Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (10): 2118-2125.doi: 10.12122/j.issn.1673-4254.2025.10.08
Yuan MI1( ), Xuzhe LI2, Zhanpeng WANG3, Yanjie LIU4, Chuntao SONG1, Lantao WANG1, Lei WANG4(
), Xuzhe LI2, Zhanpeng WANG3, Yanjie LIU4, Chuntao SONG1, Lantao WANG1, Lei WANG4( )
)
Received:2025-05-12
Online:2025-10-20
Published:2025-10-24
Contact:
Lei WANG
E-mail:miyuan_medicine@163.com;wangleidoctor@ hebmu.edu.cn
Yuan MI, Xuzhe LI, Zhanpeng WANG, Yanjie LIU, Chuntao SONG, Lantao WANG, Lei WANG. LINC00261 suppresses esophageal squamous cell carcinoma proliferation, invasion, and metastasis by targeting the miR-23a-3p/ZNF292 axis[J]. Journal of Southern Medical University, 2025, 45(10): 2118-2125.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.10.08
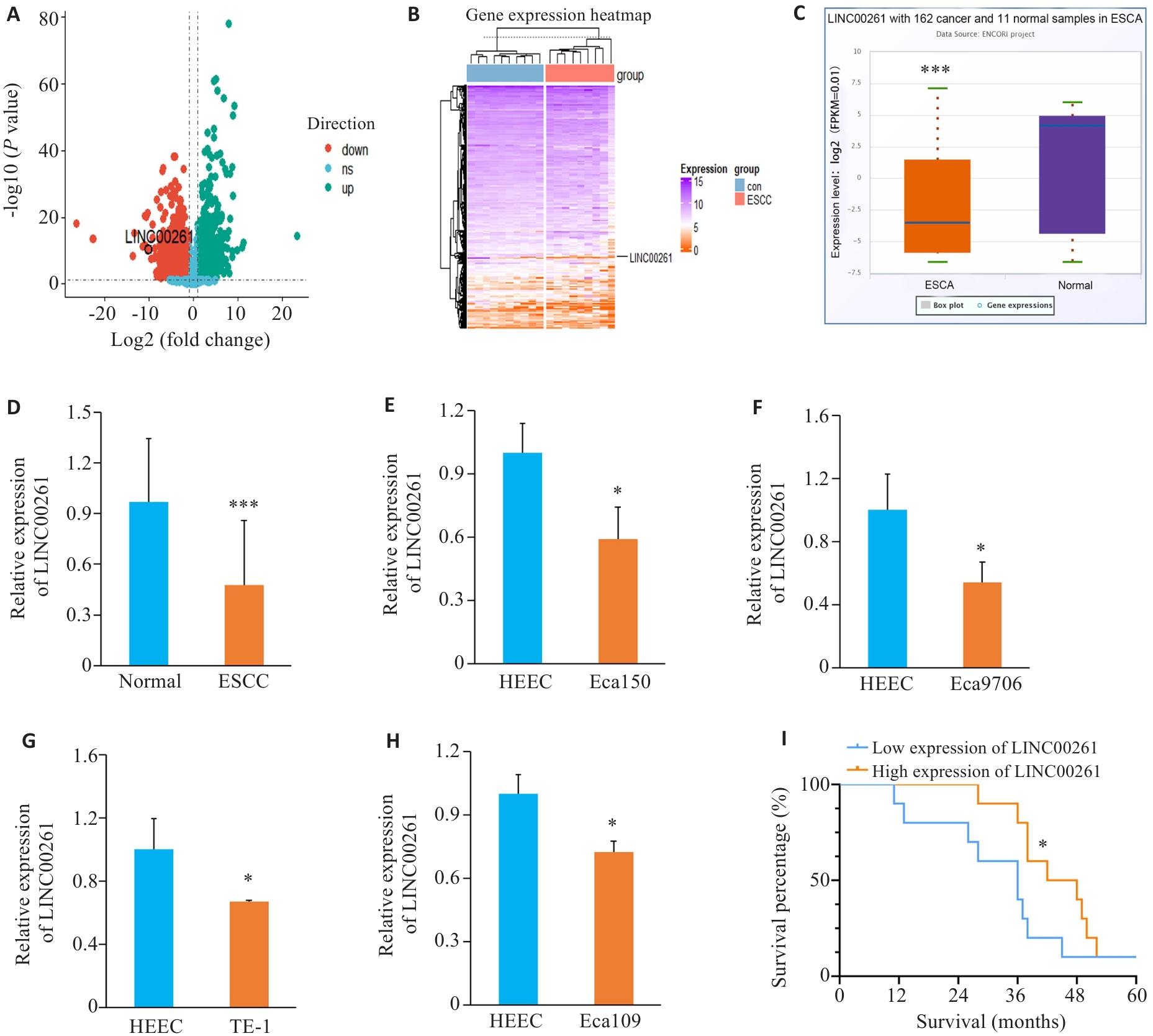
Fig.1 LINC00261 is lowly expressed in esophageal squamous cell carcinoma (ESCC) tissues and cells. A: Changes of each gene in the GSE149612 dataset. B: Expression of each gene in each tissue in the GSE149612 dataset. C: Expression of LINC00261 in ENCORI. D: Expression changes of LINC00261 in clinical ESCC and normal esophageal tissues. E-H: Expression of LINC00261 in ESCC cells and normal esophageal cells. I: Survival analysis of ESCC patients with high-level or low-level LINC00261 expression. *P<0.05 vs HEEC, ***P<0.001 vs Normal.

Fig.2 Overexpression of LINC00261 inhibits proliferation, invasion and metastasis of ESCC cells and promotes cell apoptosis. A: qRT-PCR for detecting expression changes of LINC00261 after overexpression. B: CCK-8 assay fir assessing changes in cell proliferation ability. C: Clone formation assay for assessing cell proliferation ability. D: Transwell assay for assessing changes in cell invasion and metastasis abilities (×100). E: Detection of changes in cell apoptosis by flow cytometry. **P<0.01, ***P<0.001 vs pcDNA.
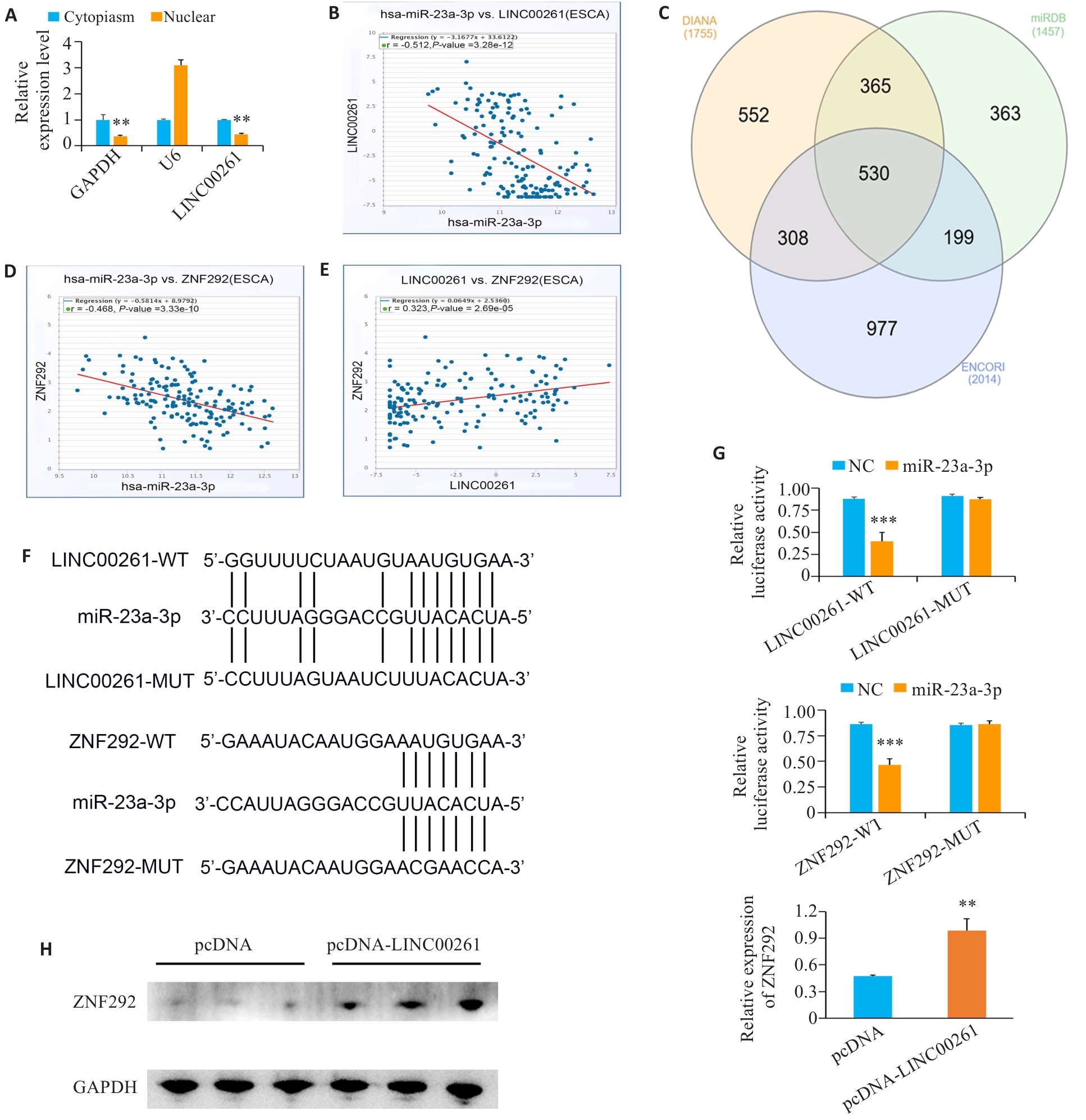
Fig.3 LINC00261 targets miR-23a-3p/ZNF292 in ESCC. A: Location of LINC00261 in ESCC cells. B: Correlation analysis of LINC00261 and miR-23a-3p in the ENCORI database. C: Prediction of the targeted genes of miR-23a-3p by ENCORI, DIANA and miRDB databases. D: Correlation analysis of miR-23a-3p and ZNF292 in the ENCORI database. E: Correlation analysis of LINC00261 and ZNF292 in the ENCORI database. F: Bioinformatics websites predict the potential binding sequences of LINC00261 with miR-23a-3p and miR-23a-3p with ZNF292. G: Binding of LINC00261 with miR-23a-3p, and miR-23a-3p and ZNF292 verified by luciferase reporter gene assay. H: Changes in the expression of ZNF292 after overexpression of LINC00261. **P<0.01, ***P<0.001 vs pcDNA.
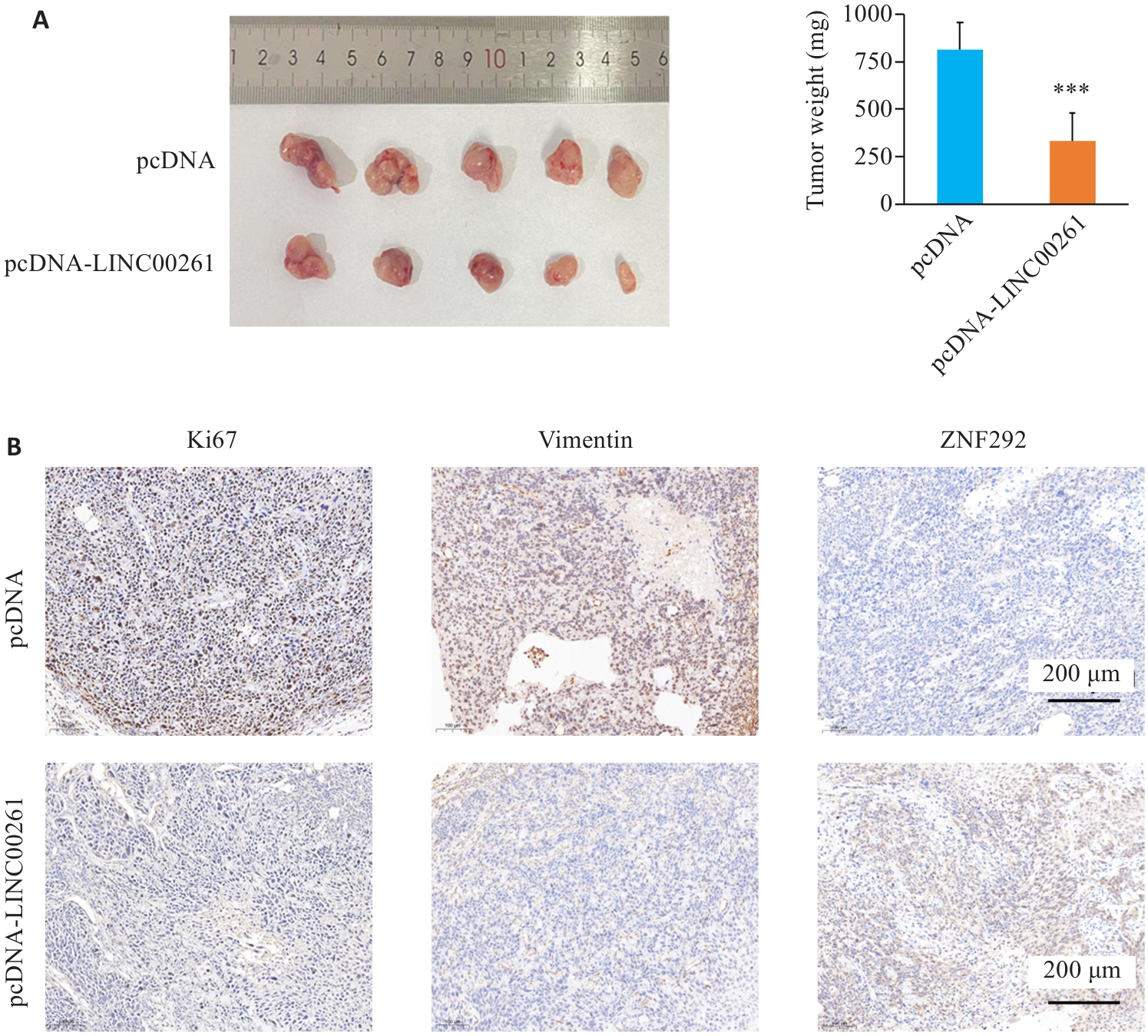
Fig.4 LINC00261 affects proliferation and metastasis of ESCC xenografts in nude mice. A: Weight changes of subcutaneous transplanted tumors in each group. B: Immunohistochemistry for Ki67, Vimentin and ZNF292 in the tumor tissues in each group. *** P<0.001 vs pcDNA.
| [1] | Zhao DY, Guo YP, Wei HF, et al. Multi-omics characterization of esophageal squamous cell carcinoma identifies molecular subtypes and therapeutic targets[J]. JCI Insight, 2024, 9(10): e171916. doi:10.1172/jci.insight.171916 |
| [2] | Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-49. doi:10.3322/caac.21660 |
| [3] | Guo HY, Shu JH, Hu GB, et al. Downregulation of RCN1 inhibits esophageal squamous cell carcinoma progression and M2 macrophage polarization[J]. PLoS One, 2024, 19(5): e0302780. doi:10.1371/journal.pone.0302780 |
| [4] | Wang LJ, Wang XJ, Yan PW, et al. LINC00261 suppresses cisplatin resistance of esophageal squamous cell carcinoma through miR-545-3p/MT1M axis[J]. Front Cell Dev Biol, 2021, 9: 687788. doi:10.3389/fcell.2021.687788 |
| [5] | Liu XJ, Zhao SM, Wang KK, et al. Spatial transcriptomics analysis of esophageal squamous precancerous lesions and their progression to esophageal cancer[J]. Nat Commun, 2023, 14(1): 4779. doi:10.1038/s41467-023-40343-5 |
| [6] | Yang ZL, Tian H, Chen XW, et al. Single-cell sequencing reveals immune features of treatment response to neoadjuvant immunochemotherapy in esophageal squamous cell carcinoma[J]. Nat Commun, 2024, 15(1): 9097. doi:10.1038/s41467-024-52977-0 |
| [7] | Xia T, Meng L, Xu GX, et al. TRIM33 promotes glycolysis through regulating P53 K48-linked ubiquitination to promote esophageal squamous cell carcinoma growth[J]. Cell Death Dis, 2024, 15(10): 740. doi:10.1038/s41419-024-07137-z |
| [8] | Zhou LT, Yao N, Yang L, et al. DUSP4 promotes esophageal squamous cell carcinoma progression by dephosphorylating HSP90β[J]. Cell Rep, 2023, 42(5): 112445. doi:10.1016/j.celrep.2023.112445 |
| [9] | Chen DY, Chen TY, Guo YX, et al. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo[J]. Exp Cell Res, 2020, 396(1): 112281. doi:10.1016/j.yexcr.2020.112281 |
| [10] | Yan QH, Wong W, Gong L, et al. Roles of long non-coding RNAs in esophageal cell squamous carcinoma (Review)[J]. Int J Mol Med, 2024, 54(2): 72. doi:10.3892/ijmm.2024.5396 |
| [11] | Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm[J]. Cancer Res, 2017, 77(15): 3965-81. doi:10.1158/0008-5472.can-16-2634 |
| [12] | Smolarz B, Zadrożna-Nowak A, Romanowicz H. The role of lncRNA in the development of tumors, including breast cancer[J]. Int J Mol Sci, 2021, 22(16): 8427. doi:10.3390/ijms22168427 |
| [13] | Dai ZT, Wu YL, Xu T, et al. The role of lncRNA SNHG14 in gastric cancer: enhancing tumor cell proliferation and migration, and mechanisms of CDH2 expression[J]. Cell Cycle, 2023, 22(23/24): 2522-37. doi:10.1080/15384101.2023.2289745 |
| [14] | Mehmandar-Oskuie A, Jahankhani K, Rostamlou A, et al. Molecular landscape of LncRNAs in bladder cancer: from drug resistance to novel LncRNA-based therapeutic strategies[J]. Biomed Pharmacother, 2023, 165: 115242. doi:10.1016/j.biopha.2023.115242 |
| [15] | Liu JQ, Deng M, Xue NN, et al. lncRNA KLF3-AS1 suppresses cell migration and invasion in ESCC by impairing miR-185-5p-targeted KLF3 inhibition[J]. Mol Ther Nucleic Acids, 2020, 20: 231-41. doi:10.1016/j.omtn.2020.01.020 |
| [16] | Tao QY, Li XJ, Xia YY, et al. LINC00261 triggers DNA damage via the miR-23a-3p/CELF2 axis to mitigate the malignant characteristics of 131I-resistant papillary thyroid carcinoma cells[J]. Biochem Biophys Rep, 2024, 40: 101858. doi:10.1016/j.bbrep.2024.101858 |
| [17] | Li XY, Li JY, Lu P, et al. LINC00261 relieves the progression of sepsis-induced acute kidney injury by inhibiting NF-κB activation through targeting the miR-654-5p/SOCS3 axis[J]. J Bioenerg Biomembr, 2021, 53(2): 129-37. doi:10.1007/s10863-021-09874-8 |
| [18] | Fang QJ, Sang L, Du SH. Long noncoding RNA LINC00261 regulates endometrial carcinoma progression by modulating miRNA/FOXO1 expression[J]. Cell Biochem Funct, 2018, 36(6): 323-30. doi:10.1002/cbf.3352 |
| [19] | Li Y, Wu CH. LINC00261/microRNA-550a-3p/SDPR axis affects the biological characteristics of breast cancer stem cells[J]. IUBMB Life, 2021, 73(1): 188-201. doi:10.1002/iub.2416 |
| [20] | Zou J, Pei XZ, Xing D, et al. LINC00261 elevation inhibits angiogenesis and cell cycle progression of pancreatic cancer cells by upregulating SCP2 via targeting FOXP3[J]. J Cell Mol Med, 2021, 25(20): 9826-36. doi:10.1111/jcmm.16930 |
| [21] | Liao J, Dong LP. Linc00261 suppresses growth and metastasis of non-small cell lung cancer via repressing epithelial-mesenchymal transition[J]. Eur Rev Med Pharmacol Sci, 2019, 23(9): 3829-37. |
| [22] | Lin WY, Zhou QY, Wang CQ, et al. LncRNAs regulate metabolism in cancer[J]. Int J Biol Sci, 2020, 16(7): 1194-206. doi:10.7150/ijbs.40769 |
| [23] | Xue ST, Zheng B, Cao SQ, et al. Long non-coding RNA LINC00680 functions as a CeRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis[J]. Mol Cancer, 2022, 21(1): 69. doi:10.1186/s12943-022-01539-3 |
| [24] | Han G, Guo QY, Ma N, et al. Apatinib inhibits cell proliferation and migration of osteosarcoma via activating LINC00261/miR-620/PTEN axis[J]. Cell Cycle, 2021, 20(18): 1785-98. doi:10.1080/15384101.2021.1949132 |
| [25] | Yan DS, Liu WD, Liu YL, et al. LINC00261 suppresses human colon cancer progression via sponging miR-324-3p and inactivating the Wnt/β‑catenin pathway[J]. J Cell Physiol, 2019, 234(12): 22648-56. doi:10.1002/jcp.28831 |
| [26] | Zuo H, Liu S, Li X, et al. miR-23a-3p promotes the development of colon cancer by inhibiting the expression of NDRG4[J]. Clin Transl Oncol, 2023, 25(4): 933-40. doi:10.1007/s12094-022-02996-4 |
| [27] | Lu YJ, Chan YT, Tan HY, et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma[J]. J Exp Clin Cancer Res, 2022, 41(1): 3. doi:10.1186/s13046-021-02208-x |
| [28] | Ma Y, Gao JB, Guo HN. miR-23a-3p regulates Runx2 to inhibit the proliferation and metastasis of oral squamous cell carcinoma[J]. J Oncol, 2022, 2022: 8719542. doi:10.1155/2022/8719542 |
| [29] | Lee JH, Song SY, Kim MS, et al. Frameshift mutations of a tumor suppressor gene ZNF292 in gastric and colorectal cancers with high microsatellite instability[J]. APMIS, 2016, 124(7): 556-60. doi:10.1111/apm.12545 |
| [30] | Gong W, Xu J, Wang G, et al. ZNF292 suppresses proliferation of ESCC cells through ZNF292/SKP2/P27 signaling axis[J]. Chin J Cancer Res, 2021, 33(6): 637-48. doi:10.21147/j.issn.1000-9604.2021.06.01 |
| [31] | Wang X, Gao XZ, Tian JX, et al. LINC00261 inhibits progression of pancreatic cancer by down-regulating miR-23a-3p[J]. Arch Biochem Biophys, 2020, 689: 108469. doi:10.1016/j.abb.2020.108469 |
| [32] | Lin K, Jiang H, Zhuang SS, et al. Long noncoding RNA LINC00261 induces chemosensitization to 5-fluorouracil by mediating methylation-dependent repression of DPYD in human esophageal cancer[J]. FASEB J, 2019, 33(2): 1972-88. doi:10.1096/fj.201800759r |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||