南方医科大学学报 ›› 2024, Vol. 44 ›› Issue (10): 2004-2014.doi: 10.12122/j.issn.1673-4254.2024.10.19
• • 上一篇
黄晓茵1( ), 陈凤莲1, 张煜1,2,3(
), 陈凤莲1, 张煜1,2,3( ), 梁淑君1,2,3(
), 梁淑君1,2,3( )
)
收稿日期:2024-05-28
出版日期:2024-10-20
发布日期:2024-10-31
通讯作者:
张煜,梁淑君
E-mail:huangxiaoyin2003@163.com;yuzhang@smu.edu.cn;lsj123@smu.edu.cn
作者简介:黄晓茵,在读本科生,E-mail: huangxiaoyin2003@163.com
基金资助:
Xiaoyin HUANG1( ), Fenglian CHEN1, Yu ZHANG1,2,3(
), Fenglian CHEN1, Yu ZHANG1,2,3( ), Shujun LIANG1,2,3(
), Shujun LIANG1,2,3( )
)
Received:2024-05-28
Online:2024-10-20
Published:2024-10-31
Contact:
Yu ZHANG, Shujun LIANG
E-mail:huangxiaoyin2003@163.com;yuzhang@smu.edu.cn;lsj123@smu.edu.cn
Supported by:摘要:
目的 探讨脑胶质瘤患者肿瘤亚区影像组学特征,评估其对患者生存期的预后价值。 方法 对388例胶质瘤患者的术前MRI多序列影像和临床数据进行回顾性分析,从瘤周水肿区域、肿瘤核心区以及全肿瘤区域提取T1、T2、T1加权对比增强(T1CE)、液体衰减反转恢复(FLAIR)序列的影像组学特征。将病例按照7∶3分为训练集(271例)和测试集(117例)。利用随机生存森林算法在训练集中筛选与总生存期相关的影像组学特征,并构建影像组学评分(Rad-score)。根据Rad-score将患者分为高、低风险组,使用Kaplan-Meier分析两组生存差异。建立瘤周水肿区、肿瘤核心区和全肿瘤区域的Cox比例风险回归模型,并通过五折交叉验证及受试者工作特征曲线下面积评估模型1年、3年生存率的预测效能,采用10例胶质瘤患者作外部验证。选择表现最优的模型进行生存期预测情况的列线图分析。 结果 肿瘤核心区、瘤周水肿区和全肿瘤区域分别筛选出的影像组学特征数量分别为5、7、5,根据Rad-score,两风险组在训练集和测试集的总生存期存在差异(P<0.05)。单因素和多因素Cox分析显示,年龄、异柠檬酸脱氢酶状态和Rad-score是总生存期的独立影响因素。联合模型在训练集和测试集中的AUC表现优于单一Rad-score模型,其中全肿瘤模型的1年、3年生存率预测AUC分别为0.750、0.778(训练集),0.764、0.800(测试集)和0.938、0.917(外部验证集)。 结论 基于术前多模态MRI影像组学特征与临床信息联合构建的预测模型能有效预测胶质瘤患者的生存期。
黄晓茵, 陈凤莲, 张煜, 梁淑君. 多参数多区域MRI影像组学特征与临床信息联合模型可有效预测脑胶质瘤患者生存期[J]. 南方医科大学学报, 2024, 44(10): 2004-2014.
Xiaoyin HUANG, Fenglian CHEN, Yu ZHANG, Shujun LIANG. A predictive model for survival outcomes of glioma patients based on multi-parametric, multi-regional MRI radiomics features and clinical features[J]. Journal of Southern Medical University, 2024, 44(10): 2004-2014.
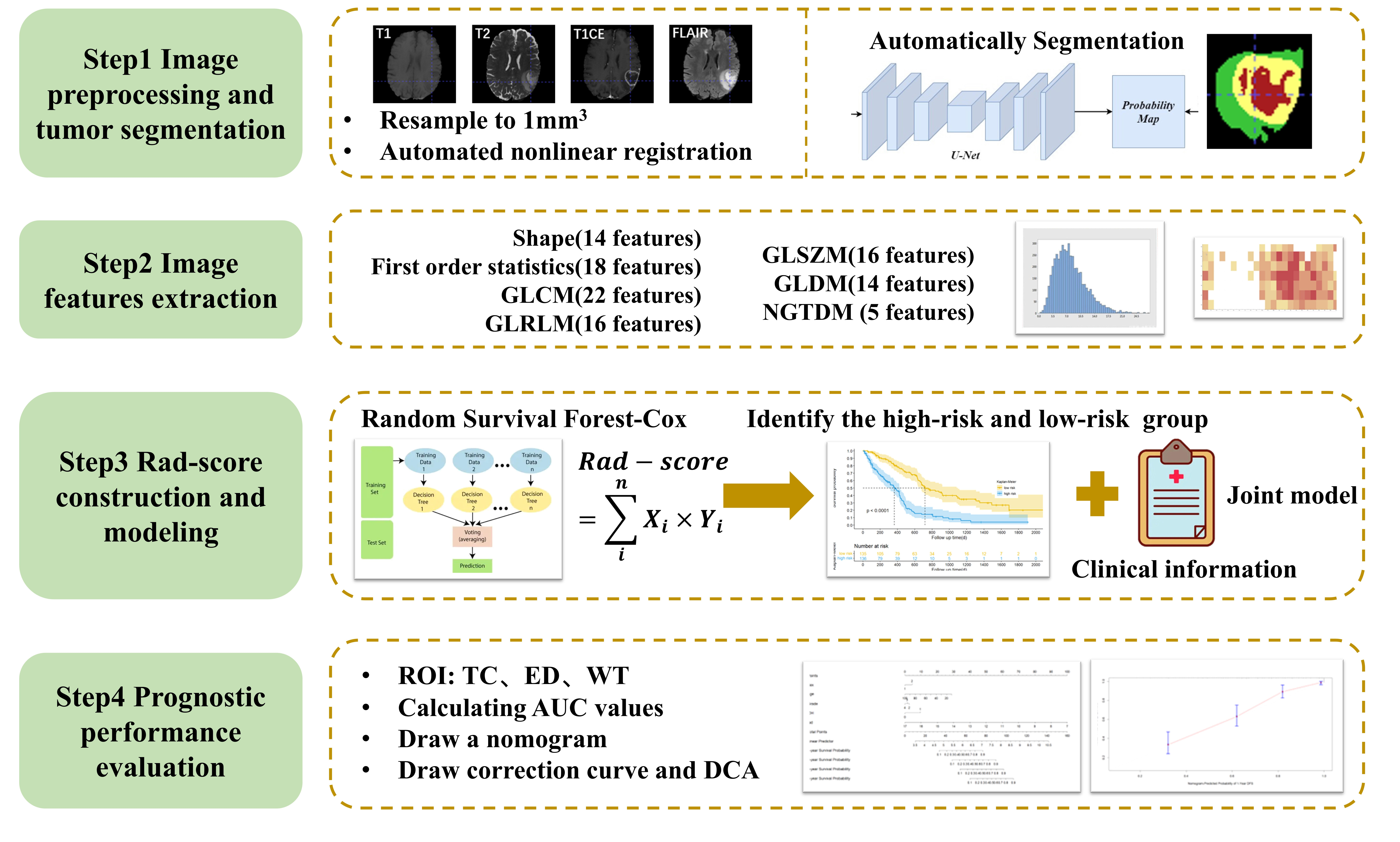
图1 研究设计流程图
Fig.1 Flowchart of the study design. T1CE: T1-weighted contrast-enhanced; FLAIR: Fluid attenuated inversion recovery; GLCM: Gray-level co-occurrence matrix; GLRLM: Gray-level run-length matrix; GLSZM: Gray-level size zone matrix; GLDM: Gray-level dependence matrix; NGTDM: Neighborhood gray tone difference matrix; TC: Tumor core; ED: Edema region; WT: Whole tumor; AUC: Area under the curve; DCA: Decision curve analysis.
| Item | Training dataset (n=271) | Test dataset (n=117) | χ2 | P |
|---|---|---|---|---|
| Age (year, Mean±SD) | 60.72±13.40 | 58.32±13.29 | 79.249 | 0.110 |
| Gender [n (%)] | 2.399 | 0.121 | ||
| Male | 158 (58.30) | 78 (66.70) | ||
| Female | 113 (41.70) | 39 (33.30) | ||
| WHO grade [n (%)] | 1.990 | 0.370 | ||
| Ⅱ | 2 (0.70) | 0 (0.00) | ||
| Ⅲ | 3 (1.10) | 3 (2.60) | ||
| Ⅳ | 266 (98.20) | 114 (97.40) | ||
| IDH [n (%)] | 1.462 | 0.227 | ||
| Wide type | 251 (92.60) | 104 (88.90) | ||
| Mutant | 20 (7.40) | 13 (11.10) | ||
| Status [n (%)] | 1.421 | 0.233 | ||
| Non-censoring | 159 (58.70) | 61 (52.10) | ||
| Censoring | 112 (41.30) | 76 (47.90) | ||
| Survival time (d, Mean±SD) | 461.57±414.48 | 558.85±463.16 | 329.837 | 0.312 |
表1 训练集与测试集患者的一般临床资料比较
Tab.1 Comparison of general clinical data of the patients between the training and test datasets
| Item | Training dataset (n=271) | Test dataset (n=117) | χ2 | P |
|---|---|---|---|---|
| Age (year, Mean±SD) | 60.72±13.40 | 58.32±13.29 | 79.249 | 0.110 |
| Gender [n (%)] | 2.399 | 0.121 | ||
| Male | 158 (58.30) | 78 (66.70) | ||
| Female | 113 (41.70) | 39 (33.30) | ||
| WHO grade [n (%)] | 1.990 | 0.370 | ||
| Ⅱ | 2 (0.70) | 0 (0.00) | ||
| Ⅲ | 3 (1.10) | 3 (2.60) | ||
| Ⅳ | 266 (98.20) | 114 (97.40) | ||
| IDH [n (%)] | 1.462 | 0.227 | ||
| Wide type | 251 (92.60) | 104 (88.90) | ||
| Mutant | 20 (7.40) | 13 (11.10) | ||
| Status [n (%)] | 1.421 | 0.233 | ||
| Non-censoring | 159 (58.70) | 61 (52.10) | ||
| Censoring | 112 (41.30) | 76 (47.90) | ||
| Survival time (d, Mean±SD) | 461.57±414.48 | 558.85±463.16 | 329.837 | 0.312 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HHH_glszm_SizeZoneNonUniformityNormalized | T1 | -2.886 |
| 2 | wavelet.HHL_glcm_Imc2 | T1 | -1.126 |
| 3 | lbp.3D.m2_firstorder_Maximum | T1 | 1.154 |
| 4 | wavelet.HHL_gldm_DependenceNonUniformityNormalized | T2 | -1.209 |
| 5 | original_shape_MajorAxisLength | FLAIR | 1.530 |
表2 预测胶质瘤预后的最优特征所属序列及其系数(肿瘤核心区)
Tab.2 Optimal features from the tumor core for predicting glioma prognosis and the corresponding sequences and coefficients (Core tumor)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HHH_glszm_SizeZoneNonUniformityNormalized | T1 | -2.886 |
| 2 | wavelet.HHL_glcm_Imc2 | T1 | -1.126 |
| 3 | lbp.3D.m2_firstorder_Maximum | T1 | 1.154 |
| 4 | wavelet.HHL_gldm_DependenceNonUniformityNormalized | T2 | -1.209 |
| 5 | original_shape_MajorAxisLength | FLAIR | 1.530 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | original_shape_Maximum2DDiameterSlice | T1 | 0.988 |
| 2 | original_shape_Flatness | T1 | -0.915 |
| 3 | wavelet.HHH_gldm_DependenceNonUniformityNormalized | T1CE | 0.902 |
| 4 | wavelet.HHL_glszm_SizeZoneNonUniformity | T1CE | 0.526 |
| 5 | lbp.3D.m2_firstorder_90Percentile | T1CE | 0.956 |
| 6 | lbp.3D.k_firstorder_Minimum | FLAIR | -3.017 |
| 7 | log.sigma.4.0.mm.3D_glrlm_LongRunLowGrayLevelEmphasis | FLAIR | 2.411 |
表3 预测胶质瘤预后的最优特征所属序列及其系数(瘤周水肿区)
Tab.3 Optimal features from the peritumor edema region for predicting glioma prognosis and their corresponding sequences and coefficients (Edema region)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | original_shape_Maximum2DDiameterSlice | T1 | 0.988 |
| 2 | original_shape_Flatness | T1 | -0.915 |
| 3 | wavelet.HHH_gldm_DependenceNonUniformityNormalized | T1CE | 0.902 |
| 4 | wavelet.HHL_glszm_SizeZoneNonUniformity | T1CE | 0.526 |
| 5 | lbp.3D.m2_firstorder_90Percentile | T1CE | 0.956 |
| 6 | lbp.3D.k_firstorder_Minimum | FLAIR | -3.017 |
| 7 | log.sigma.4.0.mm.3D_glrlm_LongRunLowGrayLevelEmphasis | FLAIR | 2.411 |
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HLL_firstorder_Mean | T1 | 1.787 |
| 2 | log.sigma.5.0.mm.3D_firstorder_90Percentile | T1 | 2.131 |
| 3 | original_shape_Sphericity | FLAIR | -1.247 |
| 4 | wavelet.LLH_firstorder_Skewness | FLAIR | -0.228 |
| 5 | wavelet.LHL_firstorder_Skewness | FLAIR | -1.360 |
表4 预测胶质瘤预后的最优特征所属序列及其系数(全肿瘤区域)
Tab.4 Optimal features from the whole tumor for predicting glioma prognosis and their corresponding sequences and coefficients (The whole tumor)
| No. | Feature name | Sequence | Coefficient |
|---|---|---|---|
| 1 | wavelet.HLL_firstorder_Mean | T1 | 1.787 |
| 2 | log.sigma.5.0.mm.3D_firstorder_90Percentile | T1 | 2.131 |
| 3 | original_shape_Sphericity | FLAIR | -1.247 |
| 4 | wavelet.LLH_firstorder_Skewness | FLAIR | -0.228 |
| 5 | wavelet.LHL_firstorder_Skewness | FLAIR | -1.360 |
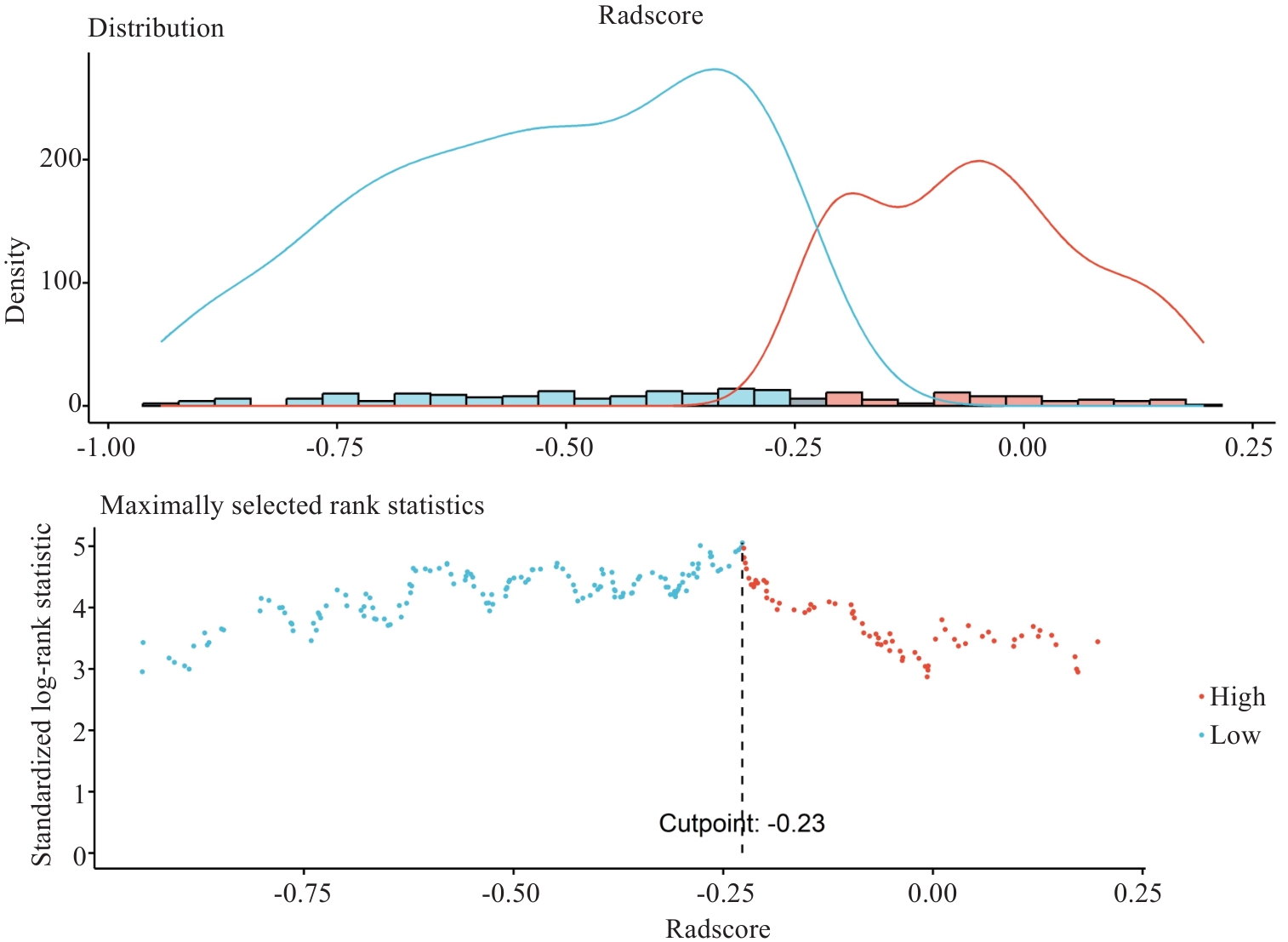
图2 Rad-score最佳阈值的确定(全肿瘤区域)
Fig.2 Optimal cutoff value of Rad-score for the whole tumor. Red lines or dots represent patients with high-risk prognosis and blue ones the low-risk patients. The optimal cutoff value is -0.2274 (shown by the vertical line).
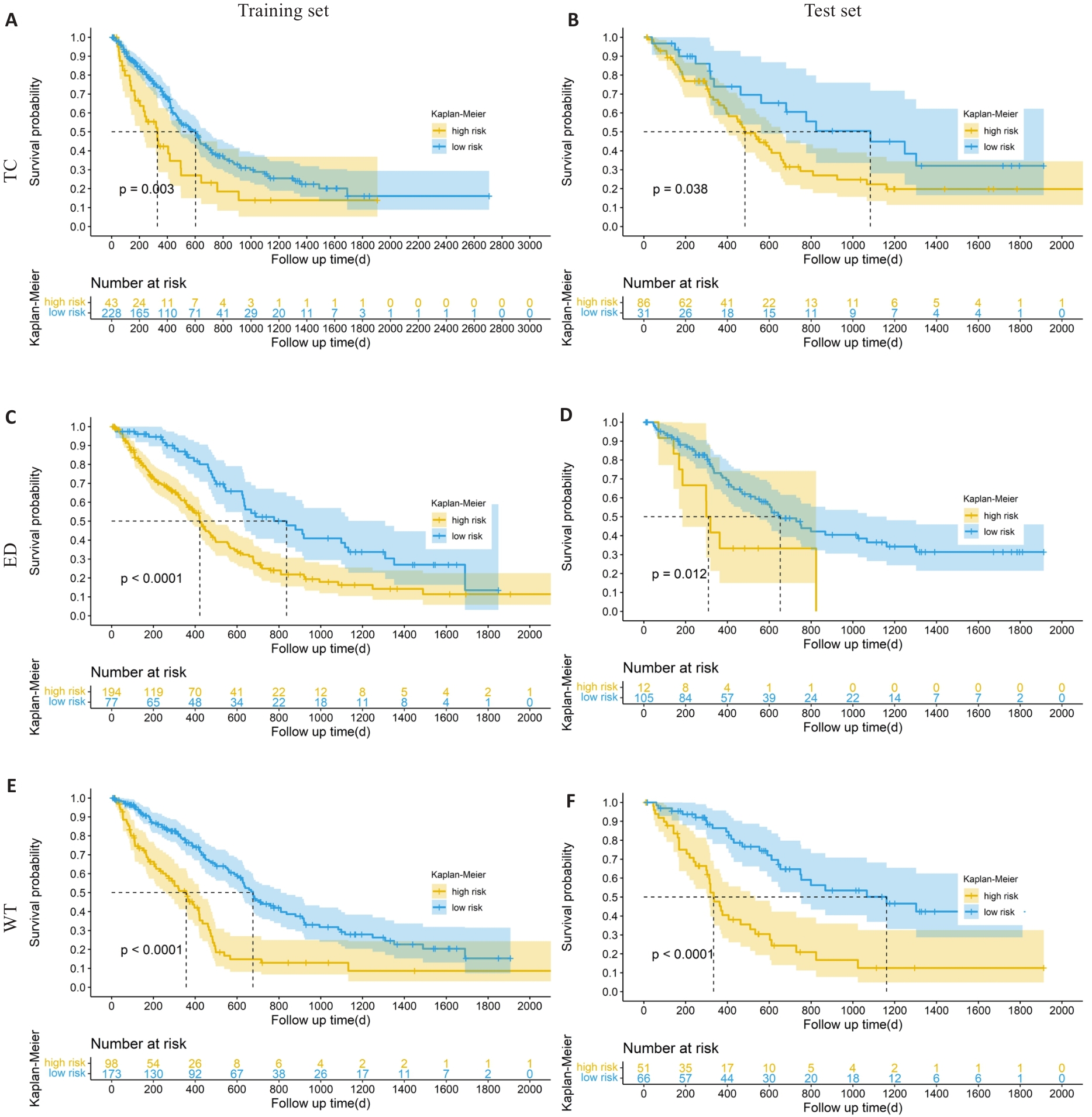
图3 训练集和测试集不同分组的Kaplan-Meier分析
Fig.3 Kaplan-Meier survival analysis of the patients in the high- and low-risk groups in the training and test sets. A, B: Kaplan-Meier analysis of the core tumor area in training set and test set. C, D: Kaplan-Meier analysis of peritumoral edema in the training set and test set. E, F: Kaplan-Meier analysis of the whole tumor in the training set and test set.
| Item | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.04 | 1.03-1.06 | <0.001 | 1.03 | 1.02-1.05 | <0.001 |
| Gender | 0.96 | 0.73-1.25 | 0.748 | - | - | - |
| WHO grade | 3.16 | 2.27-4.40 | <0.001 | 1.30 | 0.92-1.84 | 0.138 |
| IDH | 0.13 | 0.06-0.33 | <0.001 | 0.23 | 0.09-0.56 | 0.001 |
| Rad-score (TC) | 1.51 | 1.15-1.98 | 0.003 | 1.43 | 1.08-1.91 | 0.013 |
| Rad-score (ED) | 1.48 | 1.18-1.86 | <0.001 | 1.49 | 1.18-1.88 | 0.001 |
| Rad-score (WT) | 2.79 | 2.15-3.61 | <0.001 | 2.31 | 1.77-3.03 | <0.001 |
表5 胶质瘤患者总生存期预测的单因素和多因素分析结果
Tab.5 Univariate and multivariate analysis of Cox proportional hazards of overall survival of the glioma patients
| Item | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.04 | 1.03-1.06 | <0.001 | 1.03 | 1.02-1.05 | <0.001 |
| Gender | 0.96 | 0.73-1.25 | 0.748 | - | - | - |
| WHO grade | 3.16 | 2.27-4.40 | <0.001 | 1.30 | 0.92-1.84 | 0.138 |
| IDH | 0.13 | 0.06-0.33 | <0.001 | 0.23 | 0.09-0.56 | 0.001 |
| Rad-score (TC) | 1.51 | 1.15-1.98 | 0.003 | 1.43 | 1.08-1.91 | 0.013 |
| Rad-score (ED) | 1.48 | 1.18-1.86 | <0.001 | 1.49 | 1.18-1.88 | 0.001 |
| Rad-score (WT) | 2.79 | 2.15-3.61 | <0.001 | 2.31 | 1.77-3.03 | <0.001 |
| Tumor area | Model | Training set (Cross validation) | Test set | ||
|---|---|---|---|---|---|
1-year overall survival AUC | 3-year overall survival AUC | 1-year overall survival AUC | 3-year overall survival AUC | ||
| TC | Clinical information | 0.716 | 0.758 | 0.682 | 0.743 |
| Rad-score | 0.583 | 0.613 | 0.503 | 0.637 | |
| Joint model | 0.727 | 0.792 | 0.684 | 0.707 | |
| ED | Clinical information | 0.716 | 0.767 | 0.682 | 0.743 |
| Rad-score | 0.678 | 0.728 | 0.582 | 0.613 | |
| Joint model | 0.750 | 0.816 | 0.678 | 0.747 | |
| WT | Clinical information | 0.722 | 0.752 | 0.682 | 0.743 |
| Rad-score | 0.707 | 0.711 | 0.711 | 0.739 | |
| Joint model | 0.750 | 0.778 | 0.764 | 0.800 | |
表6 不同模型的效能
Tab.6 Performance of different models
| Tumor area | Model | Training set (Cross validation) | Test set | ||
|---|---|---|---|---|---|
1-year overall survival AUC | 3-year overall survival AUC | 1-year overall survival AUC | 3-year overall survival AUC | ||
| TC | Clinical information | 0.716 | 0.758 | 0.682 | 0.743 |
| Rad-score | 0.583 | 0.613 | 0.503 | 0.637 | |
| Joint model | 0.727 | 0.792 | 0.684 | 0.707 | |
| ED | Clinical information | 0.716 | 0.767 | 0.682 | 0.743 |
| Rad-score | 0.678 | 0.728 | 0.582 | 0.613 | |
| Joint model | 0.750 | 0.816 | 0.678 | 0.747 | |
| WT | Clinical information | 0.722 | 0.752 | 0.682 | 0.743 |
| Rad-score | 0.707 | 0.711 | 0.711 | 0.739 | |
| Joint model | 0.750 | 0.778 | 0.764 | 0.800 | |
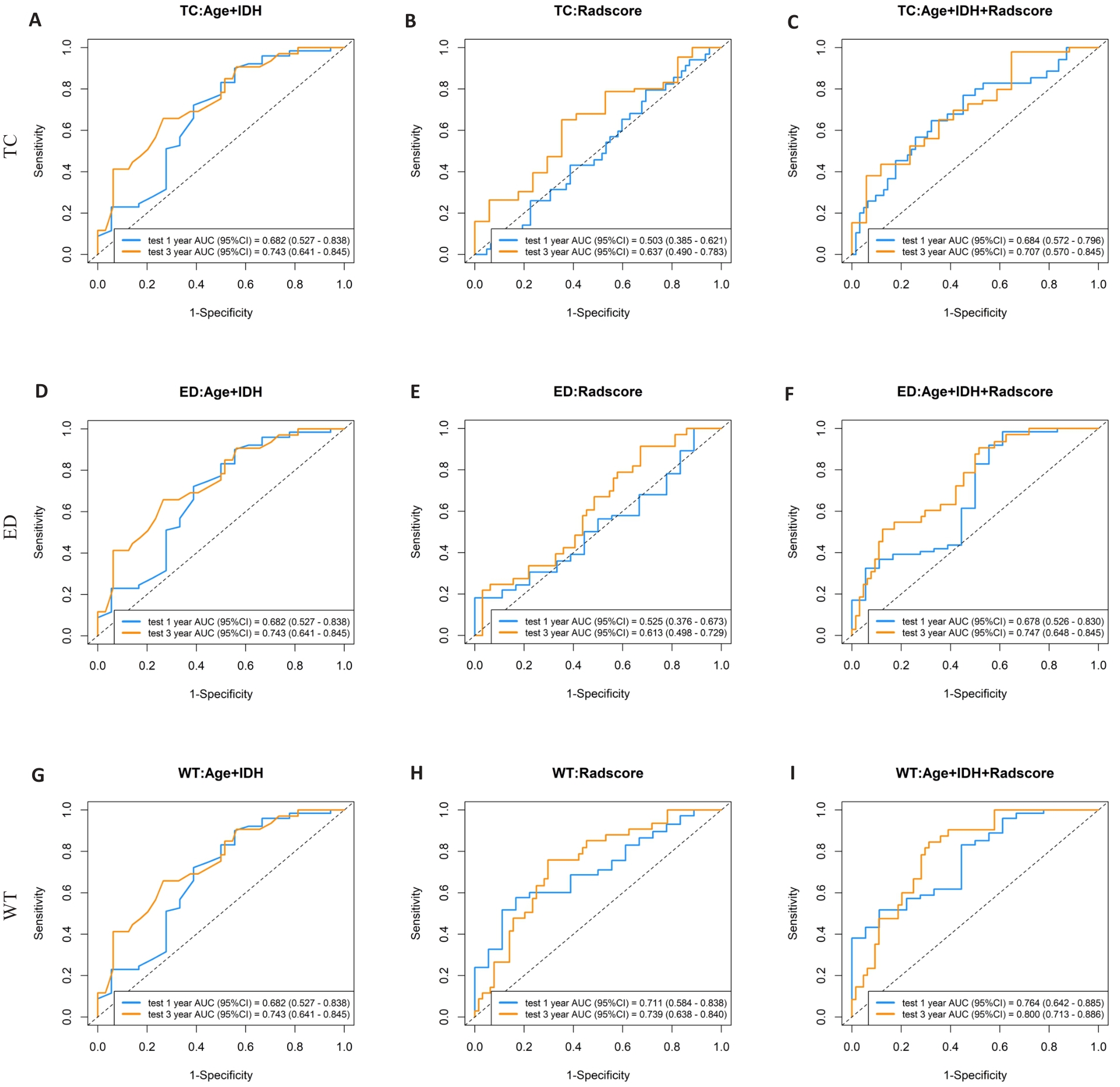
图4 肿瘤核心区、瘤周水肿区、全肿瘤区域的测试集中不同模型之间1、3年生存率受试者工作特征曲线
Fig.4 Receiver-operating characteristic curves of 1-year and 3-year survival rates predicted by the clinical information model, Rad-score model, and joint model in the test set for core tumor area (A-C), peritumoral edema area (D-F), and whole tumor (G-I).
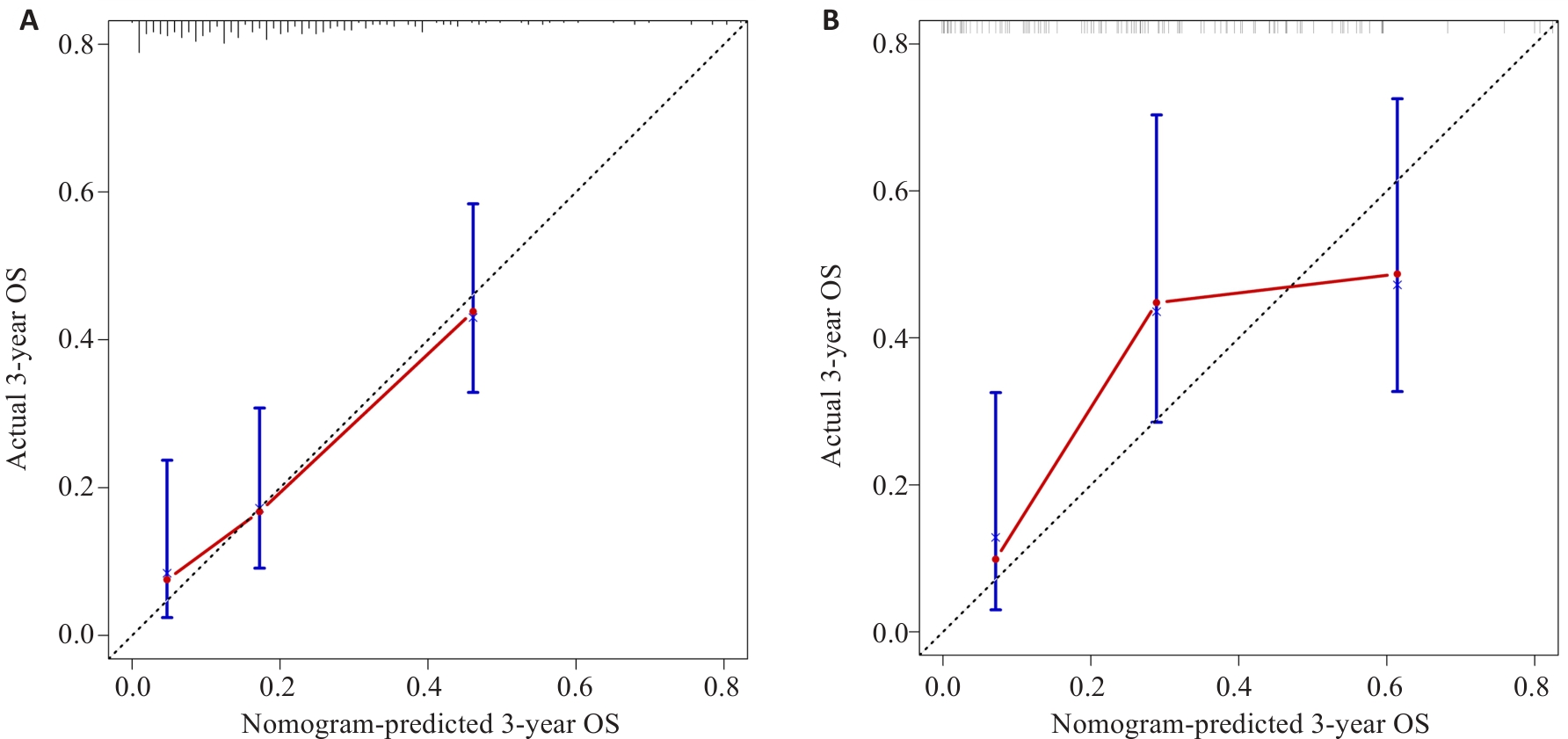
图6 训练集(A)和测试集(B)中胶质瘤患者预测3年生存率列线图的校准曲线
Fig.6 Nomogram for predicting 3-year OS and PFS in patients with glioma in the training set (A) and test set (B).
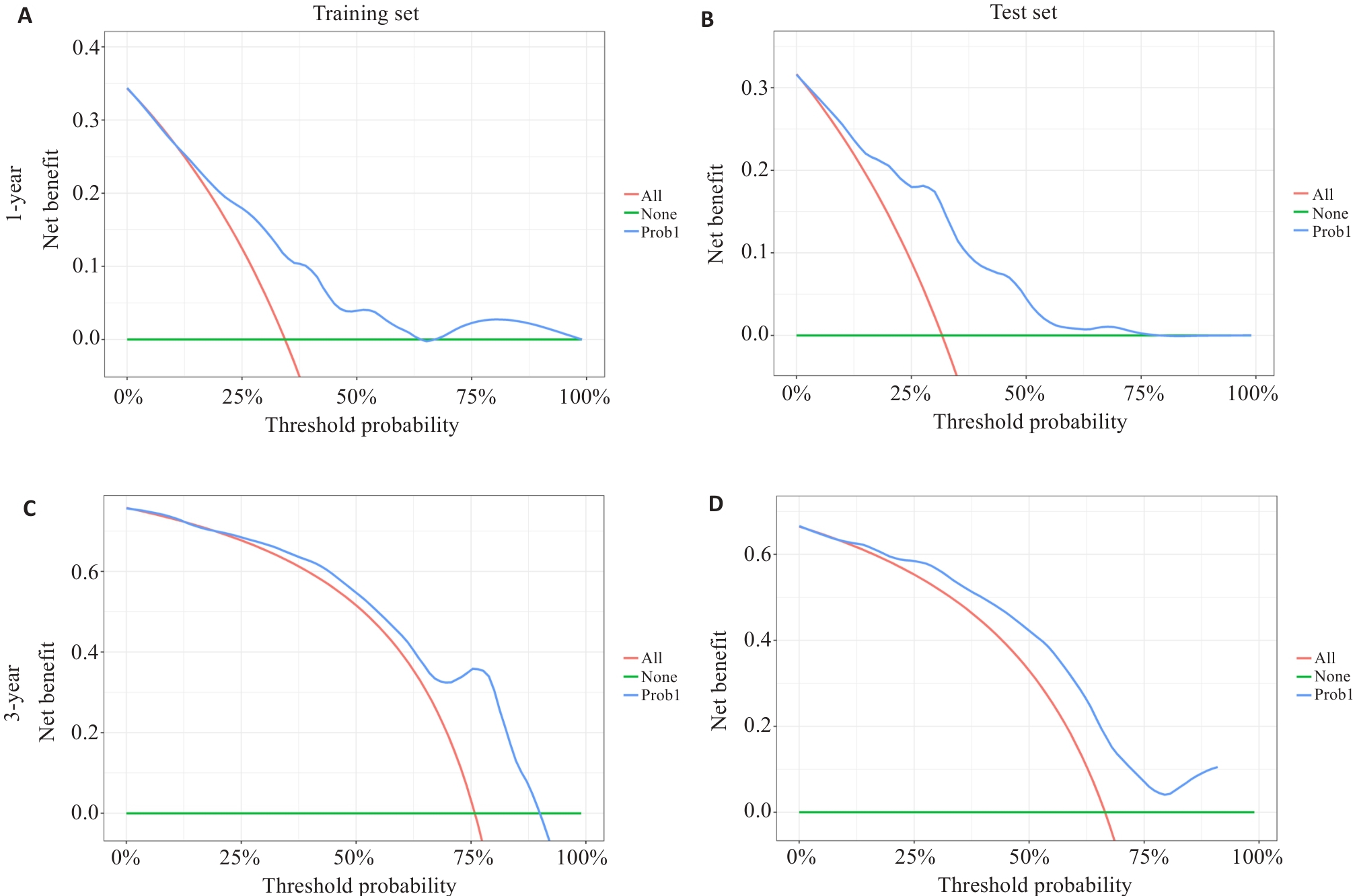
图7 训练集(A、C)和测试集(B、D)中列线图模型分别预测胶质瘤1、3年总生存率的决策曲线
Fig.7 Decision curve analysis of survival prediction nomograms for 1-year and 3-year overall survival rates of glioma patients in the training set (A, C) and test set (B, D).
| 1 | Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016[J]. Neuro Oncol, 2019, 21(): v1-10. |
| 2 | Xu HS, Zhang AK, Han XY, et al. ITGB2 as a prognostic indicator and a predictive marker for immunotherapy in gliomas[J]. Cancer Immunol Immunother, 2022, 71(3): 645-60. |
| 3 | 李绍山, 付 强, 秦 虎, 等. 脑胶质瘤在CT平扫及三期增强中的图像表现特征分析[J]. 中国CT和MRI杂志, 2019, 17(7): 5-7. DOI: 10.3969/j.issn.1672-5131.2019.07.002 |
| 4 | Rasmussen BK, Hansen S, Laursen RJ, et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry[J]. J Neurooncol, 2017, 135(3): 571-9. |
| 5 | Lin ZY, Yang RW, Li KS, et al. Establishment of age group classification for risk stratification in glioma patients[J]. BMC Neurol, 2020, 20(1): 310. |
| 6 | Zhang BQ, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas[J]. Neuro Oncol, 2017, 19(1): 109-17. |
| 7 | Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials[J]. Neuro Oncol, 2015, 17(9): 1188-98. |
| 8 | Park YW, Kim S, Park CJ, et al. Adding radiomics to the 2021 WHO updates may improve prognostic prediction for current IDH-wildtype histological lower-grade gliomas with known EGFR amplification and TERT promoter mutation status[J]. Eur Radiol, 2022, 32(12): 8089-98. |
| 9 | 杜 鹏, 耿道颖. 多模态MRI影像组学在胶质瘤精准诊疗中的研究进展[J]. 国际医学放射学杂志, 2023, 46(6): 672-9. |
| 10 | Bakas S, Akbari H, Pisapia J, et al. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-index[J]. Clin Cancer Res, 2017, 23(16): 4724-34. |
| 11 | Weller M, Weber RG, Willscher E, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups[J]. Acta Neuropathol, 2015, 129(5): 679-93. |
| 12 | Calabrese E, Villanueva-Meyer JE, Rudie JD, et al. The university of California San francisco preoperative diffuse glioma MRI dataset[J]. Radiol Artif Intell, 2022, 4(6): e220058. |
| 13 | Bale TA, Rosenblum MK. The 2021 WHO Classification of Tumors of the Central Nervous System: an update on pediatric low-grade gliomas and glioneuronal tumors[J]. Brain Pathol, 2022, 32(4): e13060. |
| 14 | Bakas S, Sako C, Akbari H, et al. The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: advanced MRI, clinical, genomics, & radiomics[J]. Sci Data, 2022, 9(1): 453. |
| 15 | 邓景景. 生存分析模型风险预测能力评价方法及其应用研究[D]. 南京: 东南大学, 2016. |
| 16 | Kamarudin AN, Cox T, Kolamunnage-Dona R. Time-dependent ROC curve analysis in medical research: current methods and applications[J]. BMC Med Res Methodol, 2017, 17(1): 53. |
| 17 | Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves[J]. Biometrics, 2005, 61(1): 92-105. |
| 18 | Foltyn-Dumitru M, Schell M, Rastogi A, et al. Impact of signal intensity normalization of MRI on the generalizability of radiomic-based prediction of molecular glioma subtypes[J]. Eur Radiol, 2024, 34(4): 2782-90. |
| 19 | Foltyn-Dumitru M, Schell M, Sahm F, et al. Advancing noninvasive glioma classification with diffusion radiomics: exploring the impact of signal intensity normalization[J]. Neurooncol Adv, 2024, 6(1): vdae043. |
| 20 | Huang DC, Gao TY, Zhang Y, et al. A study on prognosis of diffuse glioma based on clinical factors and magnetic resonance imaging radiomics[J]. World Neurosurg, 2024, 186: e514-30. |
| 21 | Usuzaki T, Takahashi K, Inamori R, et al. Identifying key factors for predicting O6-Methylguanine-DNA methyltransferase status in adult patients with diffuse glioma: a multimodal analysis of demographics, radiomics, and MRI by variable Vision Transformer[J]. Neuroradiology, 2024, 66(5): 761-73. |
| 22 | Truong NCD, Bangalore Yogananda CG, Wagner BC, et al. Two-stage training framework using multicontrast MRI radiomics for IDH mutation status prediction in glioma[J]. Radiol Artif Intell, 2024, 6(4): e230218. |
| 23 | Markwell SM, Ross JL, Olson CL, et al. Necrotic reshaping of the glioma microenvironment drives disease progression[J]. Acta Neuropathol, 2022, 143(3): 291-310. |
| 24 | Williams BJ, Frieboes H, Baxter M, et al. Metabolomic differentiation of tumor core vs. edge in glioma via machine learning[J]. Neurosurgery, 2024, 70(): 210. |
| 25 | Zhou Y, Gu HL, Zhang XL, et al. Multiparametric magnetic resonance imaging-derived radiomics for the prediction of disease-free survival in early-stage squamous cervical cancer[J]. Eur Radiol, 2022, 32(4): 2540-51. |
| 26 | Lemée JM, Clavreul A, Menei P. Intratumoral heterogeneity in glioblastoma: don't forget the peritumoral brain zone[J]. Neuro-oncology, 2015, 17(10): 1322-32. |
| 27 | Saadoun S, Papadopoulos MC, Davies DC, et al. Increased aquaporin 1 water channel expression in human brain tumours[J]. Br J Cancer, 2002, 87(6): 621-3. |
| 28 | Yang Y, Han Y, Zhao SJ, et al. Spatial heterogeneity of edema region uncovers survival-relevant habitat of Glioblastoma[J]. Eur J Radiol, 2022, 154: 110423. |
| 29 | 侯秋阳, 叶成坤, 刘 畅, 等. 基于多参数磁共振瘤周区域的影像组学模型在脑胶质瘤预后评估中的应用价值[J]. 安徽医科大学学报, 2024, 59(1): 154-61. |
| 30 | 卢明君, 屈耀铭, 马安东,等. 多模态MRI影像组学可预测弥漫性较低级别胶质瘤的1p/19q共缺失状态[J]. 南方医科大学学报, 2023, 43(6): 1023-8. DOI: 10.12122/j.issn.1673-4254.2023.06.19 |
| 31 | Fan ZC, Zhang L, Yang GQ, et al. MRI radiomics for predicting poor disease-free survival in muscle invasive bladder cancer: the results of the retrospective cohort study[J]. Abdom Radiol, 2024, 49(1): 151-62. |
| 32 | Ahrari S, Zaragori T, Zinsz A, et al. Application of PET imaging delta radiomics for predicting progression-free survival in rare high-grade glioma[J]. Sci Rep, 2024, 14(1): 3256. |
| 33 | Ye LG, Gu LG, Zheng ZY, et al. An online survival predictor in glioma patients using machine learning based on WHO CNS5 data[J]. Front Neurol, 2023, 14: 1179761. |
| 34 | Zhao R, Zhuge Y, Camphausen K, et al. Machine learning based survival prediction in Glioma using large-scale registry data[J]. Health Informatics J, 2022, 28(4): 14604582221135427. |
| 35 | 李新宇, 尚洵杰, 夏 彤, 等. 原发性人脑胶质瘤的预后影响因素: 基于SEER数据库的分析[J]. 中国临床神经外科杂志, 2021, 26(10): 764-8. DOI: 10.13798/j.issn.1009-153X.2020.10.007 |
| 36 | Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary[J]. Acta Neuropathol, 2016, 131(6): 803-20. |
| 37 | Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas[J]. J Clin Oncol, 2009, 27(25): 4150-4. |
| 38 | 张 宁, 马辉辉, 王 凡, 等. 脑胶质瘤患者 MGMT 启动子甲基化和 IDH1 突变与临床预后相关[J]. 基础医学与临床, 2021, 41(10): 1476-80. DOI: 10.3969/j.issn.1001-6325.2021.10.014 |
| 39 | 胡 哲,王玉红,刘晓龙, 等. 基于术前MRI图像构建影像组学与深度学习的机器学习模型预测胶质瘤IDH-1基因表达的研究[J]. 临床神经外科杂志, 2024, 21(2): 187-92. DOI: 10.3969/j.issn.1672-7770.2024.02.013 |
| 40 | Choi Y, Nam Y, Jang J, et al. Radiomics may increase the prognostic value for survival in glioblastoma patients when combined with conventional clinical and genetic prognostic models[J]. Eur Radiol, 2021, 31(4): 2084-93. |
| 41 | Park CJ, Kim S, Han K, et al. Diffusion- and perfusion-weighted MRI radiomics for survival prediction in patients with lower-grade gliomas[J]. Yonsei Med J, 2024, 65(5): 283-92. |
| [1] | 吴垂杏, 钟伟雄, 谢金城, 杨蕊梦, 吴元魁, 许乙凯, 王琳婧, 甄鑫. 基于序列缺失的MRI多序列特征填补与融合互助模型:鉴别高低级别胶质瘤[J]. 南方医科大学学报, 2024, 44(8): 1561-1570. |
| [2] | 刘科, 马振岩, 付磊, 张丽萍, 阿鑫, 肖少波, 张震, 张洪博, 赵蕾, 钱赓. 心脏磁共振成像整体纵向应变对急性ST段抬高型心肌梗死后左心室重构的预测价值:403例前瞻性研究[J]. 南方医科大学学报, 2024, 44(6): 1033-1039. |
| [3] | 陈莉莉, 吴天宇, 张铭, 丁子夏, 张妍, 杨依清, 郑佳倩, 张小楠. 类风湿关节炎的潜在生物标志物及其免疫调控机制:基于GEO数据库[J]. 南方医科大学学报, 2024, 44(6): 1098-1108. |
| [4] | 申采玉, 王帅, 周锐盈, 汪雨贺, 高琴, 陈兴智, 杨枢. 慢性心力衰竭合并肺部感染患者院内死亡风险预测:基于可解释性机器学习方法[J]. 南方医科大学学报, 2024, 44(6): 1141-1148. |
| [5] | 左志威, 孟庆良, 崔家康, 郭克磊, 卞华. 基于硬皮病线粒体相关基因的人工神经网络模型的构建[J]. 南方医科大学学报, 2024, 44(5): 920-929. |
| [6] | 黄秋虎, 周 建, 王子珍, 杨 堃, 陈政纲. miR-26b-3p 靶向 CREB1 调控神经胶质瘤细胞的增殖、迁移及侵袭[J]. 南方医科大学学报, 2024, 44(3): 578-584. |
| [7] | 刘云泽, 李宬润, 郭俊唐, 刘 阳. 基于临床-影像组学列线图模型鉴别局灶性机化性肺炎与肺腺癌[J]. 南方医科大学学报, 2024, 44(2): 397-404. |
| [8] | 何慧珊, 郭二嘉, 蒙文仪, 王 彧, 王 雯, 何文乐, 吴元魁, 阳 维. 基于磁共振图像机器学习放射组学模型预测脑胶质瘤的强化[J]. 南方医科大学学报, 2024, 44(1): 194-200. |
| [9] | 张振阳, 谢金城, 钟伟雄, 梁芳蓉, 杨蕊梦, 甄 鑫. 基于距匹配及判别表征学习的多模态特征融合分类模型研究:高级别胶质瘤与单发性脑转移瘤的鉴别诊断[J]. 南方医科大学学报, 2024, 44(1): 138-145. |
| [10] | 于正涛, 李佳梦, 蒋俊文, 李 由, 林 珑, 夏 鹰, 王 磊. miRNA-128-3p通过下调KLHDC8A的表达抑制胶质瘤细胞的恶性生物学行为[J]. 南方医科大学学报, 2023, 43(9): 1447-1459. |
| [11] | 楚智钦, 屈耀铭, 钟 涛, 梁淑君, 温志波, 张 煜. 磁共振酰胺质子转移模态的胶质瘤IDH基因分型识别:基于深度学习的Dual-Aware框架[J]. 南方医科大学学报, 2023, 43(8): 1379-1387. |
| [12] | 刘羽轩, 楚智钦, 张 煜. 基于物理模型的级联生成对抗网络加速定量多参数磁共振成像[J]. 南方医科大学学报, 2023, 43(8): 1402-1409. |
| [13] | 罗 枭, 程 义, 吴 骋, 贺 佳. 预测重症缺血性脑卒中死亡风险的模型:基于内在可解释性机器学习方法[J]. 南方医科大学学报, 2023, 43(7): 1241-1247. |
| [14] | 叶静静, 徐文琴, 奚邦生, 王能乾, 陈天兵. 乳酸诱导PLEKHA4的上调参与胶质瘤细胞增殖和凋亡的生物学进程[J]. 南方医科大学学报, 2023, 43(7): 1071-1080. |
| [15] | 高凯绩, 王一豪, 曹海坤, 贾建光. 机器学习模型和Cox回归模型预测食管胃结合部腺癌预后的效能[J]. 南方医科大学学报, 2023, 43(6): 952-963. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||