 52. doi:10.1186/s13073-024-01323-6 |
| [17] | Jiménez A, Vlacho B, Mata-Cases M, et al. Sex and age significantly modulate cardiovascular disease presentation in type 2 diabetes: a large population-based cohort study[J]. Front Endocrinol: Lausanne, 2024, 15: 1344007. doi:10.3389/fendo.2024.1344007 |
| [18] | Liu C, Yin Q, Li M, et al. ACTB methylation in blood as a potential marker for the pre-clinical detection of stroke: a prospective nested case-control study[J]. Front Neurosci, 2021, 15: 644943. doi:10.3389/fnins.2021.644943 |
| [19] | Schott S, Yang R, Stöcker S, et al. HYAL2 methylation in peripheral blood as a potential marker for the detection of pancreatic cancer: a case control study[J]. Oncotarget, 2017, 8(40): 67614-25. doi:10.18632/oncotarget.18757 |
| [20] | Ramezankhani A, Azizi F, Hadaegh F. Sex differences in risk factors for coronary heart disease events: a prospective cohort study in Iran[J]. Sci Rep, 2023, 13(1): 22398. doi:10.1038/s41598-023-50028-0 |
| [21] | Lee GB, Nam GE, Kim W, et al. Association between premature menopause and cardiovascular diseases and all-cause mortality in Korean women[J]. J Am Heart Assoc, 2023, 12(22): e030117. doi:10.1161/jaha.123.030117 |
| [22] | Goldsborough E, Osuji N, Blaha MJ. Assessment of cardiovascular disease risk a 2022 update[J]. Endocrinol Metab Clin N Am, 2022, 51(3): 483-509. doi:10.1016/j.ecl.2022.02.005 |
| [23] | Andersson C, Nayor M, Tsao CW, et al. Framingham heart study: JACC focus seminar, 1/8[J]. J Am Coll Cardiol, 2021, 77(21): 2680-92. doi:10.1016/j.jacc.2021.01.059 |
| [24] | Marrugat J, Vila J, Baena-Díez JM, et al. Relative validity of the 10-year cardiovascular risk estimate in a population cohort of the REGICOR study[J]. Rev Esp Cardiol, 2011, 64(5): 385-94. doi:10.1016/j.rec.2010.12.017 |
| [25] | Erdmann J, van der Laan SW. Unfolding and disentangling coronary vascular disease through genome-wide association studies[J]. Eur Heart J, 2021, 42(9): 934-7. doi:10.1093/eurheartj/ehaa1089 |
| [26] | Hartiala JA, Han Y, Jia Q, et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction[J]. Eur Heart J, 2021, 42(9): 919-33. |
| [27] | Shah S, Henry A, Roselli C, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure[J]. Nat Commun, 2020, 11(1): 163. |
| [28] | Tcheandjieu C, Zhu X, Hilliard AT, et al. Large-scale genome-wide association study of coronary artery disease in genetically diverse populations[J]. Nat Med, 2022, 28(8): 1679-92. |
| [29] | Joáo Job PM, Dos Reis Lívero FA, Junior AG. Epigenetic control of hypertension by DNA methylation: a real possibility[J]. Curr Pharm Des, 2021, 27(35): 3722-8. doi:10.2174/1381612827666210322141703 |
| [30] | Nadiger N, Veed JK, Chinya Nataraj P, et al. DNA methylation and type 2 diabetes: a systematic review[J]. Clin Epigenetics, 2024, 16(1): 67. doi:10.1186/s13148-024-01670-6 |
| [31] | Wang S, Zha L, Cui X, et al. Epigenetic regulation of hepatic lipid metabolism by DNA methylation[J]. Adv Sci: Weinh, 2023, 10(20): e2206068. doi:10.1002/advs.202206068 |
| [32] | Xie M, Tang Q, Nie J, et al. BMAL1-downregulation aggravates Porphyromonas gingivalis-induced atherosclerosis by encouraging oxidative stress[J]. Circ Res, 2020, 126(6): e15-29. doi:10.1161/circresaha.119.315502 |
| [33] | Xia YJ, Brewer A, Bell JT. DNA methylation signatures of incident coronary heart disease: findings from epigenome-wide association studies[J]. Clin Epigenet, 2021, 13(1): 186. doi:10.1186/s13148-021-01175-6 |
| [34] | Uddin MDM, Nguyen NQH, Yu B, et al. Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease[J]. Nat Commun, 2022, 13(1): 5350. doi:10.1038/s41467-022-33093-3 |
| [35] | Zhang X, Wang C, He D, et al. Identification of DNA methylation-regulated genes as potential biomarkers for coronary heart disease via machine learning in the Framingham Heart Study[J]. Clin Epigenetics, 2022, 14(1): 122. doi:10.1186/s13148-022-01343-2 |
| [36] | Feng CH, Chang YM, Weng JR. Rapid methylation of valsartan in human plasma using evaporative derivatization reagent to improve its sensitivity in MALDI-TOF mass spectrometry[J]. Anal Methods, 2022, 14(37): 3694-701. doi:10.1039/d2ay01164f |
| [1] | 夏冰, 彭进, 丁九阳, 王杰, 唐国伟, 刘国杰, 王沄, 万昌武, 乐翠云. ATF3通过NF-κB信号通路调控动脉粥样硬化斑块内的炎症反应[J]. 南方医科大学学报, 2025, 45(6): 1131-1142. |
| [2] | 胡司淦, 程增为, 李敏, 高世毅, 高大胜, 康品方. 冠状动脉慢性完全闭塞病变侧支循环的建立与胰岛素抵抗的相关性[J]. 南方医科大学学报, 2024, 44(4): 780-786. |
| [3] | 武若杰, 刘 睿, 张一粟, 李晓红. 帕瑞昔布钠改善腹腔镜下直肠癌根治术患者的炎症微环境并促进患者恢复:基于下调CXCL8-CXCR1/2表达[J]. 南方医科大学学报, 2024, 44(2): 363-369. |
| [4] | 谭茹雪, 包晓樟, 韩亮, 李朝晖, 田男. 基于HOXA9 DNA甲基化的两位点联合预测模型可用于脑膜瘤进展风险的早期筛查[J]. 南方医科大学学报, 2024, 44(11): 2110-2120. |
| [5] | 姜 勇, 葛文婷, 赵 莹, 吴煜格, 霍一鸣, 潘岚婷, 曹 爽. LINC00926通过募集ELAVL1促进低氧诱导的人脐静脉血管内皮细胞焦亡[J]. 南方医科大学学报, 2023, 43(5): 807-814. |
| [6] | 臧紫珊, 乔 荣, 朱 强, 周夏婕, 顾万建, 韩宝惠, 杨蓉西. 外周血中KCNMA1基因甲基化水平与肺癌的发生和发展相关[J]. 南方医科大学学报, 2023, 43(3): 349-359. |
| [7] | 陆 琦, 鲍迎春, 陈旭姣, 王春明. 白兔局部动脉粥样硬化不稳定斑块对其余血管斑块形成的影响[J]. 南方医科大学学报, 2023, 43(1): 117-121. |
| [8] | 韩艳艳, 费 翔, 任 玲, 汪晶晶, 陈 韬, 郭 军, 汪 奇. 颈动脉斑块内新生血管与PCI术后患者再次血运重建具有相关性[J]. 南方医科大学学报, 2022, 42(6): 892-898. |
| [9] | 殷益飞, 李 红, 杨春生, 张敏敏, 黄选东, 李梦夏, 杨蓉西, 张正东. HYAL2基因 DNA 甲基化水平可用于甲状腺良恶性肿瘤的鉴别诊断[J]. 南方医科大学学报, 2022, 42(1): 123-129. |
| [10] | 王子贤, 赖伟华, 钟诗龙. 两样本孟德尔随机化方法分析血液代谢物与冠心病的因果关系[J]. 南方医科大学学报, 2021, 41(2): 272-278. |
| [11] | 周夏婕, 雷水芳, 黎立喜, 徐 天, 顾万建, 马 飞, 杨蓉西. 中国女性外周血EMR3基因甲基化水平与乳腺癌的相关性[J]. 南方医科大学学报, 2021, 41(10): 1456-1463. |
| [12] | 高亚莉, 罗小玲, 孟 婷, 朱敏娟, 田渼雯, 陆晓和. DNMT1蛋白通过沉默MEG3基因促进视网膜母细胞瘤增殖[J]. 南方医科大学学报, 2020, 40(09): 1239-1245. |
| [13] | 黄建玉,马 骏,郭 琰,王 瑶,张群辉,钟俊达,姜文才,李雁卓,徐 琳. 高血压患者外周血红蛋白水平与动脉硬化程度显著相关[J]. 南方医科大学学报, 2020, 40(02): 240-245. |
| [14] | 乔香瑞,刘军辉,花 蕊,卓小桢. 循环单核细胞和血浆中GDF-15和NT-proBNP对慢性心力衰竭的诊断及心血管事件的预测价值[J]. 南方医科大学学报, 2019, 39(11): 1273-1279. |
| [15] | 郑中华,季慧慧,陈楚嘉,李银,段世伟. 健康人群CDKN2A和CDKN2B基因甲基化水平与衰老的相关性[J]. 南方医科大学学报, 2019, 39(06): 724-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||