Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (12): 2347-2358.doi: 10.12122/j.issn.1673-4254.2024.12.10
Previous Articles Next Articles
Huahua ZHANG1( ), Qingyin TA1,2(
), Qingyin TA1,2( ), Yun FENG1, Jiming HAN1(
), Yun FENG1, Jiming HAN1( )
)
Received:2024-06-27
Online:2024-12-20
Published:2024-12-26
Contact:
Jiming HAN
E-mail:yadxzhh@yau.edu.cn;1909983949@qq.com;mthgh_jm@163.com
Supported by:Huahua ZHANG, Qingyin TA, Yun FENG, Jiming HAN. Holliday junction-recognizing protein is a potential predictive and prognostic biomarker for kidney renal clear cell carcinoma[J]. Journal of Southern Medical University, 2024, 44(12): 2347-2358.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.12.10
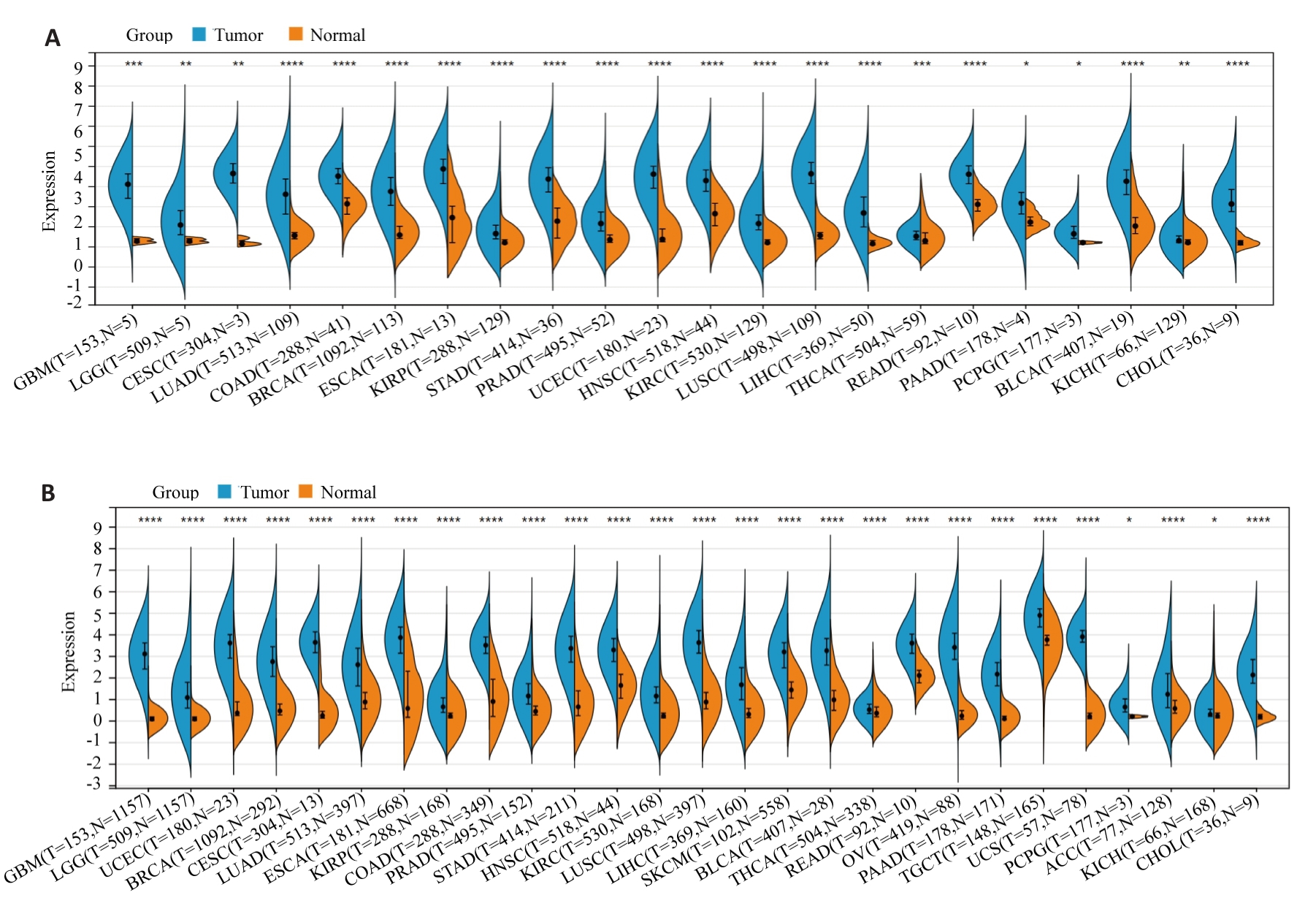
Fig.1 Expression of Holliday junction-recognizing protein (HJURP) in pan-cancer. A: Expression of HJURP in different human tumors based on TCGA database. B: HJURP expression in 26 human tumors analyzed by integrating normal tissue data from GTEx database and TCGA tumor tissue. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs Normal group.
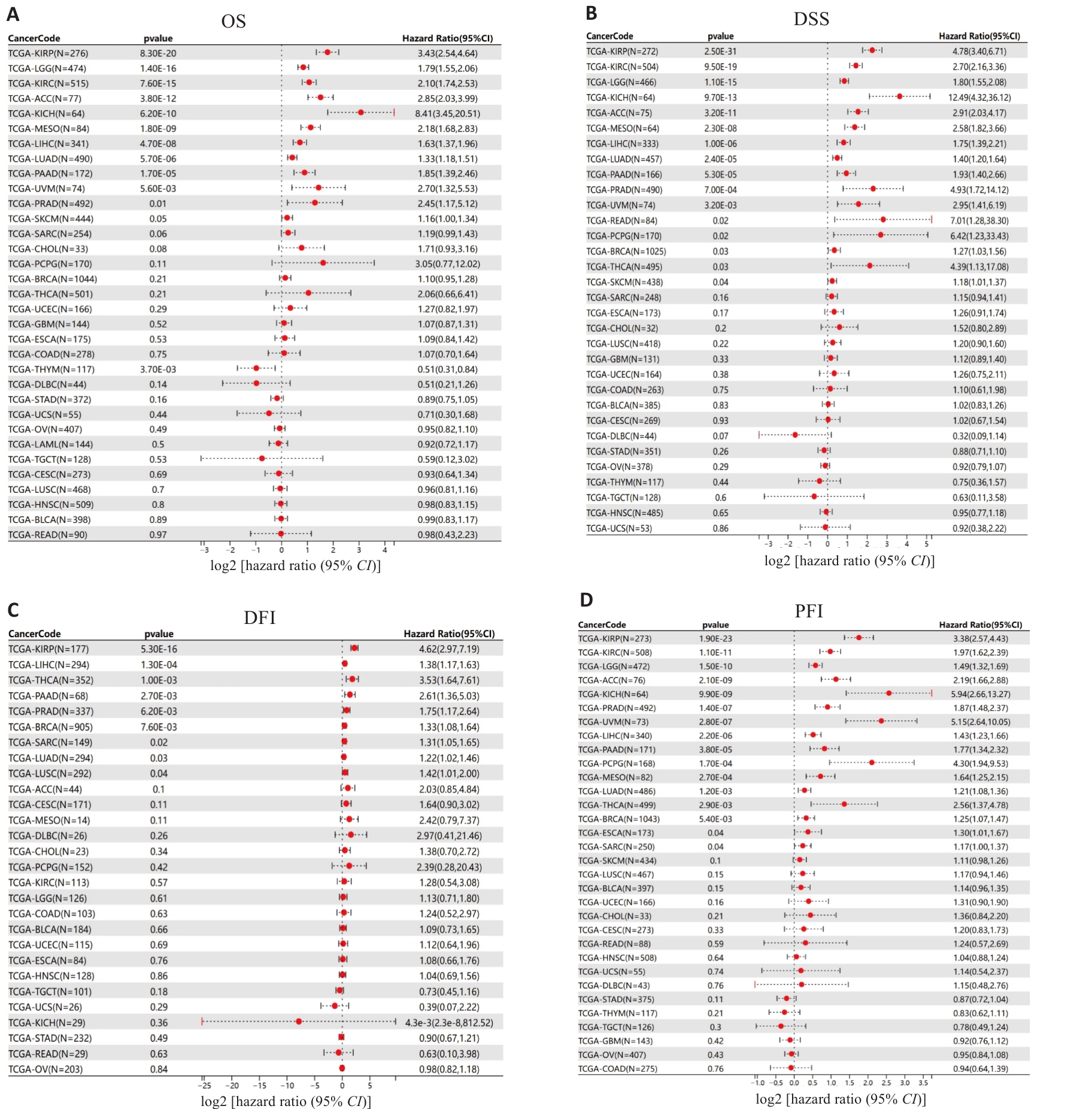
Fig.2 Survival forest map for prognostic analysis of HJURP expression in pan-cancer. OS: Overall survival; DSS: Disease-specific survival; DFI: Disease-free interval; PFI: Progression-free interval.
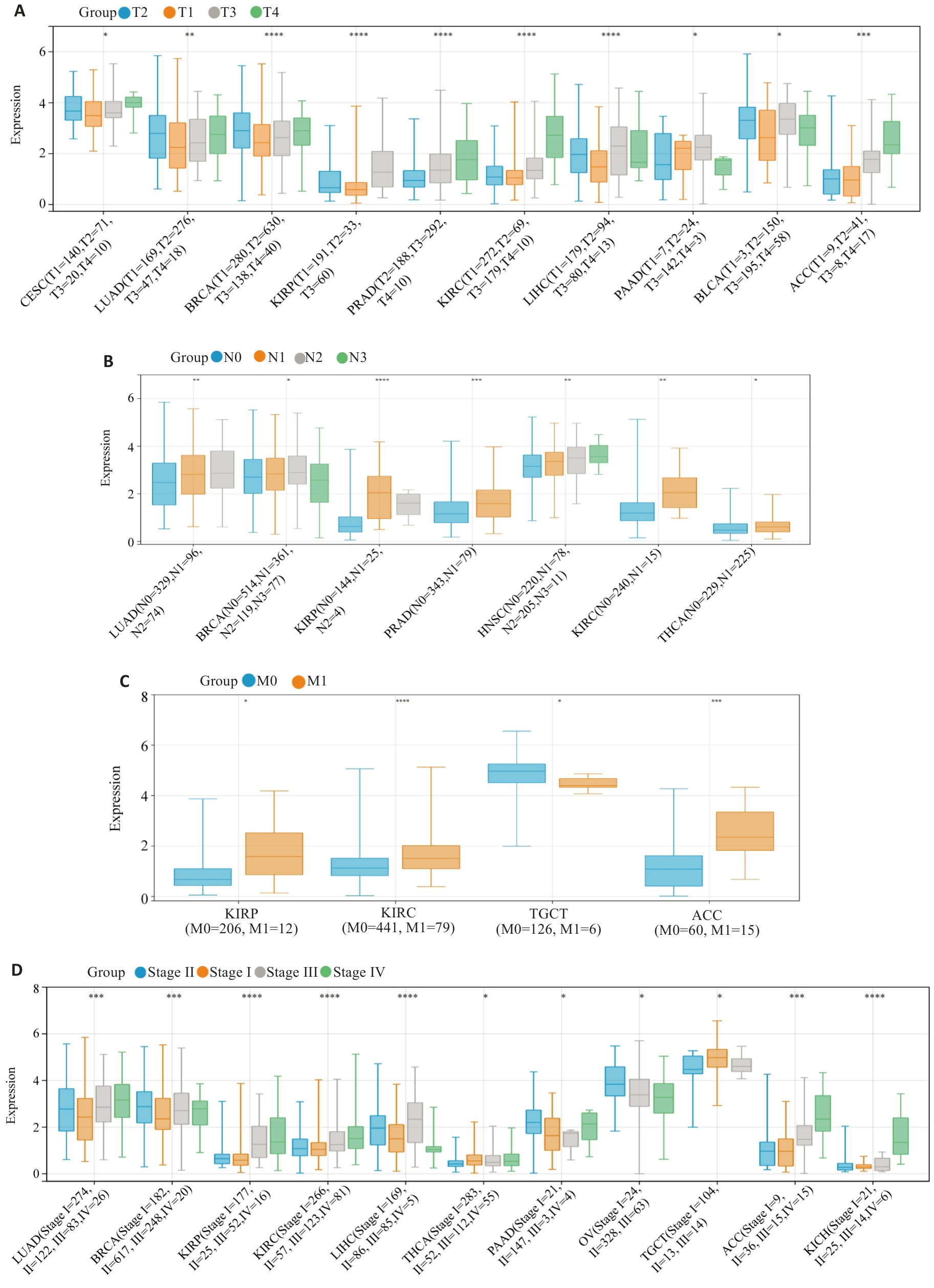
Fig.3 Associations between HJURP expression and clinicopathological characteristics of pan-cancer. A: T grade. B: N grade. C: M status. D: clinical stage. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs T1, N0, M0 or Stage I.
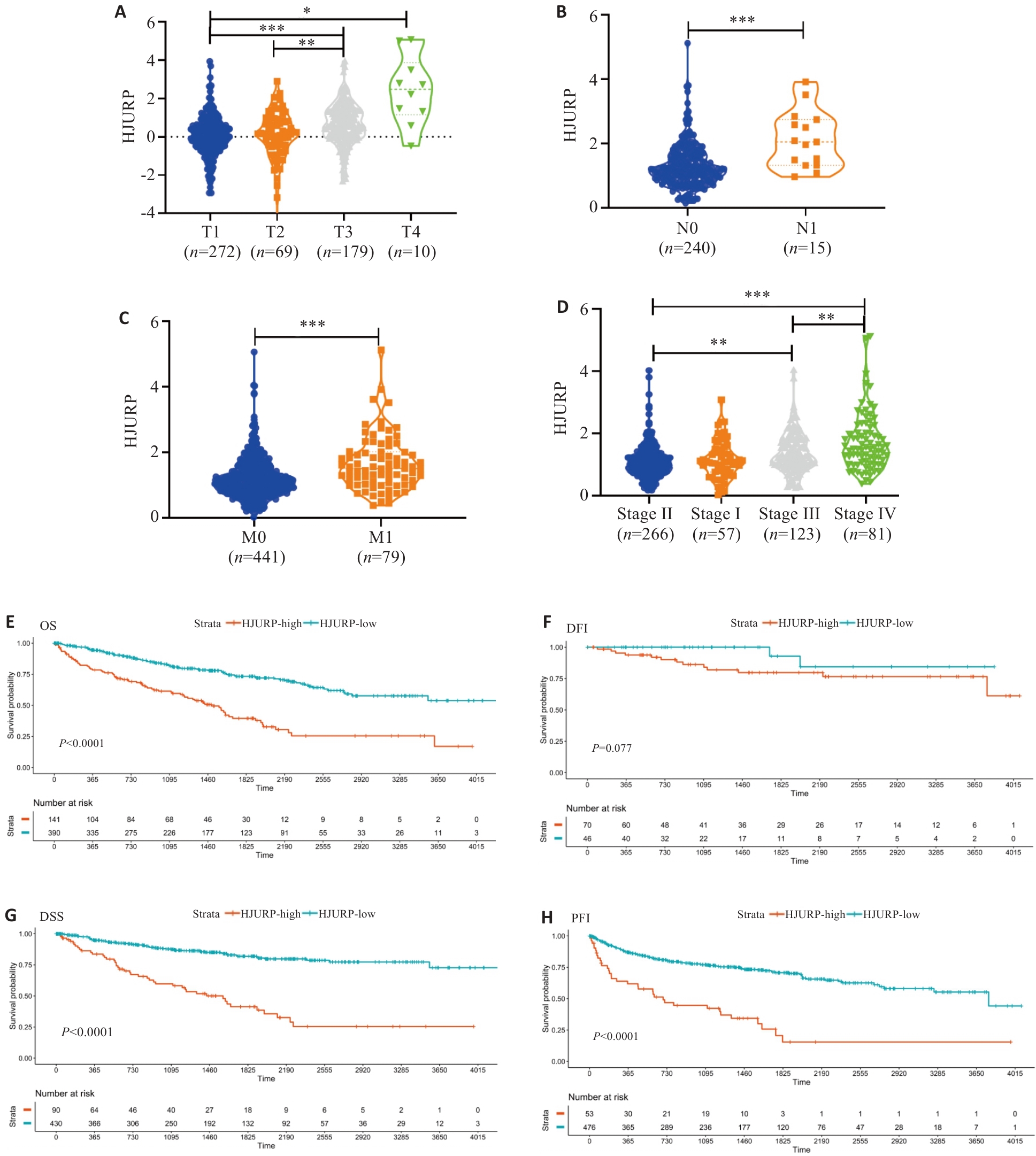
Fig.5 Correlation of HJURP expression with clinicopathological characteristics and prognosis of kidney renal clear cell carcinoma (KIRC). A: T grade. B: N grade. C: M status. D: clinical stage. E: HR (95% CI) for OS. F: HR (95% CI) for DSS. G: HR (95% CI) for DFI. H: HR (95% CI) for PFI. *P<0.05, **P<0.01, ***P<0.001 vs T1, N0, M0, or Stage I.
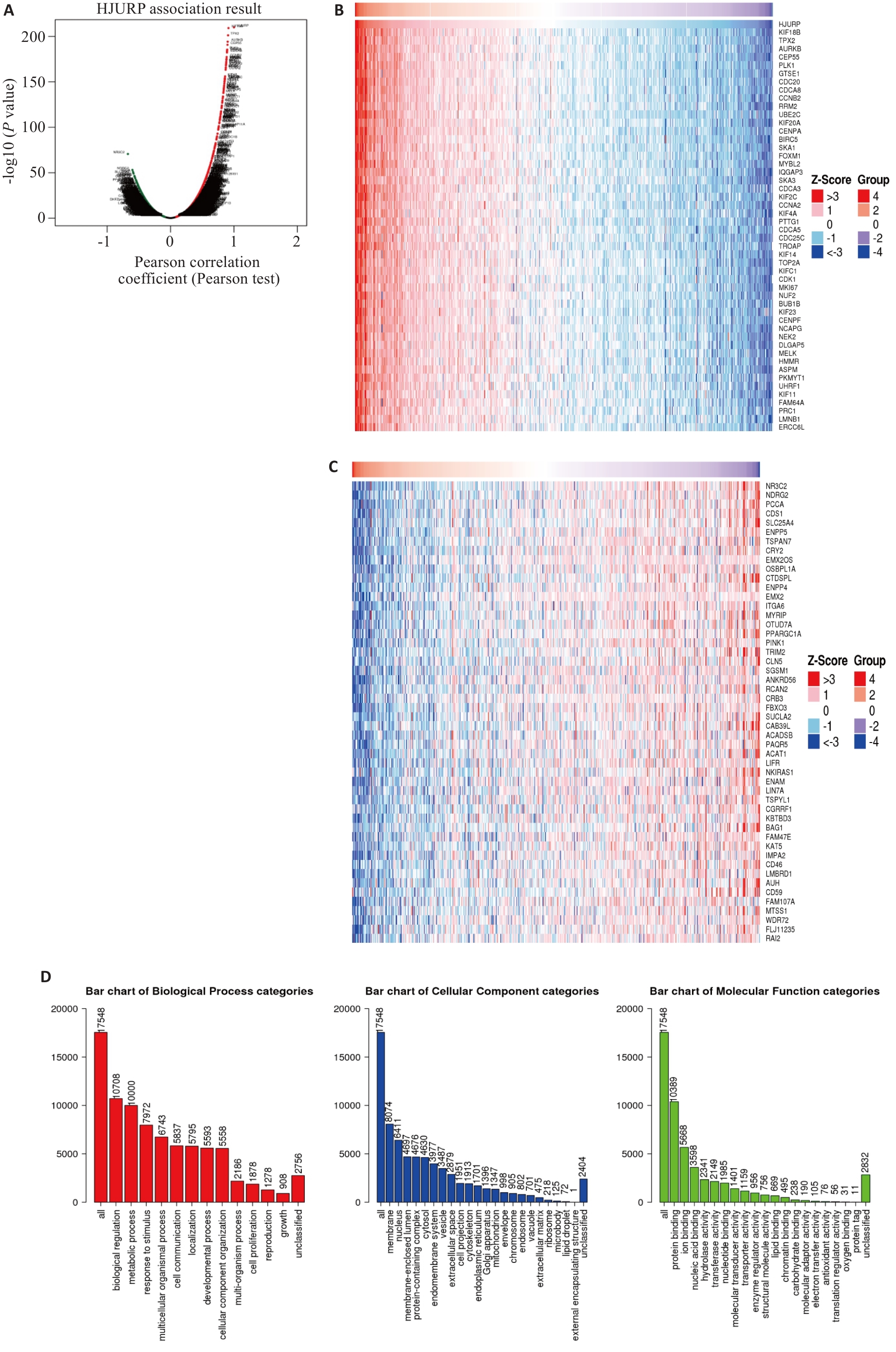
Fig.6 Analysis of HJURP-related genes in KIRC and the GO function. A: Volcanic plot of HJURP-related genes in KIRC. B: Heat map of genes positively associated with HJURP in KIRC. C: Heat map of genes negatively associated with HJURP in KIRC. D: GO function analysis of HJURP in KIRC.
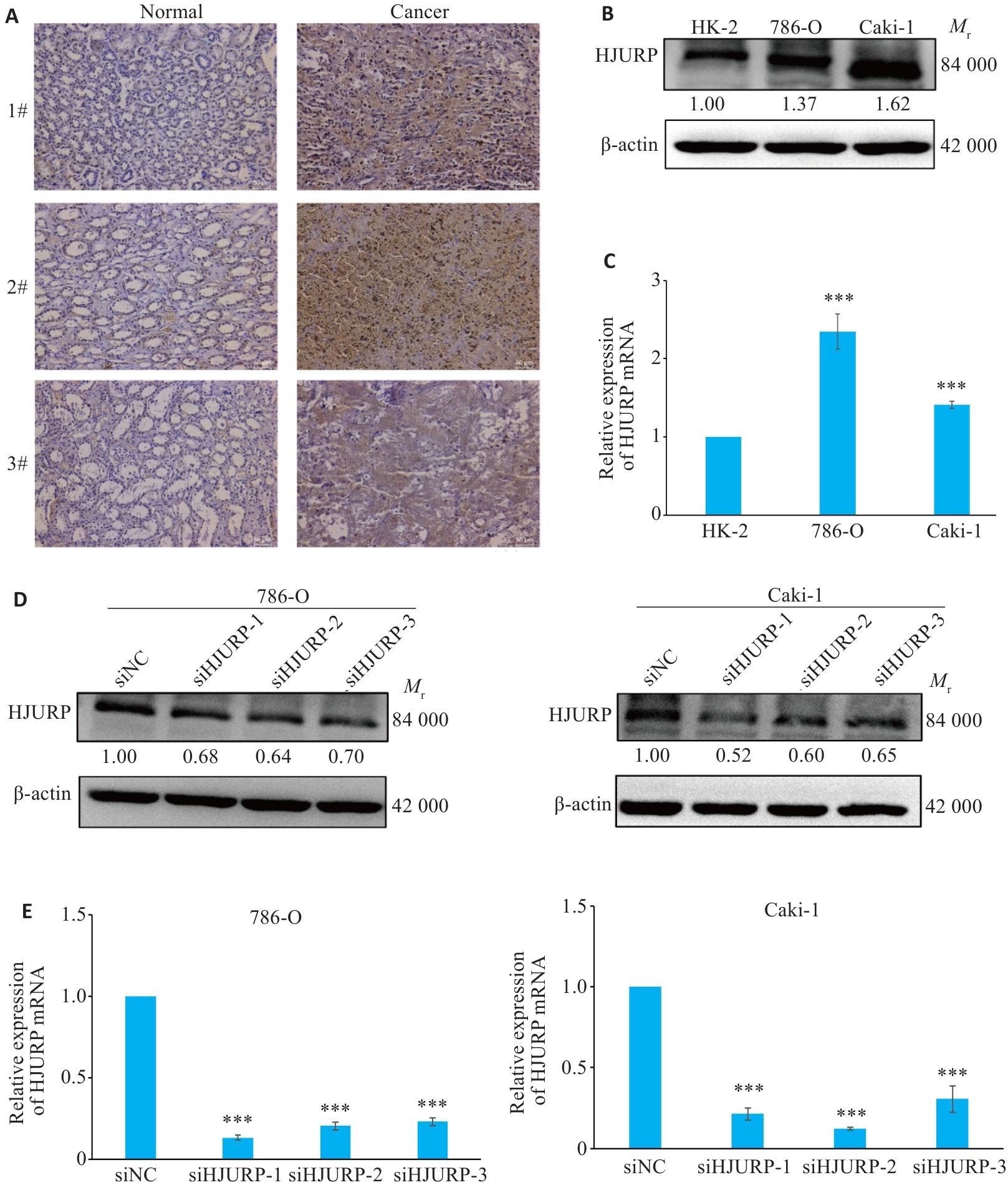
Fig.7 Expression of HJURP in KIRC and validation of the efficiency of HJURP interference. A: HJURP in adjacent and KIRC tissues detected by immunohistochemistry (×100). B: Differential expression of HJURP in KIRC verified by Western blotting. C: Expression of HJURP mRNA in KIRC cell lines detected by qRT-PCR. D, E: Transfection efficiency of si-HJURP sequences assessed by Western blotting (D) and qRT-PCR (E). ***P<0.001 vs HK-2 or siNC group.
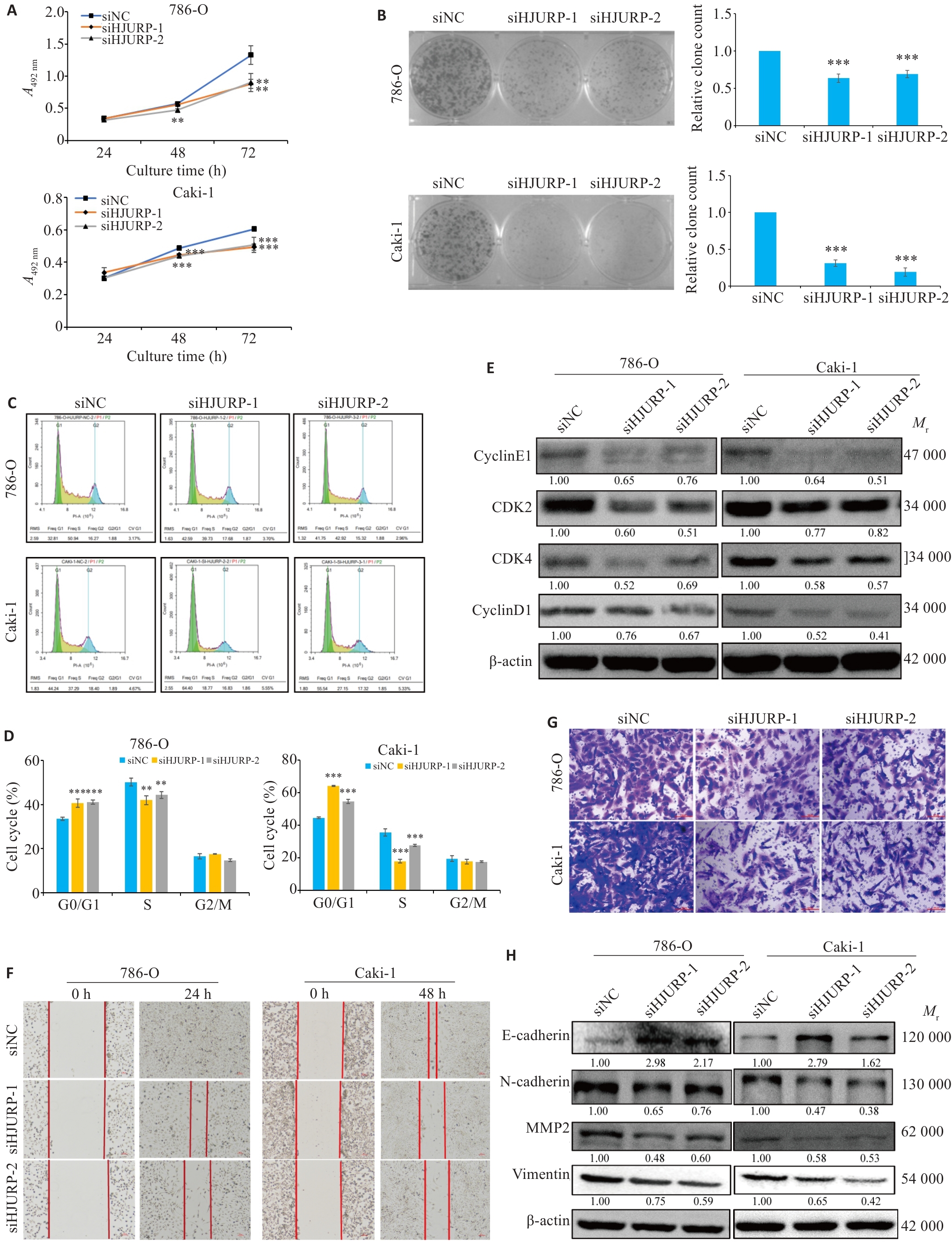
Fig.8 Silencing of HJURP suppresses proliferation and migration of KIRC cells in vitro. A:CCK-8 assays for cell viability. B: Clone formation test to detect the changes of cell clone formation ability. C, D: Flow cytometry for analyzing cell cycle distribution in 786-O and Caki-1 cells (Mean±SD, n=3). E: Changes in cell cycle-related proteins after silencing HJURP. F: Scratch test for assessing cell migration ability (×100). G: Transwell assays for assessing cell migration (×200). H: Western blotting for detecting cell migration-related protein expressions including E-cadherin, N-cadherin, MMP2 and vimentin. **P<0.01, ***P<0.001 vs siNC group.
| 1 | Zheng RS, Chen R, Han BF, et al. Cancer incidence and mortality in China, 2022[J]. Zhonghua Zhong Liu Za Zhi, 2024, 46(3): 221-31. |
| 2 | Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma[J]. N Engl J Med, 2015, 372(4): 311-9. |
| 3 | Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma[J]. N Engl J Med, 2015, 373(1): 23-34. |
| 4 | Robert C, Ribas A, Wolchok JD, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial[J]. Lancet, 2014, 384(9948): 1109-17. |
| 5 | Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer[J]. N Engl J Med, 2012, 366(26): 2455-65. |
| 6 | Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project[J]. Nat Genet, 2013, 45(10): 1113-20. |
| 7 | Kato T, Sato N, Hayama S, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells[J]. Cancer Res, 2007, 67(18): 8544-53. |
| 8 | Barnhart MC, Kuich PH, Stellfox ME, et al. HJURP is a CENP-a chromatin assembly factor sufficient to form a functional de novo kinetochore[J]. J Cell Biol, 2011, 194(2): 229-43. |
| 9 | Wei Y, Ouyang GL, Yao WX, et al. Knockdown of HJURP inhibits non-small cell lung cancer cell proliferation, migration, and invasion by repressing Wnt/β-catenin signaling[J]. Eur Rev Med Pharmacol Sci, 2019, 23(9): 3847-56. |
| 10 | Kang DH, Woo J, Kim H, et al. Prognostic relevance of HJURP expression in patients with surgically resected colorectal cancer[J]. Int J Mol Sci, 2020, 21(21): 7928. |
| 11 | Wang CJ, Li X, Shi P, et al. Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling[J]. Cell Death Dis, 2020, 11(5): 386. |
| 12 | Cao R, Wang G, Qian KY, et al. Silencing of HJURP induces dysregulation of cell cycle and ROS metabolism in bladder cancer cells via PPARγ-SIRT1 feedback loop[J]. J Cancer, 2017, 8(12): 2282-95. |
| 13 | Wang L, Qu JL, Liang Y, et al. Identification and validation of key genes with prognostic value in non-small-cell lung cancer via integrated bioinformatics analysis[J]. Thorac Cancer, 2020, 11(4): 851-66. |
| 14 | Gu Y, Li J, Guo DL, et al. Identification of 13 key genes correlated with progression and prognosis in hepatocellular carcinoma by weighted gene co-expression network analysis[J]. Front Genet, 2020, 11: 153. |
| 15 | Hu Z, Huang G, Sadanandam A, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer[J]. Breast Cancer Res, 2010, 12(2): R18. |
| 16 | Fu FQ, Zhang Y, Gao ZD, et al. Development and validation of a five-gene model to predict postoperative brain metastasis in operable lung adenocarcinoma[J]. Int J Cancer, 2020, 147(2): 584-92. |
| 17 | Shen WT, Song ZG, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform[J]. Imeta, 2022, 1(3): e36. |
| 18 | Li TW, Fan JY, Wang BB, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells[J]. Cancer Res, 2017, 77(21): e108-10. |
| 19 | Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data[J]. Nat Commun, 2013, 4: 2612. |
| 20 | Telloni SM. Tumor staging and grading: a primer[J]. Methods Mol Biol, 2017, 1606: 1-17. |
| 21 | Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types[J]. Nucleic Acids Res, 2018, 46(D1): D956-63. |
| 22 | Li L, Yuan Q, Chu YM, et al. Advances in holliday junction recognition protein (HJURP): structure, molecular functions, and roles in cancer[J]. Front Cell Dev Biol, 2023, 11: 1106638. |
| 23 | Chen TC, Zhou LF, Zhou Y, et al. HJURP promotes epithelial-to-mesenchymal transition via upregulating SPHK1 in hepatocellular carcinoma[J]. Int J Biol Sci, 2019, 15(6): 1139-47. |
| 24 | Yang Y, Yuan JY, Liu ZZ, et al. The expression, clinical relevance, and prognostic significance of HJURP in cholangiocarcinoma[J]. Front Oncol, 2022, 12: 972550. |
| 25 | Zhang F, Yuan DB, Song JK, et al. HJURP is a prognostic biomarker for clear cell renal cell carcinoma and is linked to immune infiltration[J]. Int Immunopharmacol, 2021, 99: 107899. |
| 26 | Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression[J]. Cancer Res, 2019, 79(18): 4557-66. |
| 27 | Luo DC, Liao SN, Liu Y, et al. Holliday cross-recognition protein HJURP: association with the tumor microenvironment in hepatocellular carcinoma and with patient prognosis[J]. Pathol Oncol Res, 2022, 28: 1610506. |
| 28 | Liu L, Zhang ZF, Xia XL, et al. KIF18B promotes breast cancer cell proliferation, migration and invasion by targeting TRIP13 and activating the Wnt/β‑catenin signaling pathway[J]. Oncol Lett, 2022, 23(4): 112. |
| 29 | Xie JL, Wang B, Luo WJ, et al. Upregulation of KIF18B facilitates malignant phenotype of esophageal squamous cell carcinoma by activating CDCA8/mTORC1 pathway[J]. J Clin Lab Anal, 2022, 36(10): e24633. |
| 30 | Koike Y, Yin CZ, Sato Y, et al. TPX2 is a prognostic marker and promotes cell proliferation in neuroblastoma[J]. Oncol Lett, 2022, 23(4): 136. |
| 31 | Li LZ, Jiang PC, Hu WM, et al. AURKB promotes bladder cancer progression by deregulating the p53 DNA damage response pathway via MAD2L2[J]. J Transl Med, 2024, 22(1): 295. |
| [1] | Ting XIE, Yunyun WANG, Ting GUO, Chunhua YUAN. The peptide toxin components and nucleotide metabolites in Macrothele raveni venom synergistically inhibit cancer cell proliferation by activating the pro-apoptotic pathways [J]. Journal of Southern Medical University, 2025, 45(7): 1460-1470. |
| [2] | Xiuying GONG, Shunfu HOU, Miaomiao ZHAO, Xiaona WANG, Zhihan ZHANG, Qinghua LIU, Chonggao YIN, Hongli LI. LncRNA SNHG15 promotes proliferation, migration and invasion of lung adenocarcinoma cells by regulating COX6B1 through sponge adsorption of miR-30b-3p [J]. Journal of Southern Medical University, 2025, 45(7): 1498-1505. |
| [3] | Yumei ZENG, Jike LI, Zhongxi HUANG, Yibo ZHOU. Villin-like protein VILL suppresses proliferation of nasopharyngeal carcinoma cells by interacting with LMO7 protein [J]. Journal of Southern Medical University, 2025, 45(5): 954-961. |
| [4] | Yaqing YUE, Zhaoxia MU, Xibo WANG, Yan LIU. Aurora-A overexpression promotes cervical cancer cell invasion and metastasis by activating the NF-κBp65/ARPC4 signaling axis [J]. Journal of Southern Medical University, 2025, 45(4): 837-843. |
| [5] | Yi ZHANG, Yu SHEN, Zhiqiang WAN, Song TAO, Yakui LIU, Shuanhu WANG. High expression of CDKN3 promotes migration and invasion of gastric cancer cells by regulating the p53/NF-κB signaling pathway and inhibiting cell apoptosis [J]. Journal of Southern Medical University, 2025, 45(4): 853-861. |
| [6] | Shunjie QING, Zhiyong SHEN. High expression of hexokinase 2 promotes proliferation, migration and invasion of colorectal cancer cells by activating the JAK/STAT pathway and regulating tumor immune microenvironment [J]. Journal of Southern Medical University, 2025, 45(3): 542-553. |
| [7] | Lu TAO, Zhuoli WEI, Yueyue WANG, Ping XIANG. CEACAM6 inhibits proliferation and migration of nasopharyngeal carcinoma cells by suppressing epithelial-mesenchymal transition [J]. Journal of Southern Medical University, 2025, 45(3): 566-576. |
| [8] | Qingqing HUANG, Wenjing ZHANG, Xiaofeng ZHANG, Lian WANG, Xue SONG, Zhijun GENG, Lugen ZUO, Yueyue WANG, Jing LI, Jianguo HU. High MYO1B expression promotes proliferation, migration and invasion of gastric cancer cells and is associated with poor patient prognosis [J]. Journal of Southern Medical University, 2025, 45(3): 622-631. |
| [9] | Jinhua ZOU, Hui WANG, Dongyan ZHANG. SLC1A5 overexpression accelerates progression of hepatocellular carcinoma by promoting M2 polarization of macrophages [J]. Journal of Southern Medical University, 2025, 45(2): 269-284. |
| [10] | Yu BIN, Ziwen LI, Suwei ZUO, Sinuo SUN, Min LI, Jiayin SONG, Xu LIN, Gang XUE, Jingfang WU. High expression of apolipoprotein C1 promotes proliferation and inhibits apoptosis of papillary thyroid carcinoma cells by activating the JAK2/STAT3 signaling pathway [J]. Journal of Southern Medical University, 2025, 45(2): 359-370. |
| [11] | Zhoufang CAO, Yuan WANG, Mengna WANG, Yue SUN, Feifei LIU. LINC00837/miR-671-5p/SERPINE2 functional axis promotes pathological processes of fibroblast-like synovial cells in rheumatoid arthritis [J]. Journal of Southern Medical University, 2025, 45(2): 371-378. |
| [12] | Yaobin WANG, Liuyan CHEN, Yiling LUO, Jiqing SHEN, Sufang ZHOU. Predictive value of NUF2 for prognosis and immunotherapy responses in pan-cancer [J]. Journal of Southern Medical University, 2025, 45(1): 137-149. |
| [13] | Xiaohua CHEN, Hui LU, Ziliang WANG, Lian WANG, Yongsheng XIA, Zhijun GENG, Xiaofeng ZHANG, Xue SONG, Yueyue WANG, Jing LI, Jianguo HU, Lugen ZUO. Role of Abelson interactor 2 in progression and prognosis of gastric cancer and its regulatory mechanisms [J]. Journal of Southern Medical University, 2024, 44(9): 1653-1661. |
| [14] | Liangjun XUE, Qiuyu TAN, Jingwen XU, Lu FENG, Wenjin LI, Liang YAN, Yulei LI. MiR-6838-5p overexpression inhibits proliferation of breast cancer MCF-7 cells by downregulating DDR1 expression [J]. Journal of Southern Medical University, 2024, 44(9): 1677-1684. |
| [15] | Kai JI, Guanyu YU, Leqi ZHOU, Tianshuai ZHANG, Qianlong LING, Wenjiang MAN, Bing ZHU, Wei ZHANG. HNRNPA1 gene is highly expressed in colorectal cancer: its prognostic implications and potential as a therapeutic target [J]. Journal of Southern Medical University, 2024, 44(9): 1685-1695. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||