Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (10): 2004-2014.doi: 10.12122/j.issn.1673-4254.2024.10.19
Previous Articles Next Articles
Xiaoyin HUANG1( ), Fenglian CHEN1, Yu ZHANG1,2,3(
), Fenglian CHEN1, Yu ZHANG1,2,3( ), Shujun LIANG1,2,3(
), Shujun LIANG1,2,3( )
)
Received:2024-05-28
Online:2024-10-20
Published:2024-10-31
Contact:
Yu ZHANG, Shujun LIANG
E-mail:huangxiaoyin2003@163.com;yuzhang@smu.edu.cn;lsj123@smu.edu.cn
Supported by:Xiaoyin HUANG, Fenglian CHEN, Yu ZHANG, Shujun LIANG. A predictive model for survival outcomes of glioma patients based on multi-parametric, multi-regional MRI radiomics features and clinical features[J]. Journal of Southern Medical University, 2024, 44(10): 2004-2014.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.10.19
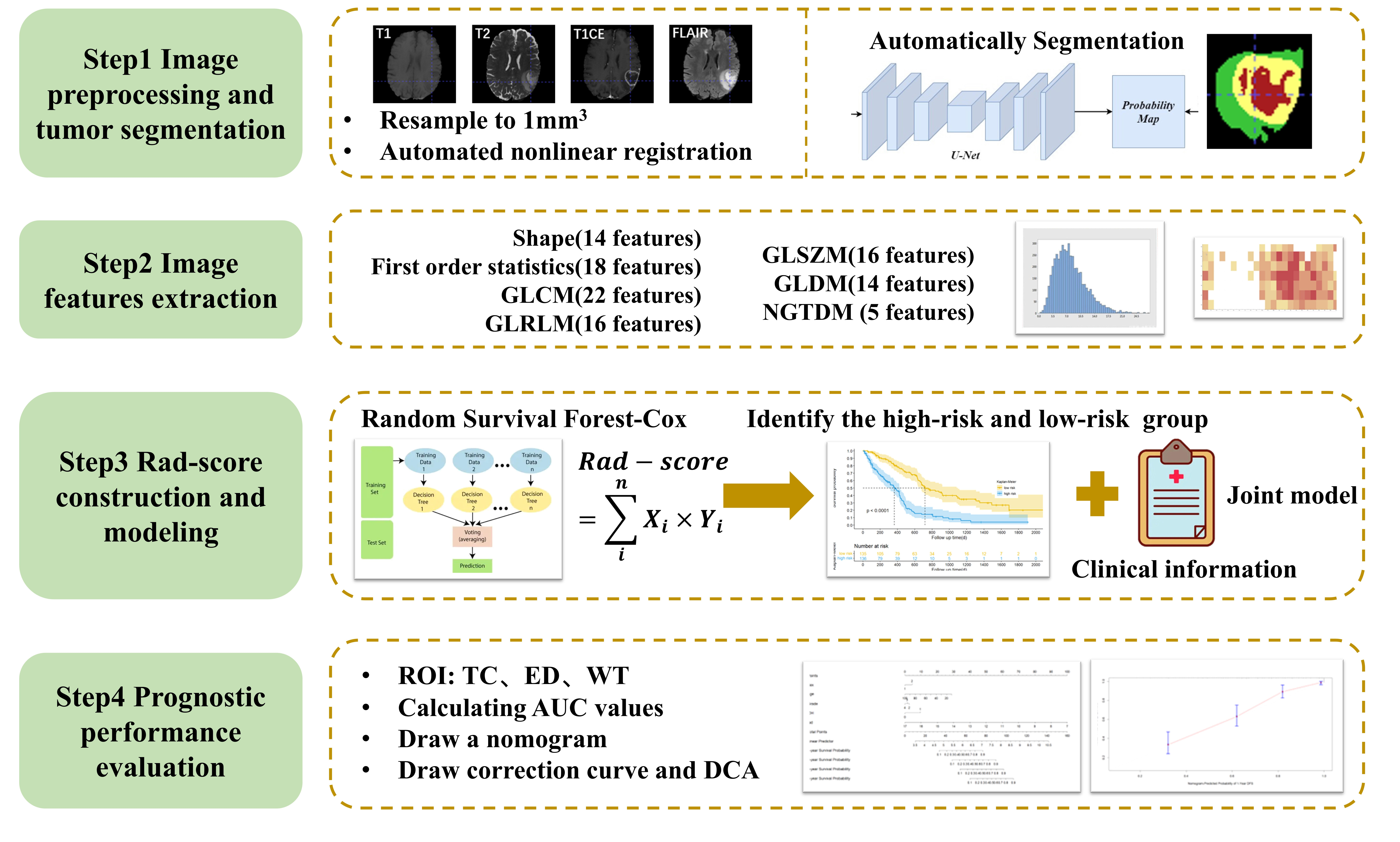
Fig.1 Flowchart of the study design. T1CE: T1-weighted contrast-enhanced; FLAIR: Fluid attenuated inversion recovery; GLCM: Gray-level co-occurrence matrix; GLRLM: Gray-level run-length matrix; GLSZM: Gray-level size zone matrix; GLDM: Gray-level dependence matrix; NGTDM: Neighborhood gray tone difference matrix; TC: Tumor core; ED: Edema region; WT: Whole tumor; AUC: Area under the curve; DCA: Decision curve analysis.
| Item | Training dataset (n=271) | Test dataset (n=117) | χ2 | P |
|---|---|---|---|---|
| Age (year, Mean±SD) | 60.72±13.40 | 58.32±13.29 | 79.249 | 0.110 |
| Gender [n (%)] | 2.399 | 0.121 | ||
| Male | 158 (58.30) | 78 (66.70) | ||
| Female | 113 (41.70) | 39 (33.30) | ||
| WHO grade [n (%)] | 1.990 | 0.370 | ||
| Ⅱ | 2 (0.70) | 0 (0.00) | ||
| Ⅲ | 3 (1.10) | 3 (2.60) | ||
| Ⅳ | 266 (98.20) | 114 (97.40) | ||
| IDH [n (%)] | 1.462 | 0.227 | ||
| Wide type | 251 (92.60) | 104 (88.90) | ||
| Mutant | 20 (7.40) | 13 (11.10) | ||
| Status [n (%)] | 1.421 | 0.233 | ||
| Non-censoring | 159 (58.70) | 61 (52.10) | ||
| Censoring | 112 (41.30) | 76 (47.90) | ||
| Survival time (d, Mean±SD) | 461.57±414.48 | 558.85±463.16 | 329.837 | 0.312 |
Tab.1 Comparison of general clinical data of the patients between the training and test datasets
| Item | Training dataset (n=271) | Test dataset (n=117) | χ2 | P |
|---|---|---|---|---|
| Age (year, Mean±SD) | 60.72±13.40 | 58.32±13.29 | 79.249 | 0.110 |
| Gender [n (%)] | 2.399 | 0.121 | ||
| Male | 158 (58.30) | 78 (66.70) | ||
| Female | 113 (41.70) | 39 (33.30) | ||
| WHO grade [n (%)] | 1.990 | 0.370 | ||
| Ⅱ | 2 (0.70) | 0 (0.00) | ||
| Ⅲ | 3 (1.10) | 3 (2.60) | ||
| Ⅳ | 266 (98.20) | 114 (97.40) | ||
| IDH [n (%)] | 1.462 | 0.227 | ||
| Wide type | 251 (92.60) | 104 (88.90) | ||
| Mutant | 20 (7.40) | 13 (11.10) | ||
| Status [n (%)] | 1.421 | 0.233 | ||
| Non-censoring | 159 (58.70) | 61 (52.10) | ||
| Censoring | 112 (41.30) | 76 (47.90) | ||
| Survival time (d, Mean±SD) | 461.57±414.48 | 558.85±463.16 | 329.837 | 0.312 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HHH_glszm_SizeZoneNonUniformityNormalized | T1 | -2.886 |
| 2 | wavelet.HHL_glcm_Imc2 | T1 | -1.126 |
| 3 | lbp.3D.m2_firstorder_Maximum | T1 | 1.154 |
| 4 | wavelet.HHL_gldm_DependenceNonUniformityNormalized | T2 | -1.209 |
| 5 | original_shape_MajorAxisLength | FLAIR | 1.530 |
Tab.2 Optimal features from the tumor core for predicting glioma prognosis and the corresponding sequences and coefficients (Core tumor)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HHH_glszm_SizeZoneNonUniformityNormalized | T1 | -2.886 |
| 2 | wavelet.HHL_glcm_Imc2 | T1 | -1.126 |
| 3 | lbp.3D.m2_firstorder_Maximum | T1 | 1.154 |
| 4 | wavelet.HHL_gldm_DependenceNonUniformityNormalized | T2 | -1.209 |
| 5 | original_shape_MajorAxisLength | FLAIR | 1.530 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | original_shape_Maximum2DDiameterSlice | T1 | 0.988 |
| 2 | original_shape_Flatness | T1 | -0.915 |
| 3 | wavelet.HHH_gldm_DependenceNonUniformityNormalized | T1CE | 0.902 |
| 4 | wavelet.HHL_glszm_SizeZoneNonUniformity | T1CE | 0.526 |
| 5 | lbp.3D.m2_firstorder_90Percentile | T1CE | 0.956 |
| 6 | lbp.3D.k_firstorder_Minimum | FLAIR | -3.017 |
| 7 | log.sigma.4.0.mm.3D_glrlm_LongRunLowGrayLevelEmphasis | FLAIR | 2.411 |
Tab.3 Optimal features from the peritumor edema region for predicting glioma prognosis and their corresponding sequences and coefficients (Edema region)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | original_shape_Maximum2DDiameterSlice | T1 | 0.988 |
| 2 | original_shape_Flatness | T1 | -0.915 |
| 3 | wavelet.HHH_gldm_DependenceNonUniformityNormalized | T1CE | 0.902 |
| 4 | wavelet.HHL_glszm_SizeZoneNonUniformity | T1CE | 0.526 |
| 5 | lbp.3D.m2_firstorder_90Percentile | T1CE | 0.956 |
| 6 | lbp.3D.k_firstorder_Minimum | FLAIR | -3.017 |
| 7 | log.sigma.4.0.mm.3D_glrlm_LongRunLowGrayLevelEmphasis | FLAIR | 2.411 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HLL_firstorder_Mean | T1 | 1.787 |
| 2 | log.sigma.5.0.mm.3D_firstorder_90Percentile | T1 | 2.131 |
| 3 | original_shape_Sphericity | FLAIR | -1.247 |
| 4 | wavelet.LLH_firstorder_Skewness | FLAIR | -0.228 |
| 5 | wavelet.LHL_firstorder_Skewness | FLAIR | -1.360 |
Tab.4 Optimal features from the whole tumor for predicting glioma prognosis and their corresponding sequences and coefficients (The whole tumor)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HLL_firstorder_Mean | T1 | 1.787 |
| 2 | log.sigma.5.0.mm.3D_firstorder_90Percentile | T1 | 2.131 |
| 3 | original_shape_Sphericity | FLAIR | -1.247 |
| 4 | wavelet.LLH_firstorder_Skewness | FLAIR | -0.228 |
| 5 | wavelet.LHL_firstorder_Skewness | FLAIR | -1.360 |
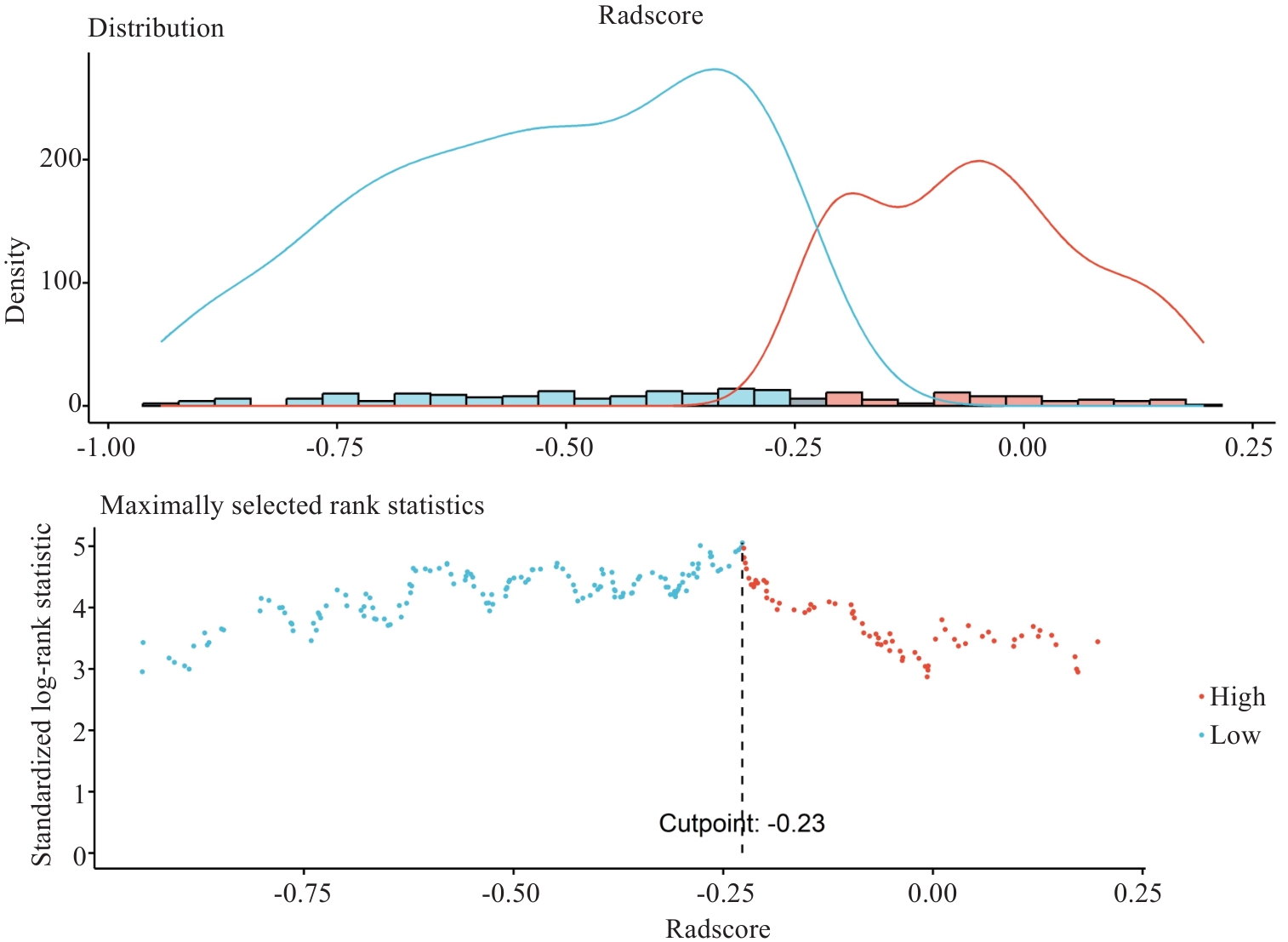
Fig.2 Optimal cutoff value of Rad-score for the whole tumor. Red lines or dots represent patients with high-risk prognosis and blue ones the low-risk patients. The optimal cutoff value is -0.2274 (shown by the vertical line).
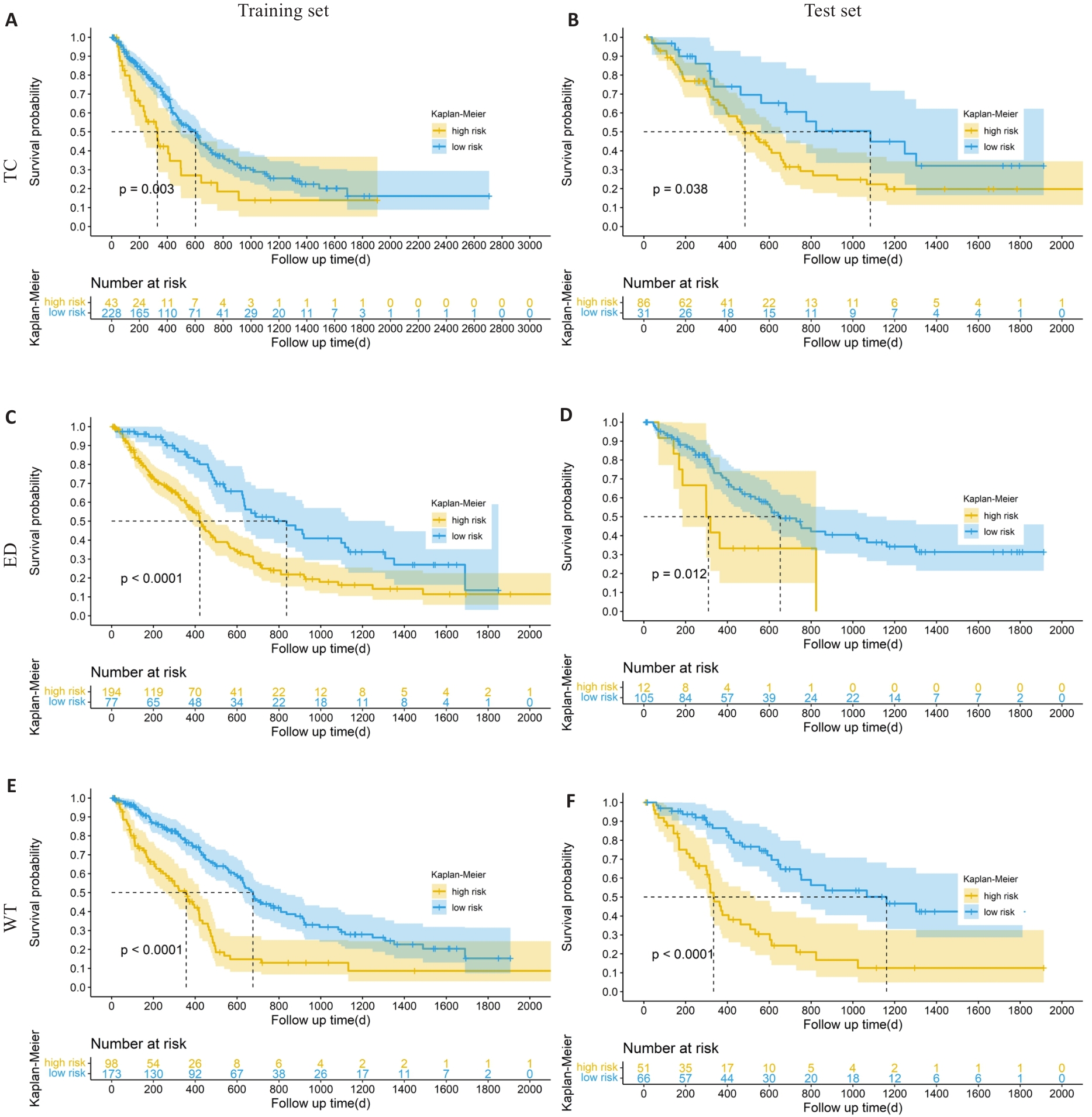
Fig.3 Kaplan-Meier survival analysis of the patients in the high- and low-risk groups in the training and test sets. A, B: Kaplan-Meier analysis of the core tumor area in training set and test set. C, D: Kaplan-Meier analysis of peritumoral edema in the training set and test set. E, F: Kaplan-Meier analysis of the whole tumor in the training set and test set.
| Item | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.04 | 1.03-1.06 | <0.001 | 1.03 | 1.02-1.05 | <0.001 |
| Gender | 0.96 | 0.73-1.25 | 0.748 | - | - | - |
| WHO grade | 3.16 | 2.27-4.40 | <0.001 | 1.30 | 0.92-1.84 | 0.138 |
| IDH | 0.13 | 0.06-0.33 | <0.001 | 0.23 | 0.09-0.56 | 0.001 |
| Rad-score (TC) | 1.51 | 1.15-1.98 | 0.003 | 1.43 | 1.08-1.91 | 0.013 |
| Rad-score (ED) | 1.48 | 1.18-1.86 | <0.001 | 1.49 | 1.18-1.88 | 0.001 |
| Rad-score (WT) | 2.79 | 2.15-3.61 | <0.001 | 2.31 | 1.77-3.03 | <0.001 |
Tab.5 Univariate and multivariate analysis of Cox proportional hazards of overall survival of the glioma patients
| Item | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.04 | 1.03-1.06 | <0.001 | 1.03 | 1.02-1.05 | <0.001 |
| Gender | 0.96 | 0.73-1.25 | 0.748 | - | - | - |
| WHO grade | 3.16 | 2.27-4.40 | <0.001 | 1.30 | 0.92-1.84 | 0.138 |
| IDH | 0.13 | 0.06-0.33 | <0.001 | 0.23 | 0.09-0.56 | 0.001 |
| Rad-score (TC) | 1.51 | 1.15-1.98 | 0.003 | 1.43 | 1.08-1.91 | 0.013 |
| Rad-score (ED) | 1.48 | 1.18-1.86 | <0.001 | 1.49 | 1.18-1.88 | 0.001 |
| Rad-score (WT) | 2.79 | 2.15-3.61 | <0.001 | 2.31 | 1.77-3.03 | <0.001 |
| Tumor area | Model | Training set (Cross validation) | Test set | ||
|---|---|---|---|---|---|
1-year overall survival AUC | 3-year overall survival AUC | 1-year overall survival AUC | 3-year overall survival AUC | ||
| TC | Clinical information | 0.716 | 0.758 | 0.682 | 0.743 |
| Rad-score | 0.583 | 0.613 | 0.503 | 0.637 | |
| Joint model | 0.727 | 0.792 | 0.684 | 0.707 | |
| ED | Clinical information | 0.716 | 0.767 | 0.682 | 0.743 |
| Rad-score | 0.678 | 0.728 | 0.582 | 0.613 | |
| Joint model | 0.750 | 0.816 | 0.678 | 0.747 | |
| WT | Clinical information | 0.722 | 0.752 | 0.682 | 0.743 |
| Rad-score | 0.707 | 0.711 | 0.711 | 0.739 | |
| Joint model | 0.750 | 0.778 | 0.764 | 0.800 | |
Tab.6 Performance of different models
| Tumor area | Model | Training set (Cross validation) | Test set | ||
|---|---|---|---|---|---|
1-year overall survival AUC | 3-year overall survival AUC | 1-year overall survival AUC | 3-year overall survival AUC | ||
| TC | Clinical information | 0.716 | 0.758 | 0.682 | 0.743 |
| Rad-score | 0.583 | 0.613 | 0.503 | 0.637 | |
| Joint model | 0.727 | 0.792 | 0.684 | 0.707 | |
| ED | Clinical information | 0.716 | 0.767 | 0.682 | 0.743 |
| Rad-score | 0.678 | 0.728 | 0.582 | 0.613 | |
| Joint model | 0.750 | 0.816 | 0.678 | 0.747 | |
| WT | Clinical information | 0.722 | 0.752 | 0.682 | 0.743 |
| Rad-score | 0.707 | 0.711 | 0.711 | 0.739 | |
| Joint model | 0.750 | 0.778 | 0.764 | 0.800 | |
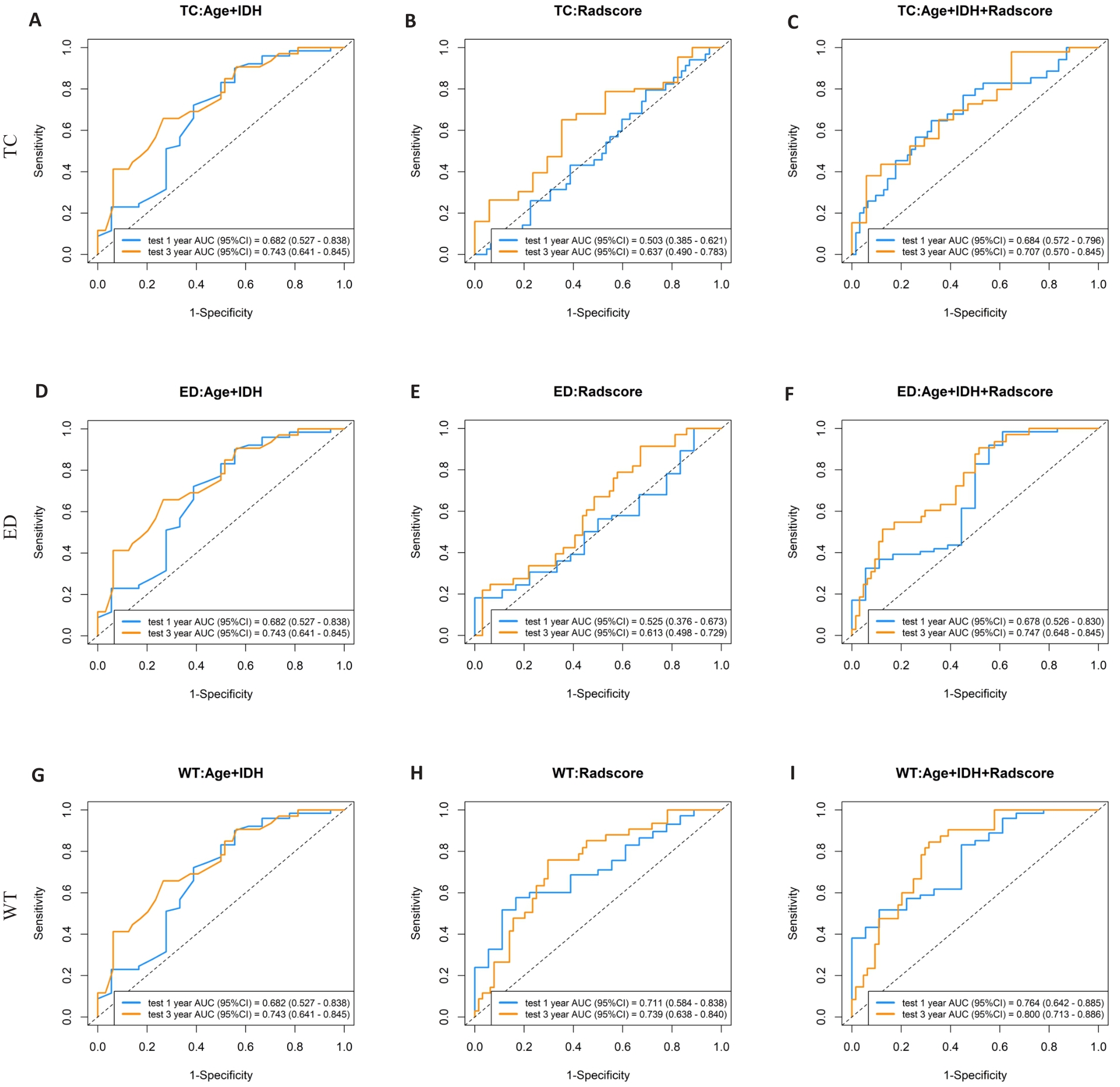
Fig.4 Receiver-operating characteristic curves of 1-year and 3-year survival rates predicted by the clinical information model, Rad-score model, and joint model in the test set for core tumor area (A-C), peritumoral edema area (D-F), and whole tumor (G-I).
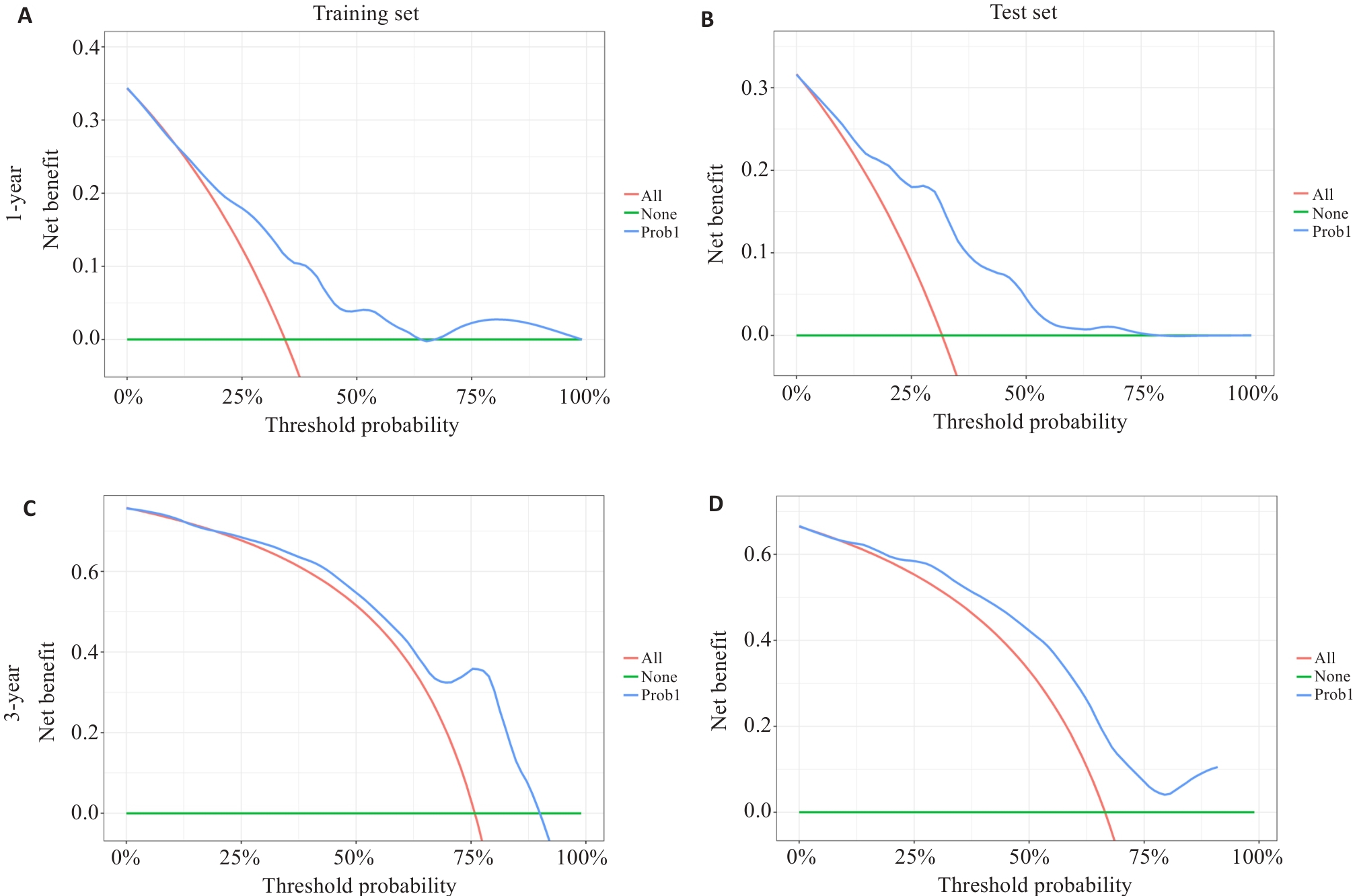
Fig.7 Decision curve analysis of survival prediction nomograms for 1-year and 3-year overall survival rates of glioma patients in the training set (A, C) and test set (B, D).
| 1 | Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016[J]. Neuro Oncol, 2019, 21(): v1-10. |
| 2 | Xu HS, Zhang AK, Han XY, et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas[J]. Cancer Immunol Immunother, 2022, 71(3): 645-60. |
| 3 | 李绍山, 付 强, 秦 虎, 等. 脑胶质瘤在CT平扫及三期增强中的图像表现特征分析[J]. 中国CT和MRI杂志, 2019, 17(7): 5-7. DOI: 10.3969/j.issn.1672-5131.2019.07.002 |
| 4 | Rasmussen BK, Hansen S, Laursen RJ, et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry[J]. J Neurooncol, 2017, 135(3): 571-9. |
| 5 | Lin ZY, Yang RW, Li KS, et al. Establishment of age group classification for risk stratification in glioma patients[J]. BMC Neurol, 2020, 20(1): 310. |
| 6 | Zhang BQ, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas[J]. Neuro Oncol, 2017, 19(1): 109-17. |
| 7 | Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials[J]. Neuro Oncol, 2015, 17(9): 1188-98. |
| 8 | Park YW, Kim S, Park CJ, et al. Adding radiomics to the 2021 WHO updates may improve prognostic prediction for current IDH-wildtype histological lower-grade gliomas with known EGFR amplification and TERT promoter mutation status[J]. Eur Radiol, 2022, 32(12): 8089-98. |
| 9 | 杜 鹏, 耿道颖. 多模态MRI影像组学在胶质瘤精准诊疗中的研究进展[J]. 国际医学放射学杂志, 2023, 46(6): 672-9. |
| 10 | Bakas S, Akbari H, Pisapia J, et al. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index[J]. Clin Cancer Res, 2017, 23(16): 4724-34. |
| 11 | Weller M, Weber RG, Willscher E, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups[J]. Acta Neuropathol, 2015, 129(5): 679-93. |
| 12 | Calabrese E, Villanueva-Meyer JE, Rudie JD, et al. The university of California San francisco preoperative diffuse glioma MRI dataset[J]. Radiol Artif Intell, 2022, 4(6): e220058. |
| 13 | Bale TA, Rosenblum MK. The 2021 WHO Classification of Tumors of the Central Nervous System: an update on pediatric low-grade gliomas and glioneuronal tumors[J]. Brain Pathol, 2022, 32(4): e13060. |
| 14 | Bakas S, Sako C, Akbari H, et al. The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: advanced MRI, clinical, genomics, & radiomics[J]. Sci Data, 2022, 9(1): 453. |
| 15 | 邓景景. 生存分析模型风险预测能力评价方法及其应用研究[D]. 南京: 东南大学, 2016. |
| 16 | Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications[J]. BMC Med Res Methodol, 2017, 17(1): 53. |
| 17 | Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves[J]. Biometrics, 2005, 61(1): 92-105. |
| 18 | Foltyn-Dumitru M, Schell M, Rastogi A, et al. Impact of signal intensity normalization of MRI on the generalizability of radiomic-based prediction of molecular glioma subtypes[J]. Eur Radiol, 2024, 34(4): 2782-90. |
| 19 | Foltyn-Dumitru M, Schell M, Sahm F, et al. Advancing noninvasive glioma classification with diffusion radiomics: exploring the impact of signal intensity normalization[J]. Neurooncol Adv, 2024, 6(1): vdae043. |
| 20 | Huang DC, Gao TY, Zhang Y, et al. A study on prognosis of diffuse glioma based on clinical factors and magnetic resonance imaging radiomics[J]. World Neurosurg, 2024, 186: e514-30. |
| 21 | Usuzaki T, Takahashi K, Inamori R, et al. Identifying key factors for predicting O6-Methylguanine-DNA methyltransferase status in adult patients with diffuse glioma: a multimodal analysis of demographics, radiomics, and MRI by variable Vision Transformer[J]. Neuroradiology, 2024, 66(5): 761-73. |
| 22 | Truong NCD, Bangalore Yogananda CG, Wagner BC, et al. Two-stage training framework using multicontrast MRI radiomics for IDH mutation status prediction in glioma[J]. Radiol Artif Intell, 2024, 6(4): e230218. |
| 23 | Markwell SM, Ross JL, Olson CL, et al. Necrotic reshaping of the glioma microenvironment drives disease progression[J]. Acta Neuropathol, 2022, 143(3): 291-310. |
| 24 | Williams BJ, Frieboes H, Baxter M, et al. Metabolomic differentiation of tumor core vs. edge in glioma via machine learning[J]. Neurosurgery, 2024, 70(): 210. |
| 25 | Zhou Y, Gu HL, Zhang XL, et al. Multiparametric magnetic resonance imaging-derived radiomics for the prediction of disease-free survival in early-stage squamous cervical cancer[J]. Eur Radiol, 2022, 32(4): 2540-51. |
| 26 | Lemée JM, Clavreul A, Menei P. Intratumoral heterogeneity in glioblastoma: don't forget the peritumoral brain zone[J]. Neuro-oncology, 2015, 17(10): 1322-32. |
| 27 | Saadoun S, Papadopoulos MC, Davies DC, et al. Increased aquaporin 1 water channel expression in human brain tumours[J]. Br J Cancer, 2002, 87(6): 621-3. |
| 28 | Yang Y, Han Y, Zhao SJ, et al. Spatial heterogeneity of edema region uncovers survival-relevant habitat of Glioblastoma[J]. Eur J Radiol, 2022, 154: 110423. |
| 29 | 侯秋阳, 叶成坤, 刘 畅, 等. 基于多参数磁共振瘤周区域的影像组学模型在脑胶质瘤预后评估中的应用价值[J]. 安徽医科大学学报, 2024, 59(1): 154-61. |
| 30 | 卢明君, 屈耀铭, 马安东,等. 多模态MRI影像组学可预测弥漫性较低级别胶质瘤的1p/19q共缺失状态[J]. 南方医科大学学报, 2023, 43(6): 1023-8. DOI: 10.12122/j.issn.1673-4254.2023.06.19 |
| 31 | Fan ZC, Zhang L, Yang GQ, et al. MRI radiomics for predicting poor disease-free survival in muscle invasive bladder cancer: the results of the retrospective cohort study[J]. Abdom Radiol, 2024, 49(1): 151-62. |
| 32 | Ahrari S, Zaragori T, Zinsz A, et al. Application of PET imaging delta radiomics for predicting progression-free survival in rare high-grade glioma[J]. Sci Rep, 2024, 14(1): 3256. |
| 33 | Ye LG, Gu LG, Zheng ZY, et al. An online survival predictor in glioma patients using machine learning based on WHO CNS5 data[J]. Front Neurol, 2023, 14: 1179761. |
| 34 | Zhao R, Zhuge Y, Camphausen K, et al. Machine learning based survival prediction in Glioma using large-scale registry data[J]. Health Informatics J, 2022, 28(4): 14604582221135427. |
| 35 | 李新宇, 尚洵杰, 夏 彤, 等. 原发性人脑胶质瘤的预后影响因素: 基于SEER数据库的分析[J]. 中国临床神经外科杂志, 2021, 26(10): 764-8. DOI: 10.13798/j.issn.1009-153X.2020.10.007 |
| 36 | Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary[J]. Acta Neuropathol, 2016, 131(6): 803-20. |
| 37 | Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas[J]. J Clin Oncol, 2009, 27(25): 4150-4. |
| 38 | 张 宁, 马辉辉, 王 凡, 等. 脑胶质瘤患者 MGMT 启动子甲基化和 IDH1 突变与临床预后相关[J]. 基础医学与临床, 2021, 41(10): 1476-80. DOI: 10.3969/j.issn.1001-6325.2021.10.014 |
| 39 | 胡 哲,王玉红,刘晓龙, 等. 基于术前MRI图像构建影像组学与深度学习的机器学习模型预测胶质瘤IDH-1基因表达的研究[J]. 临床神经外科杂志, 2024, 21(2): 187-92. DOI: 10.3969/j.issn.1672-7770.2024.02.013 |
| 40 | Choi Y, Nam Y, Jang J, et al. Radiomics may increase the prognostic value for survival in glioblastoma patients when combined with conventional clinical and genetic prognostic models[J]. Eur Radiol, 2021, 31(4): 2084-93. |
| 41 | Park CJ, Kim S, Han K, et al. Diffusion- and perfusion-weighted MRI radiomics for survival prediction in patients with lower-grade gliomas[J]. Yonsei Med J, 2024, 65(5): 283-92. |
| [1] | Zhenxiang DONG, Yihao GUO, Qiang LIU, Yizhe ZHANG, Qianyi QIU, Xiaodong ZHANG, Yanqiu FENG. A single repetition time quantitative magnetic susceptibility imaging method for the lumbar spine using bipolar readout gradient [J]. Journal of Southern Medical University, 2025, 45(6): 1336-1342. |
| [2] | Yuxiang LIAO, Jingping LIU, Bo LIU, Xiyun FEI, Chen JIN. Circ_EPHB4 regulates temozolomide sensitivity in glioma cells through the miR-424-5p/Wnt3 axis [J]. Journal of Southern Medical University, 2025, 45(5): 942-953. |
| [3] | Meimei CHEN, Yang WANG, Huangwei LEI, Fei ZHANG, Ruina HUANG, Zhaoyang YANG. Construction of recognition models for subthreshold depression based on multiple machine learning algorithms and vocal emotional characteristics [J]. Journal of Southern Medical University, 2025, 45(4): 711-717. |
| [4] | Weiyang FANG, Hui XIAO, Shuang WANG, Xiaoming LIN, Chaomin CHEN. A deep learning model based on magnetic resonance imaging and clinical feature fusion for predicting preoperative cytokeratin 19 status in hepatocellular carcinoma [J]. Journal of Southern Medical University, 2024, 44(9): 1738-1751. |
| [5] | Chuixing WU, Weixiong ZHONG, Jincheng XIE, Ruimeng YANG, Yuankui WU, Yikai XU, Linjing WANG, Xin ZHEN. An MRI multi-sequence feature imputation and fusion mutual-aid model based on sequence deletion for differentiation of high-grade from low-grade glioma [J]. Journal of Southern Medical University, 2024, 44(8): 1561-1570. |
| [6] | Lili CHEN, Tianyu WU, Ming ZHANG, Zixia DING, Yan ZHANG, Yiqing YANG, Jiaqian ZHENG, Xiaonan ZHANG. Identification of potential biomarkers and immunoregulatory mechanisms of rheumatoid arthritis based on multichip co-analysis of GEO database [J]. Journal of Southern Medical University, 2024, 44(6): 1098-1108. |
| [7] | Caiyu SHEN, Shuai WANG, Ruiying ZHOU, Yuhe WANG, Qin GAO, Xingzhi CHEN, Shu YANG. Prediction of risk of in-hospital death in patients with chronic heart failure complicated by lung infections using interpretable machine learning [J]. Journal of Southern Medical University, 2024, 44(6): 1141-1148. |
| [8] | Zhiwei ZUO, Qingliang MENG, Jiakang CUI, Kelei GUO, Hua BIAN. An artificial neural network diagnostic model for scleroderma and immune cell infiltration analysis based on mitochondria-associated genes [J]. Journal of Southern Medical University, 2024, 44(5): 920-929. |
| [9] | HUANG Qiuhu, ZHOU Jian, WANG Zizhen, YANG Kun, CHEN Zhenggang. MiR-26-3p regulates proliferation, migration, invasion and apoptosis of glioma cells by targeting CREB1 [J]. Journal of Southern Medical University, 2024, 44(3): 578-584. |
| [10] | LIU Yunze, LI Chengrun, GUO Juntang, LIU Yang. A clinical-radiomics nomogram for differentiating focal organizing pneumonia and lung adenocarcinoma [J]. Journal of Southern Medical University, 2024, 44(2): 397-404. |
| [11] | HE Huishan, GUO Erjia, MENG Wenyi, WANG Yu, WANG Wen, HE Wenle, WU Yuankui, YANG Wei. Predicting cerebral glioma enhancement pattern using a machine learning-based magnetic resonance imaging radiomics model [J]. Journal of Southern Medical University, 2024, 44(1): 194-200. |
| [12] | ZHANG Zhenyang, XIE Jincheng, ZHONG Weixiong, LIANG Fangrong, YANG Ruimeng, ZHEN Xin. A multi-modal feature fusion classification model based on distance matching and discriminative representation learning for differentiation of high-grade glioma from solitary brain metastasis [J]. Journal of Southern Medical University, 2024, 44(1): 138-145. |
| [13] | YU Zhengtao, LI Jiameng, JIANG Junwen, LI You, LIN Long, XIA Ying, WANG Lei. miRNA-128-3p inhibits malignant behavior of glioma cells by downregulating KLHDC8A expression [J]. Journal of Southern Medical University, 2023, 43(9): 1447-1459. |
| [14] | CHU Zhiqin, QU Yaoming, ZHONG Tao, LIANG Shujun, WEN Zhibo, ZHANG Yu. A Dual-Aware deep learning framework for identification of glioma isocitrate dehydrogenase genotype using magnetic resonance amide proton transfer modalities [J]. Journal of Southern Medical University, 2023, 43(8): 1379-1387. |
| [15] | LIU Yuxuan, CHU Zhiqin, ZHANG Yu. Physical model-based cascaded generative adversarial networks for accelerating quantitative multi-parametric magnetic resonance imaging [J]. Journal of Southern Medical University, 2023, 43(8): 1402-1409. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||