Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (11): 2466-2474.doi: 10.12122/j.issn.1673-4254.2025.11.19
Previous Articles Next Articles
Zhiqiang GAO( ), Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN(
), Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN( ), Guanghui WEI
), Guanghui WEI
Received:2024-05-19
Online:2025-11-20
Published:2025-11-28
Contact:
Xiaomao TIAN
E-mail:1815234622@qq.com;xiao-mao@ hospital.cqmu.edu.cn
Supported by:Zhiqiang GAO, Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN, Guanghui WEI. High-throughput circular RNA sequencing reveals tumor-specific high expression of hsa_circ_0001900 in Wilms tumor in association with poor prognosis[J]. Journal of Southern Medical University, 2025, 45(11): 2466-2474.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.11.19
| Samples | Output of data (Gb) | Q30 (%) |
|---|---|---|
| Tumor tissue-1 | 15.060 | 88.84 |
| Tumor tissue-2 | 13.910 | 88.83 |
| Tumor tissue-3 | 14.581 | 89.52 |
| Tumor tissue-4 | 13.873 | 89.72 |
| Adjacent control-1 | 13.325 | 90.70 |
| Adjacent control-2 | 14.272 | 90.45 |
| Adjacent control-3 | 15.289 | 90.44 |
| Adjacent control-4 | 11.826 | 90.67 |
Tab.1 Quality control for sequencing results
| Samples | Output of data (Gb) | Q30 (%) |
|---|---|---|
| Tumor tissue-1 | 15.060 | 88.84 |
| Tumor tissue-2 | 13.910 | 88.83 |
| Tumor tissue-3 | 14.581 | 89.52 |
| Tumor tissue-4 | 13.873 | 89.72 |
| Adjacent control-1 | 13.325 | 90.70 |
| Adjacent control-2 | 14.272 | 90.45 |
| Adjacent control-3 | 15.289 | 90.44 |
| Adjacent control-4 | 11.826 | 90.67 |
| Genes | Pre-primer (5'→3') | Post-primer (3'→5') |
|---|---|---|
| hsa_circ_0001900 | CGTTCAGTGCCTCGAAAGAAC | CTGGTCCCCTTTCAGGATGAG |
| hsa_circ_0004425 | AAGTGTGGACCCTGCAAATA | CCATTTGATCCACAGACAGGA |
| hsa_circ_0005230 | CTCTTTGTTTTGCACACTAGGGA | ACCAGGTGAGCAGTCAAGAA |
| hsa_circ_0075828 | GGAAGTGGCTAATGGATCTG | ATGGAGAACAGCCATCCATG |
| hsa_circ_0008122 | TGTCTCAAAGCCAGACCTGAT | ATTCTGTTGTTCGGTGTCCAG |
| hsa_circ_0006151 | GGCACCAGCATAGCCAGTAGT | GATGACCAGGCGCTTTTCACA |
| GAPDH | CCTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGTG |
Tab.2 Primer sequences for q-PCR
| Genes | Pre-primer (5'→3') | Post-primer (3'→5') |
|---|---|---|
| hsa_circ_0001900 | CGTTCAGTGCCTCGAAAGAAC | CTGGTCCCCTTTCAGGATGAG |
| hsa_circ_0004425 | AAGTGTGGACCCTGCAAATA | CCATTTGATCCACAGACAGGA |
| hsa_circ_0005230 | CTCTTTGTTTTGCACACTAGGGA | ACCAGGTGAGCAGTCAAGAA |
| hsa_circ_0075828 | GGAAGTGGCTAATGGATCTG | ATGGAGAACAGCCATCCATG |
| hsa_circ_0008122 | TGTCTCAAAGCCAGACCTGAT | ATTCTGTTGTTCGGTGTCCAG |
| hsa_circ_0006151 | GGCACCAGCATAGCCAGTAGT | GATGACCAGGCGCTTTTCACA |
| GAPDH | CCTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGTG |
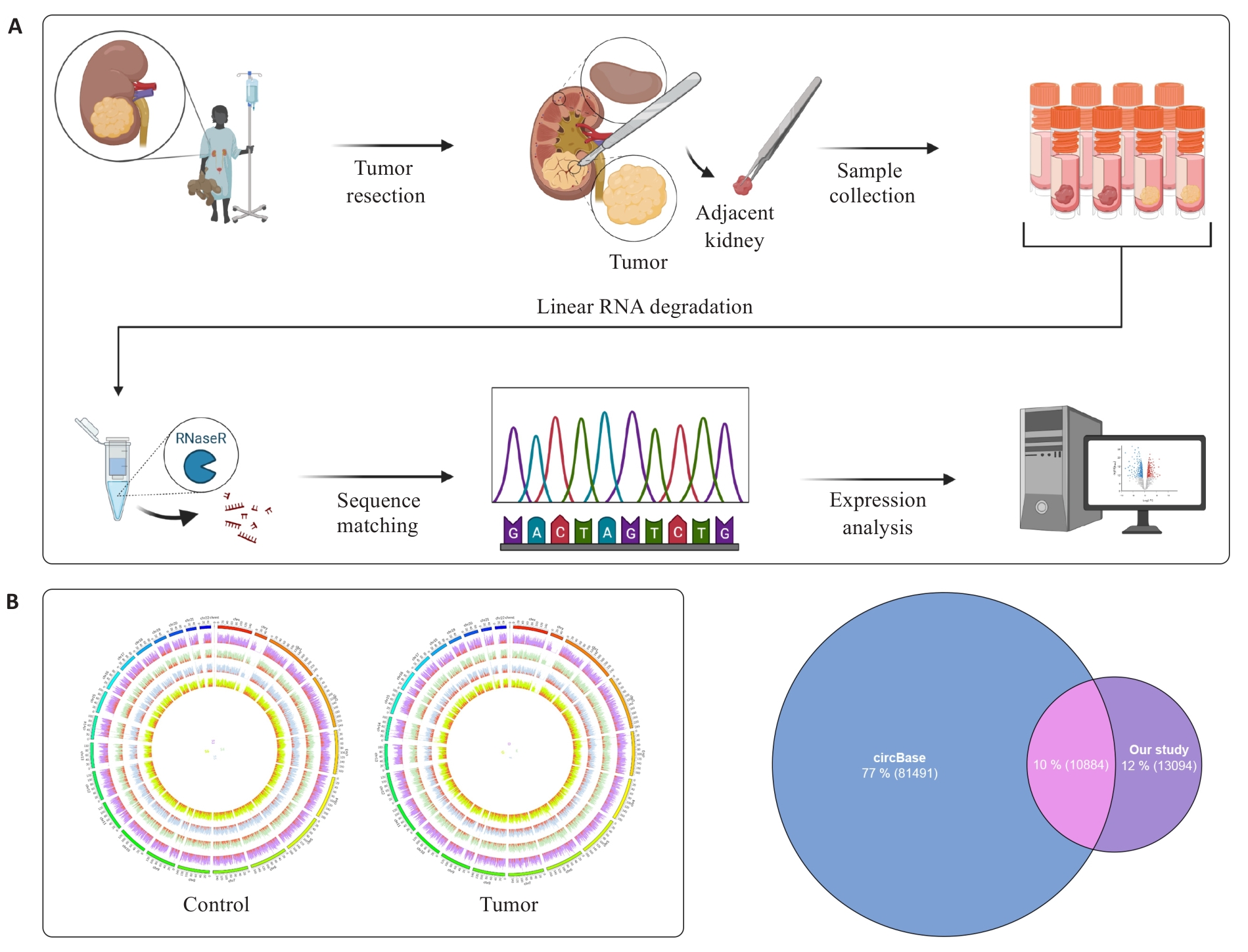
Fig.1 Identification and annotation of circRNAs A: Schematic diagram of Wilms tumor (WT) circular RNA sequencing analysis. B: Circos showing the full view of the circRNAs, with the outer circles representing chromosomes, the inner circles representing samples, and the red part of each sample representing "spliced" readings of the circRNAs. C: Venn diagram of circRNA sequencing data annotated with circBase database.

Fig.2 Classification and differential expression patterns of circRNAs. A: Circular RNA classification. B: Principal component analysis (PCA). C: Volcano plot of the differentially expressed circRNAs. Red dots indicate upregulation and blue dots indicate downregulation. The horizontal dashed line represents the truncated P value of 0.05, and the vertical dashed line represents the 2-fold logFC value.
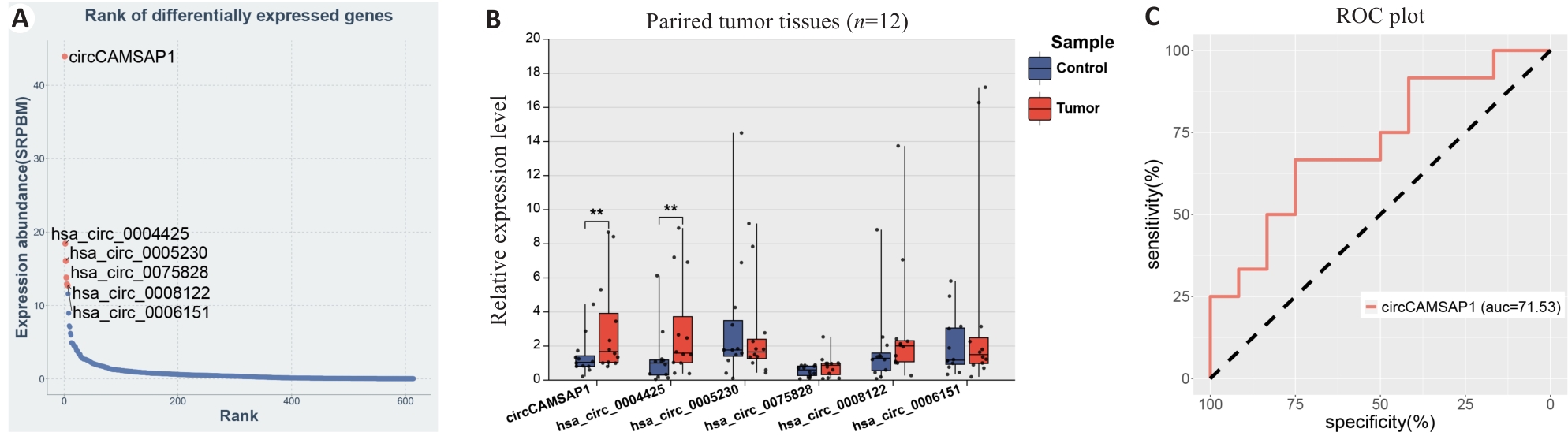
Fig.3 Validation of tumor-specific expression pattern of hsa_circ_0001900 in clinical samples of WT A: Ranking map of circRNA expression abundance. B: Expression validation of the top 6 circRNAs in clinical samples. C: ROC diagnostic curve of hsa_circ_0001900. *P<0.05, **P<0.01 vs Tumor group.
| circRNA | Chromosomal loci | Direction of ring formation | Genome length | Splice sequence length | Gene ID | Gene of parent | Trend of expression |
|---|---|---|---|---|---|---|---|
| hsa_circ_0008122 | chr19:23134080-23136043 | + | 1964 | 223 | ENSG00000183850 | ZNF730 | up |
| hsa_circ_0001900 | chr9:135881633-135883078 | - | 1446 | 425 | ENSG00000130559 | CAMSAP1 | up |
| hsa_circ_0006151 | chr10:68468123-68470163 | - | 2041 | 367 | ENSG00000138346 | DNA2 | up |
| hsa_circ_0004425 | chr9:100498765-100516771 | + | 18007 | 364 | ENSG00000241697 | TMEFF1 | up |
| hsa_circ_0075828 | Chr6:22020339-22020542 | + | 204 | 204 | ENSG00000272168 | CASC15 | up |
| hsa_circ_0005230 | Chr1:172140480-172144437 | - | 3958 | 3958 | ENSG00000230630 | DNM3OS | up |
Tab.3 Characteristics of circRNA molecules for expression validation in clinical samples
| circRNA | Chromosomal loci | Direction of ring formation | Genome length | Splice sequence length | Gene ID | Gene of parent | Trend of expression |
|---|---|---|---|---|---|---|---|
| hsa_circ_0008122 | chr19:23134080-23136043 | + | 1964 | 223 | ENSG00000183850 | ZNF730 | up |
| hsa_circ_0001900 | chr9:135881633-135883078 | - | 1446 | 425 | ENSG00000130559 | CAMSAP1 | up |
| hsa_circ_0006151 | chr10:68468123-68470163 | - | 2041 | 367 | ENSG00000138346 | DNA2 | up |
| hsa_circ_0004425 | chr9:100498765-100516771 | + | 18007 | 364 | ENSG00000241697 | TMEFF1 | up |
| hsa_circ_0075828 | Chr6:22020339-22020542 | + | 204 | 204 | ENSG00000272168 | CASC15 | up |
| hsa_circ_0005230 | Chr1:172140480-172144437 | - | 3958 | 3958 | ENSG00000230630 | DNM3OS | up |

Fig.4 Correlation analysis of hsa_circ_0001900 expression level in WT with tumor volume and prognosis. A: Expression levels of hsa_circ_0001900 in different cancers. B: Verification of differential expression of hsa_circ_0001900 in a patient cohort. C: The expression level of hsa_circ_0001900 is positively correlated with tumor volume. D: Children with high expression of hsa_circ_0001900 had shorter survival time. *P<0.05, **P<0.01 vs Tumor group.

Fig.5 Annotation and verification of circular structure of hsa_circ_0001900. A: Sanger sequencing based on PCR products that verifies the reverse splicing sequence of hsa_circ_0001900. B: Agarose gel electrophoresis verifies the RNase R enzyme tolerance of hsa_circ_0001900 from a qualitative perspective. C: qPCR verifies the structural stability of hsa_circ_0001900 from a quantitative perspective. ****P<0.0001.
| [1] | Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression[J]. Mol Cancer, 2018, 17(1): 79. doi:10.1186/s12943-018-0827-8 |
| [2] | 田小毛, 向 彬, 刘 丰, et al. 环状RNA在儿童肾母细胞瘤中的表达谱、生物学功能和临床意义 [J]. 生物化学与生物物理进展, 2023, 50(3): 463-72. |
| [3] | Zhao Y, Li J, Li J, et al. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100[J]. Eur Rev Med Pharmacol Sci, 2020, 24(8): 4420-9. |
| [4] | Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer[J]. Nat Rev Cancer, 2018, 18(1): 5-18. doi:10.1038/nrc.2017.99 |
| [5] | Hare A, Zeng M, Rehemutula A, et al. Hsa-circ_0000064 accelerates the malignant progression of gastric cancer via sponging microRNA-621[J]. Kaohsiung J Med Sci, 2021, 37(10): 841-50. doi:10.1002/kjm2.12419 |
| [6] | Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis[J]. Cancer Res, 2017, 77(9): 2339-50. doi:10.1158/0008-5472.can-16-1883 |
| [7] | Yin WB, Yan MG, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection[J]. Clin Chim Acta, 2018, 487: 363-8. doi:10.1016/j.cca.2017.10.011 |
| [8] | Qiu M, Xia W, Chen R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma[J]. Cancer Res, 2018, 78(11): 2839-51. doi:10.1158/0008-5472.can-17-2808 |
| [9] | Zhang Y, Jiang J, Zhang J, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability[J]. Mol Cancer, 2021, 20(1): 101. doi:10.1186/s12943-021-01390-y |
| [10] | Hou L, Zhang J, Zhao F. Full-length circular RNA profiling by nanopore sequencing with CIRI-long[J]. Nat Protoc, 2023, 18(6): 1795-813. doi:10.1038/s41596-023-00815-w |
| [11] | López-Jiménez E, Rojas AM, Andrés-León E. RNA sequencing and prediction tools for circular RNAs analysis[J]. Adv Exp Med Biol, 2018, 1087: 17-33. doi:10.1007/978-981-13-1426-1_2 |
| [12] | Zhang J, Zhao F. Circular RNA discovery with emerging sequencing and deep learning technologies[J]. Nat Genet, 2025, 57(5): 1089-102. doi:10.1038/s41588-025-02157-7 |
| [13] | Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance[J]. Trends Cancer, 2020, 6(4): 319-36. doi:10.1016/j.trecan.2020.01.012 |
| [14] | Zhang J, Quan Y, Su X, et al. Circ-PRMT5 stimulates the proliferative ability in Wilms' tumor through the miR-7-5p/KLF4 axis[J]. Cell Mol Biol: Noisy-le-grand, 2023, 69(8): 232-6. doi:10.14715/cmb/2023.69.8.36 |
| [15] | 田小毛, 石秦林, 陆 鹏, 等. 生物标志物对Wilms瘤预后判断及治疗指导价值的研究进展 [J]. 现代医药卫生, 2020, 36(6): 855-9. |
| [16] | Xiang B, Chen ML, Gao ZQ, et al. CCNB1 is a novel prognostic biomarker and promotes proliferation, migration and invasion in Wilms tumor[J]. BMC Med Genomics, 2023, 16(1): 189. doi:10.1186/s12920-023-01627-3 |
| [17] | 高志强, 林 洁, 洪 鹏, 等. 基于高通量RNA测序分析Wilms瘤中关键基因对预后及免疫应答的影响 [J]. 南方医科大学学报, 2024, 44(4): 727-38. |
| [18] | Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-90. doi:10.1093/bioinformatics/bty560 |
| [19] | Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol, 2015, 16: 4. doi:10.1186/s13059-014-0571-3 |
| [20] | Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs[J]. RNA, 2014, 20(11): 1666-70. doi:10.1261/rna.043687.113 |
| [21] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome Res, 2009, 19(9): 1639-45. doi:10.1101/gr.092759.109 |
| [22] | Robic A, Kühn C. Beyond back splicing, a still poorly explored world: non-canonical circular RNAs[J]. Genes: Basel, 2020, 11(9): E1111. doi:10.3390/genes11091111 |
| [23] | Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis[J]. Cell Res, 2015, 25(8): 981-4. doi:10.1038/cr.2015.82 |
| [24] | Dome JS, Graf N, Geller JI, et al. Advances in wilms tumor treatment and biology: progress through international collaboration[J]. J Clin Oncol, 2015, 33(27): 2999-3007. doi:10.1200/jco.2015.62.1888 |
| [25] | Saltzman AF, Cost NG, Romao RLP. Wilms tumor[J]. Urol Clin North Am, 2023, 50(3): 455-64. doi:10.1016/j.ucl.2023.04.008 |
| [26] | 向 彬. CircRACGAP1/miR-486-3p/CTNNBIP1调控轴促进肾母细胞瘤增殖侵袭和迁移的机制研究 [D], 重庆医科大学, 2023. |
| [27] | Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRNAs in cancer and oncology[J]. Nat Rev Clin Oncol, 2022, 19(3): 188-206. doi:10.1038/s41571-021-00585-y |
| [28] | Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-91. doi:10.1038/s41576-019-0158-7 |
| [29] | Beilerli A, Gareev I, Beylerli O, et al. Circular RNAs as biomarkers and therapeutic targets in cancer[J]. Semin Cancer Biol, 2022, 83: 242-52. doi:10.1016/j.semcancer.2020.12.026 |
| [30] | Lasda E, Parker R. Circular RNAs: diversity of form and function[J]. RNA, 2014, 20(12): 1829-42. doi:10.1261/rna.047126.114 |
| [31] | Zhang F, Zou HM, Li XY, et al. CircRNA_0017076 acts as a sponge for miR-185-5p in the control of epithelial-to-mesenchymal transition of tubular epithelial cells during renal interstitial fibrosis[J]. Hum Cell, 2023, 36(3): 1024-40. doi:10.1007/s13577-023-00877-8 |
| [32] | Wang L, Zhou Y, Jiang L, et al. CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway[J]. Mol Cancer, 2021, 20(1): 43. doi:10.1186/s12943-021-01332-8 |
| [33] | Xu F, Xiao Q, Du WW, et al. CircRNA: functions, applications and prospects[J]. Biomolecules, 2024, 14(12): 1503. doi:10.3390/biom14121503 |
| [34] | Dong J, Zeng Z, Huang Y, et al. Challenges and opportunities for circRNA identification and delivery[J]. Crit Rev Biochem Mol Biol, 2023, 58(1): 19-35. doi:10.1080/10409238.2023.2185764 |
| [35] | Xu K, Park D, Magis AT, et al. Small molecule KRAS agonist for mutant KRAS cancer therapy[J]. Mol Cancer, 2019, 18(1): 85. doi:10.1186/s12943-019-1012-4 |
| [36] | Casagrande GMS, Silva MO, Reis RM, et al. Liquid biopsy for lung cancer: up-to-date and perspectives for screening programs[J]. Int J Mol Sci, 2023, 24(3): 2505. doi:10.3390/ijms24032505 |
| [37] | Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6[J]. Cancer Cell Int, 2018, 18: 116. doi:10.1186/s12935-018-0602-3 |
| [38] | Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer[J]. Clin Chim Acta, 2017, 466: 167-71. doi:10.1016/j.cca.2017.01.025 |
| [39] | Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer[J]. World J Gastroenterol, 2017, 23(34): 6330-8. doi:10.3748/wjg.v23.i34.6330 |
| [40] | Chen Z, Xu W, Zhang D, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression[J]. Mol Ther Nucleic Acids, 2021, 23: 1120-35. doi:10.1016/j.omtn.2020.12.013 |
| [41] | Wang Y, Li X, Wang H, et al. CircCAMSAP1 promotes non-small cell lung cancer proliferation and inhibits cell apoptosis by sponging miR-1182 and regulating BIRC5[J]. Bioengineered, 2022, 13(2): 2428-39. doi:10.1080/21655979.2021.2011639 |
| [42] | Luo Z, Lu L, Tang Q, et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway[J]. J Cell Mol Med, 2021, 25(8): 3793-802. doi:10.1111/jcmm.16254 |
| [43] | Zhou C, Liu HS, Wang FW, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis[J]. Mol Ther, 2020, 28(3): 914-28. doi:10.1016/j.ymthe.2019.12.008 |
| [44] | Wang Y, Yan Q, Mo Y, et al. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumori-genesis via a SERPINH1/c-Myc positive feedback loop[J]. Mol Cancer, 2022, 21(1): 62. doi:10.1186/s12943-022-01502-2 |
| [1] | Siyuan MA, Bochao ZHANG, Chun PU. Circ_0000437 promotes proliferation, invasion, migration and epithelial-mesenchymal transition of breast cancer cells by targeting the let-7b-5p/CTPS1 axis [J]. Journal of Southern Medical University, 2025, 45(8): 1682-1696. |
| [2] | Rui CAI, Zhuo HUANG, Wenxia HE, Tianhong AI, Xiaowei SONG, Shuting HU. The splicing factor HNRNPH1 regulates Circ-MYOCD back-splicing to modulate the course of cardiac hypertrophy [J]. Journal of Southern Medical University, 2025, 45(3): 587-594. |
| [3] | GAO Zhiqiang, LIN Jie, HONG Peng, HU Zaihong, DONG Junjun, SHI Qinlin, TIAN Xiaomao, LIU Feng, WEI Guanghui. Identification of key genes in Wilms tumor based on high- throughput RNA sequencing and their impacts on prognosis and immune responses [J]. Journal of Southern Medical University, 2024, 44(4): 727-738. |
| [4] | ZHANG Ling, ZHAO Chunyu, XU Yaoyao, CHEN Yansen, CAI Zhixiong, LIN Haowei, CAI Qiaoyan. Circular RNA hsa_circ_0006834 is a potential prognostic biomarker for hepatocellular carcinoma [J]. Journal of Southern Medical University, 2023, 43(11): 1850-1856. |
| [5] | ZHOU Peitao, CHENG Binglin, SUN Yining, WU Dehua, CHEN Yuhan. Circular RNA circRSF1 binds to HuR to promote radiation-induced inflammatory phenotype in hepatic stellate cells [J]. Journal of Southern Medical University, 2023, 43(1): 46-51. |
| [6] | ZUO Xueliang, CAI Juan, CHEN Zhiqiang, LI Yanna, ZHANG Doufeng. CircPCSK5 is highly expressed in gastric cancer and promotes cancer cell proliferation, invasion and epithelial-mesenchymal transition [J]. Journal of Southern Medical University, 2022, 42(10): 1440-1451. |
| [7] | . Effect of piRNA NU13 in regulating biological behaviors of human Wilms tumor cells in vitro [J]. Journal of Southern Medical University, 2021, 41(2): 184-192. |
| [8] | . MicroRNA-200c-3p inhibits proliferation of nephroblastoma cells by targeting CCNE2 [J]. Journal of Southern Medical University, 2020, 40(09): 1246-1252. |
| [9] | . Expression of miR-155-5p in Wilms tumor and its regulatory role in proliferation, migration and apoptosis of Wilms tumor cells in vitro [J]. Journal of Southern Medical University, 2019, 39(12): 1476-1481. |
| [10] | . CircRNA_005647 inhibits expressions of fibrosis-related genes in mouse cardiac fibroblasts via sponging miR-27b-3p [J]. Journal of Southern Medical University, 2019, 39(11): 1312-1319. |
| [11] | . Role of miRNA-340 in modulating gastric cancer cell proliferation and bioinformatic analysis [J]. Journal of Southern Medical University, 2019, 39(07): 784-. |
| [12] |
.
Establishment of a colorectal cancer SW620 cell line stably over-expressing Wilm’s tumor on X chromosome using a recombinant lentivirus vector [J]. Journal of Southern Medical University, 2015, 35(08): 1122-. |
| [13] | ZHAO Yong-bin, ZHENG Shao-bin, MAO Xiang-ming, ZHOU Hai-kuan, ZHANG Peng. Clinical effects of preoperative chemotherapy for Wilms tumor [J]. Journal of Southern Medical University, 2004, 24(06): 722-724. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||