南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (11): 2320-2329.doi: 10.12122/j.issn.1673-4254.2025.11.04
收稿日期:2025-04-02
出版日期:2025-11-20
发布日期:2025-11-28
通讯作者:
廖宇翔
E-mail:Jinchen@csu.edu.cn;lyxxysw@126.com
作者简介:金 晨,博士,主治医师,E-mail: Jinchen@csu.edu.cn
基金资助:
Chen JIN( ), Jingping LIU, Bo LIU, Xiyun FEI, Yuxiang LIAO(
), Jingping LIU, Bo LIU, Xiyun FEI, Yuxiang LIAO( )
)
Received:2025-04-02
Online:2025-11-20
Published:2025-11-28
Contact:
Yuxiang LIAO
E-mail:Jinchen@csu.edu.cn;lyxxysw@126.com
摘要:
目的 探讨环状RNA circ_EPHB4在胶质瘤中的促癌作用及其与N6-甲基腺苷(m6A)阅读蛋白YTHDF3协同调控Wnt3信号通路的分子机制。 方法 通过芯片分析筛选胶质瘤组织中差异表达的circRNA,利用划痕愈合实验、Transwell侵袭实验及皮下成瘤模型,检测circ_EPHB4对胶质瘤细胞迁移、侵袭、上皮-间质转化(EMT)及体内成瘤能力的影响。采用RNA免疫沉淀、RNA稳定性实验及基因过表达/敲低技术,验证circ_EPHB4与YTHDF3对Wnt3 表达的协同调控作用。 结果 circ_EPHB4在胶质瘤组织中较癌旁组织高表达2.3倍(|log2FC|=1.2,P<0.01),在胶质瘤细胞株U373中表达量达正常细胞的2.5倍(P<0.001)。过表达circ_EPHB4可使胶质瘤细胞划痕愈合率提升至(75.2±6.3)%,较对照组增加78.6%(P<0.01);侵袭细胞数增至215±23个/视野,为对照组2.4倍(P<0.001),并促进EMT标志物N-cadherin和Vimentin的表达(P<0.001)。体内实验显示,过表达组肿瘤体积21 d达1200±150 mm³,较对照组增加140%(P<0.001);肺转移灶数6.3±1.2个,为对照组4.2倍(P<0.01)。circ_EPHB4 通过上调Wnt3表达(P<0.01)发挥促癌作用,而YTHDF3 以m6A依赖性方式延长Wnt3 mRNA半衰期至11.5±1.2 h,较对照组增加40%(P<0.01)。二者协同调控时,同时敲低可使Wnt3 mRNA表达降至单独敲低组的47%(P<0.001)。 结论 circ_EPHB4与YTHDF3通过共同靶向Wnt3信号通路促进胶质瘤进展,靶向该协同轴可能为治疗提供新策略。
金晨, 刘景平, 刘博, 费喜云, 廖宇翔. circ_EPHB4协同YTHDF3通过N6-甲基腺苷依赖性稳定Wnt3促进胶质瘤进展[J]. 南方医科大学学报, 2025, 45(11): 2320-2329.
Chen JIN, Jingping LIU, Bo LIU, Xiyun FEI, Yuxiang LIAO. circ_EPHB4 synergizes with YTHDF3 to promote glioma progression via m6A-dependent stabilization of Wnt3[J]. Journal of Southern Medical University, 2025, 45(11): 2320-2329.
| Gene | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|
| circ_EPHB4 | CAGAGAGCGCATCCGCAATTT | AAAGAAGGTGCGCCCACGGTT |
| Zeb1 | AACAGTGAGCGACCTTCATT | AGAGCGCAGAGAAGGAGAGTT |
| GAPDH | AGAGCGCAGAGAAGGAGAUTT | AAAGAGCGAGAGAAGGAGAT |
表1 目的基因及内参引物序列
Tab.1 Primer sequences for RT-qPCR of the target and housekeeping genes
| Gene | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|
| circ_EPHB4 | CAGAGAGCGCATCCGCAATTT | AAAGAAGGTGCGCCCACGGTT |
| Zeb1 | AACAGTGAGCGACCTTCATT | AGAGCGCAGAGAAGGAGAGTT |
| GAPDH | AGAGCGCAGAGAAGGAGAUTT | AAAGAGCGAGAGAAGGAGAT |
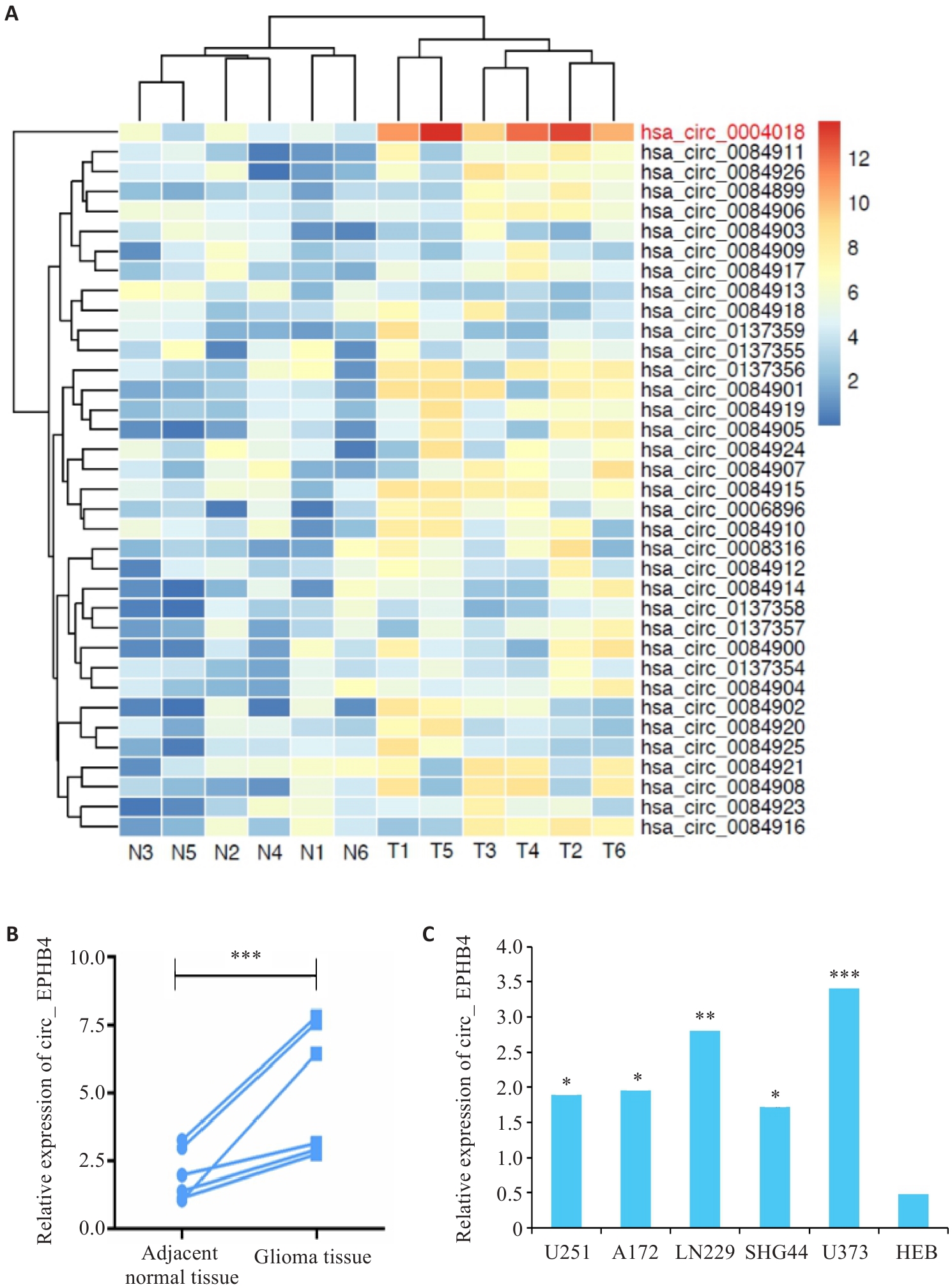
图1 circ_EPHB4在胶质瘤中表达上调
Fig.1 circ_EPHB4 is upregulated in glioma. A: Differential RNA expression analysis shows increased circ_EPHB4 expression in glioma. B: qRT-PCR confirms increased expression of circ_EPHB4 in glioma (***P<0.001). C: circ_EPHB4 expressions are significantly higher in glioma cell lines U251, A172, SHG44, LN229, and U373 than in normal human brain astrocyte HEB cells. *P<0.05, **P<0.01, ***P<0.001 vs HEB.

图2 circ_EPHB4促进胶质瘤细胞迁移、侵袭和EMT
Fig.2 circ_EPHB4 promotes glioma cell migration, invasion, and EMT. A: qRT-PCR of circ_EPHB4 expression in glioma cells after transfection with circ_EPHB4-overexpressing plasmids or circ_EPHB4 siRNA (**P<0.01). B: Scratch wound healing assay for evaluating glioma cell migration ability following circ_EPHB4 overexpression or knockdown (scale bar=100 μm). C: Transwell invasion assay for assessing glioma cell invasion ability after transfection (scale bar=50 μm). D: Western blotting of EMT-related proteins (E-cadherin, N-cadherin and vimentin) in glioma cells following circ_EPHB4 overexpression or knockdown. Data are presented as Mean±SD from 3 independent experiments. *P<0.05 vs siRNA NC; #P<0.05 vs pcDNA3.1 NC.

图3 circ_EPHB4促进胶质瘤细胞体内成瘤及转移
Fig.3 circ_EPHB4 promotes tumorigenesis and metastasis of glioma cells in nude mice. A: Number of pulmonary metastatic nodules in each group. B: Subcutaneous tumor volume at different time points in each group. C: Subcutaneous tumor weight in each group (*P<0.05, **P<0.01, ***P<0.001). D: qRT-PCR analysis of mRNA expressions of E-cadherin, N-cadherin, vimentin) in subcutaneous tumors. E: Immunohistochemicalanalysis of protein expressions of E-cadherin, N-cadherin, and vimentin in subcutaneous tumors from each group. **P<0.01, ***P<0.001 vs siRNA NC group; ##P<0.01, ### P<0.001 vs pcDNA3.1 NC group.
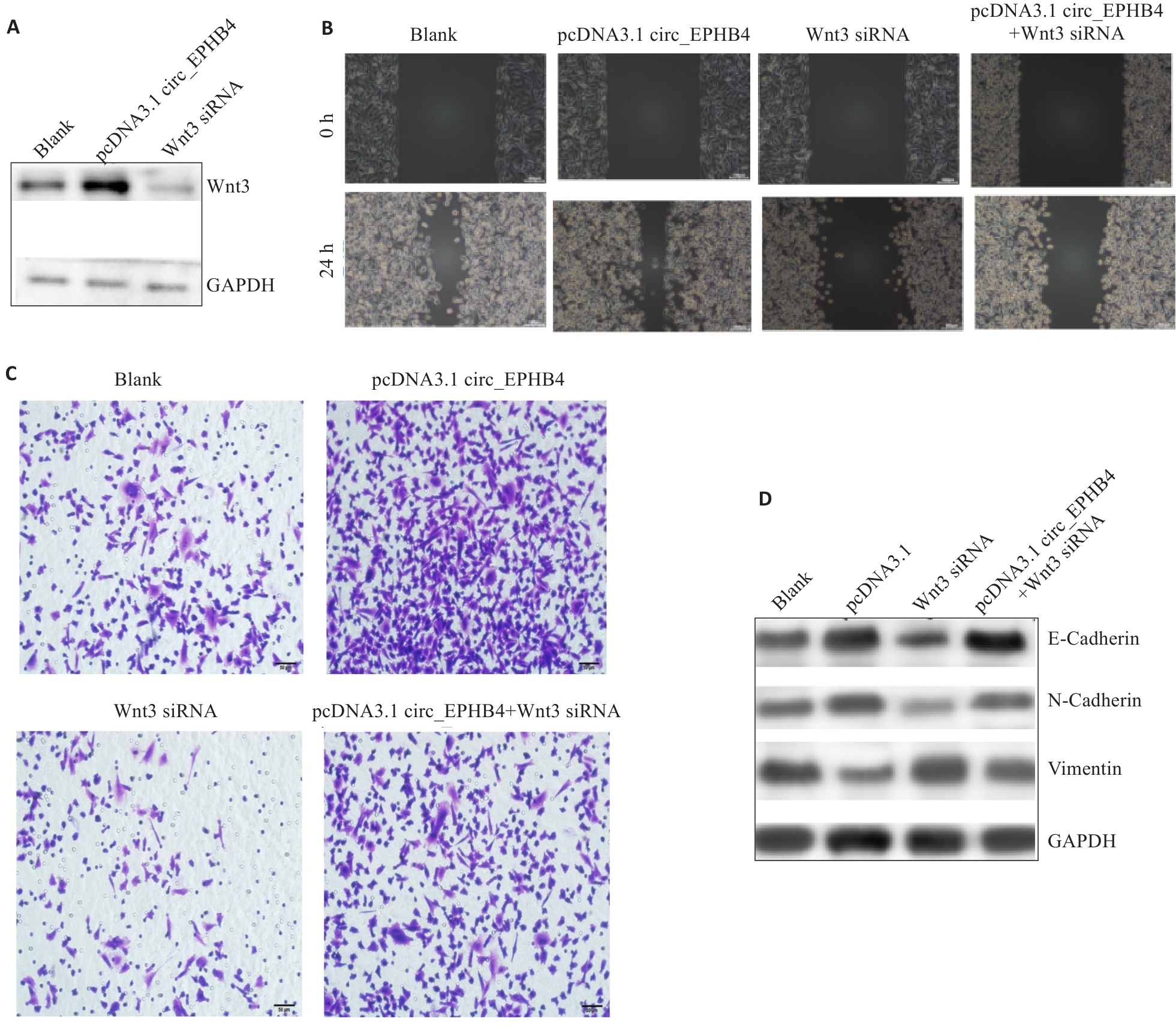
图4 circ_EPHB4通过上调Wnt3表达促进胶质瘤细胞转移
Fig.4 circ_EPHB4 promotes glioma cell metastasis by upregulating Wnt3 expression. A: Overexpression of circ_EPHB4 enhances Wnt3 expression, and circ_EPHB4 knockdown suppresses Wnt3 expression. B: Scratch wound healing assay for evaluating the effect of Wnt3 knockdown on glioma cell migration following circ_EPHB4 overexpression (scale bar=50 μm). C: Transwell invasion assay for assessing the impact of Wnt3 knockdown on invasion ability of circ_EPHB4-overexpressing glioma cells (scale bar=50 μm). D: Western blotting for evaluating the effects of Wnt3 knockdown on expressions of E-cadherin, N-cadherin and vimentin in circ_EPHB4-overexpressing glioma cells.

图5 YTHDF3通过m6A依赖性方式增强Wnt3 mRNA的稳定性
Fig.5 YTHDF3 enhances Wnt3 mRNA stability in an m6A-dependent manner. A: Direct interaction between YTHDF3 and Wnt3 mRNA in U3T3 and SHG44 cells. B: Effect of YTHDF3 knockdown on Wnt3 expression. C: Positive correlation between YTHDF3 and Wnt3 expression in the StarBase database. D: Effect of circ_EPHB4 on the interaction between YTHDF3 and Wnt3 RNA. E: Effect of combined knockdown of circ_EPHB4 and YTHDF3 on Wnt3 mRNA expression. *P<0.05, **P<0.01, ***P<0.001.
| [1] | Ludwig K, Kornblum HI. Molecular markers in glioma[J]. J Neurooncol, 2017, 134(3): 505-12. doi:10.1007/s11060-017-2379-y |
| [2] | 翟 贝, 魏 灯, 王 萍. tRNA修饰在胶质瘤中的研究进展[J]. 中国肿瘤临床, 2024,51(11): 567-71. |
| [3] | 贾金曦, 姚春旭, 刘 畅, 等. 脑胶质瘤患者术后远期预后影响因素分析[J]. 实用癌症杂志, 2024, 39(3): 514-7. |
| [4] | Liu ZX, Li LM, Sun HL, et al. Link between m6A modification and cancers[J]. Front Bioeng Biotechnol, 2018, 6: 89. doi:10.3389/fbioe.2018.00089 |
| [5] | Zhang G, Cheng C, Wang X, et al. N6-Methyladenosine methylation modification in breast cancer: current insights[J]. J Transl Med, 2024, 22(1): 971. doi:10.1186/s12967-024-05771-x |
| [6] | Rui Y, Zhang H, Yu K, et al. N6-methyladenosine regulates Cilia elongation in cancer cells by modulating HDAC6 expression[J]. Adv Sci: Weinh, 2025, 12(2): e2408488. doi:10.1002/advs.202408488 |
| [7] | Chen XH, Guo KX, Li J, et al. Regulations of m6A and other RNA modifications and their roles in cancer[J]. Front Med, 2024, 18(4): 622-48. doi:10.1007/s11684-024-1064-8 |
| [8] | Qin L, Zeng X, Qiu X, et al. The role of N6-methyladenosine modification in tumor angiogenesis[J]. Front Oncol, 2024, 14: 1467850. doi:10.3389/fonc.2024.1467850 |
| [9] | Saluja S, Ganguly S, Singh J, et al. Aberrant overexpression of m6A writer and reader genes in pediatric B-Cell Acute Lymphoblastic Leukemia (B-ALL)[J]. Transl Oncol, 2025, 56: 102403. doi:10.1016/j.tranon.2025.102403 |
| [10] | Zhao Y, Feng F, Guo QH, et al. Role of succinate dehydrogenase deficiency and oncometabolites in gastrointestinal stromal tumors[J]. World J Gastroenterol, 2020, 26(34): 5074-89. doi:10.3748/wjg.v26.i34.5074 |
| [11] | Li J, Han Y, Zhang HM, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA[J]. Biochem Biophys Res Commun, 2019, 512(3): 479-85. doi:10.1016/j.bbrc.2019.03.093 |
| [12] | Dong J, Zeng Z, Huang Y, et al. Challenges and opportunities for circRNA identification and delivery[J]. Crit Rev Biochem Mol Biol, 2023, 58(1): 19-35. doi:10.1080/10409238.2023.2185764 |
| [13] | Chen X, Lu Y. Circular RNA: biosynthesis in vitro [J]. Front Bioeng Biotechnol, 2021, 9: 787881. doi:10.3389/fbioe.2021.787881 |
| [14] | Patop IL, Kadener S. circRNAs in cancer[J]. Curr Opin Genet Dev, 2018, 48: 121-7. doi:10.1016/j.gde.2017.11.007 |
| [15] | Hanan M, Soreq H, Kadener S. CircRNAs in the brain[J]. RNA Biol, 2017, 14(8): 1028-34. doi:10.1080/15476286.2016.1255398 |
| [16] | Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2 [J]. Hepatology, 2018, 67(6): 2254-70. doi:10.1002/hep.29683 |
| [17] | Yin J, Ding F, Cheng Z, et al. METTL3-mediated m6A modification of LINC00839 maintains glioma stem cells and radiation resistance by activating Wnt/β-catenin signaling[J]. Cell Death Dis, 2023, 14(7): 417. doi:10.1038/s41419-023-05933-7 |
| [18] | Xiao L, Li X, Mu Z, et al. FTO inhibition enhances the anti-tumor effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit in glioma. Cancer Res. 2020. 80(18): canres.0132.2020. doi:10.1158/0008-5472.can-20-0132 |
| [19] | Zheng W, Li J, Zhou X, et al. The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337[J]. Arab J Gastroenterol, 2020, 21(3): 199-206. doi:10.1016/j.ajg.2020.07.010 |
| [20] | Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs[J]. EMBO J, 2019, 38(16): e100836. doi:10.15252/embj.2018100836 |
| [21] | Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review[J]. J Hematol Oncol, 2019, 12(1): 90. doi:10.1186/s13045-019-0776-8 |
| [22] | Yang R, Cui J. Advances and applications of RNA vaccines in tumor treatment[J]. Mol Cancer, 2024, 23(1): 226. doi:10.1186/s12943-024-02141-5 |
| [23] | Yi Q, Feng JG, Lan WW, et al. CircRNA and lncRNA-encoded peptide in diseases, an update review[J]. Mol Cancer, 2024, 23(1): 214. doi:10.1186/s12943-024-02131-7 |
| [24] | Zhang S, Zhang P, Wu A, et al. Downregulated M6A modification and expression of circRNA_103239 promoted the progression of glioma by regulating the miR-182-5p/MTSS1 signalling pathway[J]. J Cancer, 2023, 14(18): 3508-20. doi:10.7150/jca.85320 |
| [25] | Dai YH, Zhao SH, Chen HL, et al. RNA methylation and breast cancer: insights into m6A, m7G and m5C[J]. Mol Biol Rep, 2024, 52(1): 27. doi:10.1007/s11033-024-10138-y |
| [26] | Lin ZY, Wan AH, Sun L, et al. N6-methyladenosine demethylase FTO enhances chemo-resistance in colorectal cancer through SIVA1-mediated apoptosis[J]. Mol Ther, 2023, 31(2): 517-34. doi:10.1016/j.ymthe.2022.10.012 |
| [27] | Zhang C, Huang S, Zhuang H, et al. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation[J]. Oncogene, 2020, 39(23): 4507-18. doi:10.1038/s41388-020-1303-7 |
| [28] | Chang H, Yang J, Wang Q, et al. Role of N6-methyladenosine modification in pathogenesis of ischemic stroke[J]. Expert Rev Mol Diagn, 2022, 22(3): 295-303. doi:10.1080/14737159.2022.2049246 |
| [29] | Li L, Zhang T, Xiao M, et al. Brain macrophage senescence in glioma[J]. Semin Cancer Biol, 2024, 104/105: 46-60. doi:10.1016/j.semcancer.2024.07.005 |
| [30] | Davidson CL, Vengoji R, Jain M, et al. Biological, diagnostic and therapeutic implications of exosomes in glioma[J]. Cancer Lett, 2024, 582: 216592. doi:10.1016/j.canlet.2023.216592 |
| [31] | Yin JL, Liu G, Zhang Y, et al. Gender differences in gliomas: From epidemiological trends to changes at the hormonal and molecular levels[J]. Cancer Lett, 2024, 598: 217114. doi:10.1016/j.canlet.2024.217114 |
| [1] | 廖宇翔, 刘景平, 刘博, 费喜云, 金晨. Circ_EPHB4通过miR-424-5p/Wnt3信号轴调控胶质瘤细胞对替莫唑胺的化疗敏感性[J]. 南方医科大学学报, 2025, 45(5): 942-953. |
| [2] | 吴垂杏, 钟伟雄, 谢金城, 杨蕊梦, 吴元魁, 许乙凯, 王琳婧, 甄鑫. 基于序列缺失的MRI多序列特征填补与融合互助模型:鉴别高低级别胶质瘤[J]. 南方医科大学学报, 2024, 44(8): 1561-1570. |
| [3] | 黄秋虎, 周 建, 王子珍, 杨 堃, 陈政纲. miR-26b-3p 靶向 CREB1 调控神经胶质瘤细胞的增殖、迁移及侵袭[J]. 南方医科大学学报, 2024, 44(3): 578-584. |
| [4] | 黄晓茵, 陈凤莲, 张煜, 梁淑君. 多参数多区域MRI影像组学特征与临床信息联合模型可有效预测脑胶质瘤患者生存期[J]. 南方医科大学学报, 2024, 44(10): 2004-2014. |
| [5] | 张振阳, 谢金城, 钟伟雄, 梁芳蓉, 杨蕊梦, 甄 鑫. 基于距匹配及判别表征学习的多模态特征融合分类模型研究:高级别胶质瘤与单发性脑转移瘤的鉴别诊断[J]. 南方医科大学学报, 2024, 44(1): 138-145. |
| [6] | 于正涛, 李佳梦, 蒋俊文, 李 由, 林 珑, 夏 鹰, 王 磊. miRNA-128-3p通过下调KLHDC8A的表达抑制胶质瘤细胞的恶性生物学行为[J]. 南方医科大学学报, 2023, 43(9): 1447-1459. |
| [7] | 楚智钦, 屈耀铭, 钟 涛, 梁淑君, 温志波, 张 煜. 磁共振酰胺质子转移模态的胶质瘤IDH基因分型识别:基于深度学习的Dual-Aware框架[J]. 南方医科大学学报, 2023, 43(8): 1379-1387. |
| [8] | 叶静静, 徐文琴, 奚邦生, 王能乾, 陈天兵. 乳酸诱导PLEKHA4的上调参与胶质瘤细胞增殖和凋亡的生物学进程[J]. 南方医科大学学报, 2023, 43(7): 1071-1080. |
| [9] | 卢明君, 屈耀铭, 马安东, 朱建彬, 邹 霞, 林耕耘, 李榆欣, 刘昕孜, 温志波. 多模态MRI影像组学可预测弥漫性较低级别胶质瘤的1p/19q共缺失状态[J]. 南方医科大学学报, 2023, 43(6): 1023-1028. |
| [10] | 徐文琴, 叶静静, 王 飞, 陈天兵. 吡罗克酮乙醇胺盐通过PI3K/AKT通路破坏胶质瘤细胞的线粒体动力学[J]. 南方医科大学学报, 2023, 43(5): 764-771. |
| [11] | 孙江川, 邢家恒, 谭茹雪, 钱 颖, 田 男. 莪术醇逆转胶质瘤细胞对替莫唑胺的耐药:基于调节UTX/MGMT轴[J]. 南方医科大学学报, 2023, 43(10): 1697-1705. |
| [12] | 崔 颖, 范顺志, 潘迪迪, 巢 青. 阿托伐他汀抑制脑胶质瘤细胞的增殖和促进凋亡:基于上调miR-146a表达与抑制PI3K/Akt信号通路[J]. 南方医科大学学报, 2022, 42(6): 899-904. |
| [13] | 杨学智, 沈 红, 李 群, 代自超, 杨荣强, 黄国宾, 陈 蕊, 王 芳, 宋精玲, 华海蓉. 干扰肿瘤相关巨噬细胞的P2X4受体表达可抑制胶质瘤细胞的迁移和侵袭[J]. 南方医科大学学报, 2022, 42(5): 658-664. |
| [14] | 赵炜疆, 林佳哲. 神经调节蛋白2高表达于不同级别胶质瘤组织并通过Akt信号调控胶质瘤细胞GFAP表达[J]. 南方医科大学学报, 2021, 41(8): 1171-1176. |
| [15] | 罗奇志, 张 帆, 李 威, 王 芳, 邬力祥, 黄柏胜. LncRNA MEG3通过抑制HIF1α的表达降低胶质瘤U251细胞的增殖与侵袭能力[J]. 南方医科大学学报, 2021, 41(1): 141-145. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||