Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (7): 1217-1226.doi: 10.12122/j.issn.1673-4254.2024.07.01
Received:2024-03-13
Accepted:2024-05-20
Online:2024-07-20
Published:2024-07-25
Contact:
Rong FU
E-mail:1015001931@qq.com;834460113@qq.com
Supported by:Wenran HU, Rong FU. Trans-YOLOv5: a YOLOv5-based prior transformer network model for automated detection of abnormal cells or clumps in cervical cytology images[J]. Journal of Southern Medical University, 2024, 44(7): 1217-1226.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.07.01
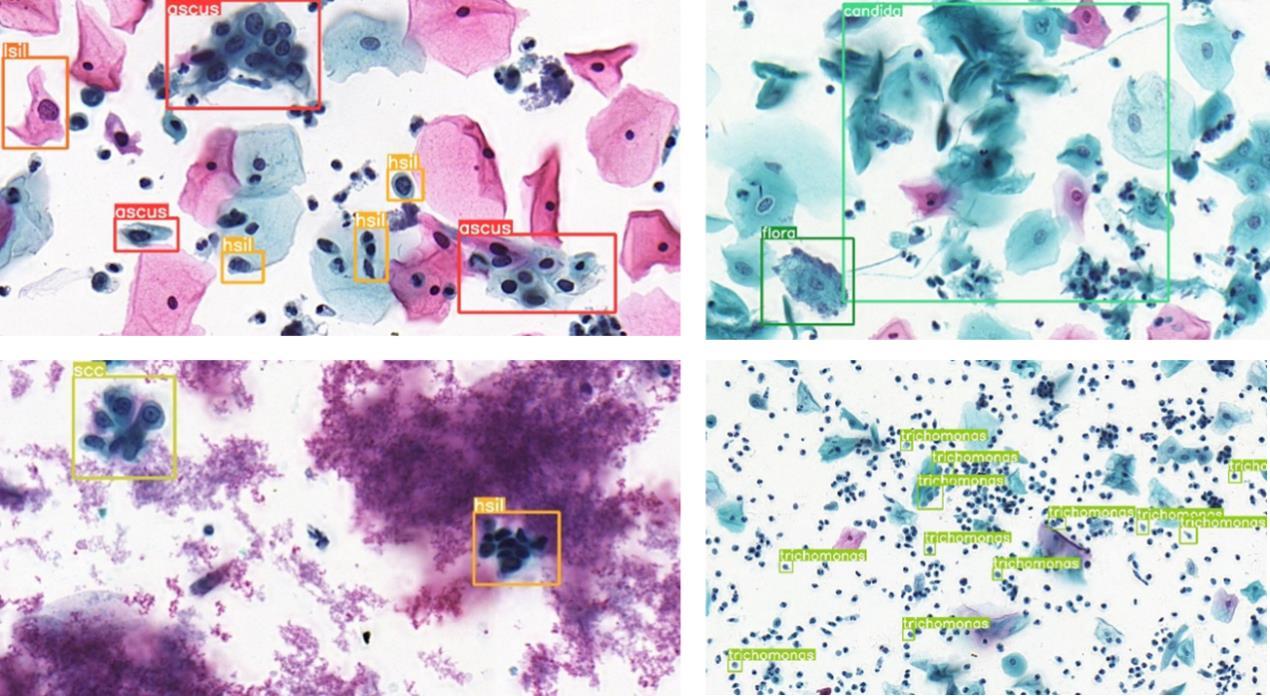
Fig.1 Representative cervical cytology images showing multiple cell types highlighted by annotation boxes from Liang's dataset[15]. The two images in the left column show abnormal cells in the categories of ascus, lsil, hsil and scc, and those in the right column show candida and trichomonas infections that cause the surrounding normal cells to have a similar appearance to atypical squamous cells.
| Dataset | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | Total | Images |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Training | 1614 | 3403 | 1220 | 23211 | 1759 | 4404 | 4413 | 288 | 104 | 235 | 119 | 40770 | 5922 |
| Validation | 166 | 470 | 196 | 2907 | 213 | 523 | 542 | 40 | 22 | 31 | 17 | 5127 | 744 |
| Test | 213 | 404 | 166 | 2803 | 228 | 662 | 477 | 26 | 24 | 36 | 18 | 5057 | 744 |
| Total | 1993 | 4277 | 1582 | 28921 | 2200 | 5589 | 5432 | 354 | 150 | 302 | 154 | 50954 | 7410 |
Tab.1 Number of cells of different pathological types in the training, validation and test datasets
| Dataset | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | Total | Images |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Training | 1614 | 3403 | 1220 | 23211 | 1759 | 4404 | 4413 | 288 | 104 | 235 | 119 | 40770 | 5922 |
| Validation | 166 | 470 | 196 | 2907 | 213 | 523 | 542 | 40 | 22 | 31 | 17 | 5127 | 744 |
| Test | 213 | 404 | 166 | 2803 | 228 | 662 | 477 | 26 | 24 | 36 | 18 | 5057 | 744 |
| Total | 1993 | 4277 | 1582 | 28921 | 2200 | 5589 | 5432 | 354 | 150 | 302 | 154 | 50954 | 7410 |
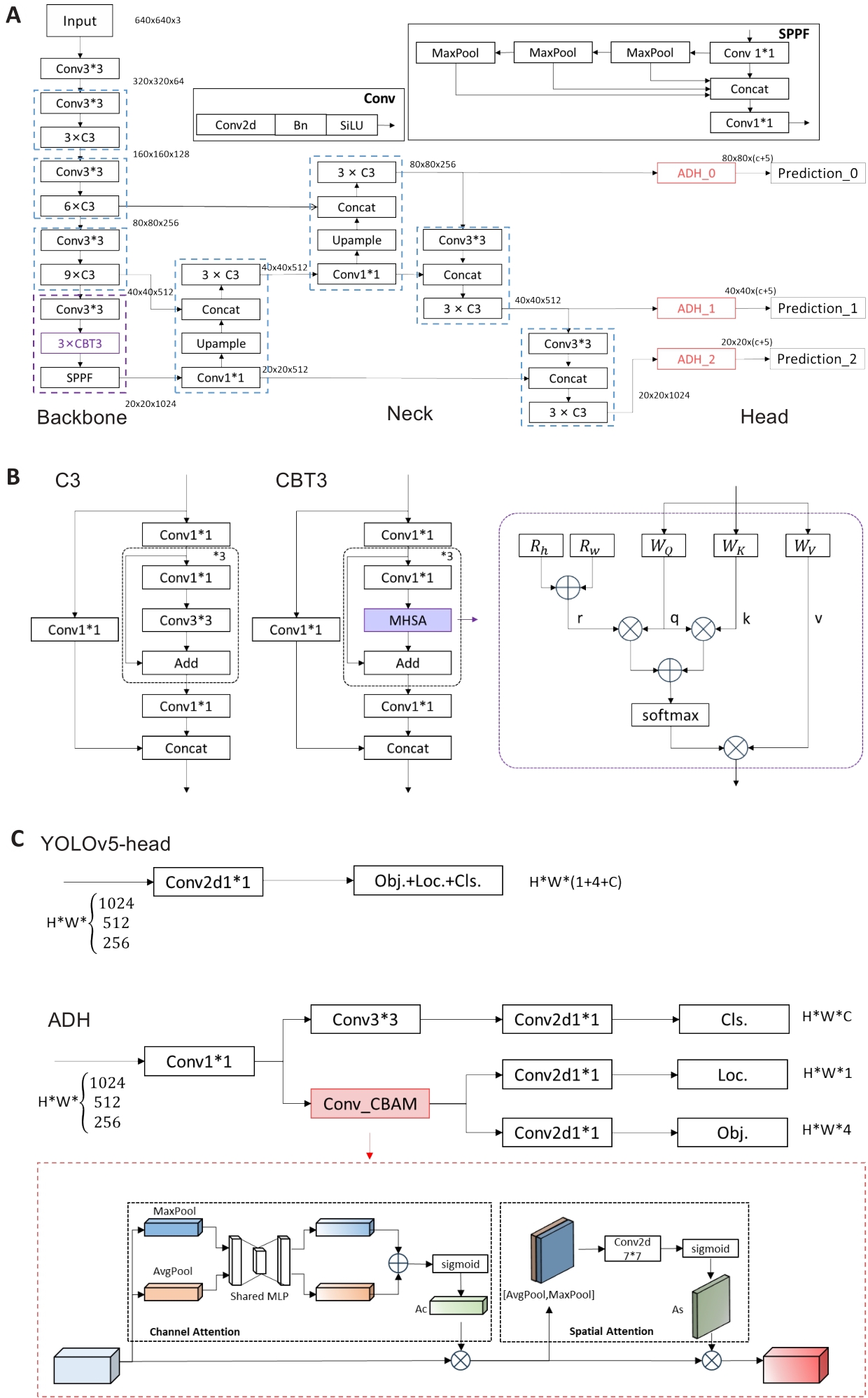
Fig.4 Architecture of the proposed Trans-YOLOv5 model. A: The details of the network structure. B: Structure of C3 and CBT3. The detailed structure of the MHSA is shown within the purple dotted box on the right side, where Rh, Rw denote the relative positional encoding of the height and the width, respectively, WQ, WK, WV are the weights of the query, the key, and the weights, respectively; + and × represent element-by-element summation and matrix multiplication, respectively. C: Structure of the original head of YOLOv5 and ADH. The detailed structure of the CBAM is shown in the red dotted box.
| Augmentation methods | Mosaic | Mixup | Rotation | flipUD | flipLR | H | S | V | Scale | Shear | Perspective |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | 0.5 | 0.5 | 5.0 | 0.5 | 0.5 | 0.5 | 0.7 | 0.4 | 0.3 | 5.0 | 0.2 |
Tab.2 Data augmentation methods and their parameters
| Augmentation methods | Mosaic | Mixup | Rotation | flipUD | flipLR | H | S | V | Scale | Shear | Perspective |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P | 0.5 | 0.5 | 5.0 | 0.5 | 0.5 | 0.5 | 0.7 | 0.4 | 0.3 | 5.0 | 0.2 |
| Models | Model type | mAP(%) | AR(%) | Params(M) | GFLOPs | FPS(img/s) |
|---|---|---|---|---|---|---|
| Faster RCNN [ | Two-stage | 57.6 | 48.1 | 41.5 | 215.8 | 18.6 |
| Cascade R-CNN[ | Two-stage | 58.1 | 47.8 | 69.2 | 243.5 | 15.9 |
| RetinaNet[ | One-stage | 46.2 | 49.1 | 37.7 | 250.3 | 16.3 |
| YOLOv3 | One-stage | 28.8 | 26.6 | 40.6 | 110.7 | 7.4 |
| YOLOv5l | One-stage | 62.6 | 52.2 | 46.2 | 108.4 | 46.9 |
| YOLOv8l | One-stage | 60.7 | 43.6 | 43.6 | 165.4 | 78.7 |
| AttFPN[ | Two-stage | 37.1 | 40.4 | 41.2 | 192.4 | 14.6 |
| Comparison Detector [ | Two-stage | 45.9 | 63.5 | - | - | - |
| YOLOv3+SSAM[ | One-stage | 33.2 | 31.9 | 61.6 | 132.8 | 6.2 |
| CR4CACD[ | Two-stage | 60.5 | 45.2 | 43.1 | 156.9 | 11.5 |
| Trans-YOLOv5 | One-stage | 64.8 | 53.3 | 49.8 | 125.3 | 29.1 |
Tab.3 Evaluation metrics of the proposed method and other state-of-the-art models for cell detection in cervical cell images
| Models | Model type | mAP(%) | AR(%) | Params(M) | GFLOPs | FPS(img/s) |
|---|---|---|---|---|---|---|
| Faster RCNN [ | Two-stage | 57.6 | 48.1 | 41.5 | 215.8 | 18.6 |
| Cascade R-CNN[ | Two-stage | 58.1 | 47.8 | 69.2 | 243.5 | 15.9 |
| RetinaNet[ | One-stage | 46.2 | 49.1 | 37.7 | 250.3 | 16.3 |
| YOLOv3 | One-stage | 28.8 | 26.6 | 40.6 | 110.7 | 7.4 |
| YOLOv5l | One-stage | 62.6 | 52.2 | 46.2 | 108.4 | 46.9 |
| YOLOv8l | One-stage | 60.7 | 43.6 | 43.6 | 165.4 | 78.7 |
| AttFPN[ | Two-stage | 37.1 | 40.4 | 41.2 | 192.4 | 14.6 |
| Comparison Detector [ | Two-stage | 45.9 | 63.5 | - | - | - |
| YOLOv3+SSAM[ | One-stage | 33.2 | 31.9 | 61.6 | 132.8 | 6.2 |
| CR4CACD[ | Two-stage | 60.5 | 45.2 | 43.1 | 156.9 | 11.5 |
| Trans-YOLOv5 | One-stage | 64.8 | 53.3 | 49.8 | 125.3 | 29.1 |
| Model | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP@50 | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Faster RCNN [ | 42.9 | 21.1 | 58.9 | 52.7 | 28.8 | 66.6 | 65.3 | 68.3 | 74.4 | 80.7 | 65.3 | 57.6 | 48.1 |
| Cascade R-CNN [ | 45.7 | 19.6 | 58.5 | 51.2 | 29.0 | 67.3 | 63.6 | 71.7 | 82.6 | 78.4 | 71.9 | 58.1 | 47.8 |
| RetinaNett [ | 36.0 | 14.2 | 57.3 | 47.3 | 3.0 | 60.4 | 61.1 | 68.7 | 22.6 | 74.5 | 62.7 | 46.2 | 49.1 |
| YOLOv3 [ | 32.5 | 2.8 | 39.8 | 42.8 | 5.8 | 59.7 | 36.2 | 26.4 | 0.0 | 46.3 | 25.0 | 28.9 | 30.2 |
| YOLOv5l | 49.5 | 24.6 | 56.4 | 53.2 | 33.1 | 70.6 | 67.9 | 82.8 | 86.9 | 86.5 | 77.6 | 62.6 | 47.6 |
| YOLOv8l | 49.9 | 28.3 | 64.5 | 56.7 | 38.4 | 70.9 | 63.6 | 81.3 | 73.5 | 74.3 | 66.6 | 60.7 | 51.8 |
| AttFPN[ | 26.5 | 19.0 | 32.1 | 45.8 | 26.2 | 55.9 | 53.1 | 32.7 | 25.7 | 55.8 | 37.1 | 37.3 | 35.8 |
| Comparison Detector [ | 27.4 | 6.7 | 41.7 | 40.1 | 21.8 | 54.5 | 45.0 | 65.5 | 63.5 | 68.1 | 70.5 | 45.9 | 33.0 |
| YOLOv3+SSAM[ | 28.6 | 10.7 | 35.6 | 39.2 | 17.6 | 52.4 | 47.3 | 19.9 | 28.2 | 56.8 | 35.3 | 33.8 | 31.1 |
| CR4CACD [ | 54.3 | 27.1 | 53.1 | 56.5 | 36.8 | 71.1 | 68.8 | 65.5 | 86.4 | 84.1 | 62.1 | 60.5 | 48.9 |
| Trans-YOLOv5 | 55.5 | 32.5 | 64.1 | 60.1 | 36.5 | 74.5 | 67.3 | 87.9 | 78.3 | 88.1 | 79.8 | 65.9 | 53.5 |
Tab.4 Comparison of the ability of the models for recognizing cervical cells in each category
| Model | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP@50 | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Faster RCNN [ | 42.9 | 21.1 | 58.9 | 52.7 | 28.8 | 66.6 | 65.3 | 68.3 | 74.4 | 80.7 | 65.3 | 57.6 | 48.1 |
| Cascade R-CNN [ | 45.7 | 19.6 | 58.5 | 51.2 | 29.0 | 67.3 | 63.6 | 71.7 | 82.6 | 78.4 | 71.9 | 58.1 | 47.8 |
| RetinaNett [ | 36.0 | 14.2 | 57.3 | 47.3 | 3.0 | 60.4 | 61.1 | 68.7 | 22.6 | 74.5 | 62.7 | 46.2 | 49.1 |
| YOLOv3 [ | 32.5 | 2.8 | 39.8 | 42.8 | 5.8 | 59.7 | 36.2 | 26.4 | 0.0 | 46.3 | 25.0 | 28.9 | 30.2 |
| YOLOv5l | 49.5 | 24.6 | 56.4 | 53.2 | 33.1 | 70.6 | 67.9 | 82.8 | 86.9 | 86.5 | 77.6 | 62.6 | 47.6 |
| YOLOv8l | 49.9 | 28.3 | 64.5 | 56.7 | 38.4 | 70.9 | 63.6 | 81.3 | 73.5 | 74.3 | 66.6 | 60.7 | 51.8 |
| AttFPN[ | 26.5 | 19.0 | 32.1 | 45.8 | 26.2 | 55.9 | 53.1 | 32.7 | 25.7 | 55.8 | 37.1 | 37.3 | 35.8 |
| Comparison Detector [ | 27.4 | 6.7 | 41.7 | 40.1 | 21.8 | 54.5 | 45.0 | 65.5 | 63.5 | 68.1 | 70.5 | 45.9 | 33.0 |
| YOLOv3+SSAM[ | 28.6 | 10.7 | 35.6 | 39.2 | 17.6 | 52.4 | 47.3 | 19.9 | 28.2 | 56.8 | 35.3 | 33.8 | 31.1 |
| CR4CACD [ | 54.3 | 27.1 | 53.1 | 56.5 | 36.8 | 71.1 | 68.8 | 65.5 | 86.4 | 84.1 | 62.1 | 60.5 | 48.9 |
| Trans-YOLOv5 | 55.5 | 32.5 | 64.1 | 60.1 | 36.5 | 74.5 | 67.3 | 87.9 | 78.3 | 88.1 | 79.8 | 65.9 | 53.5 |
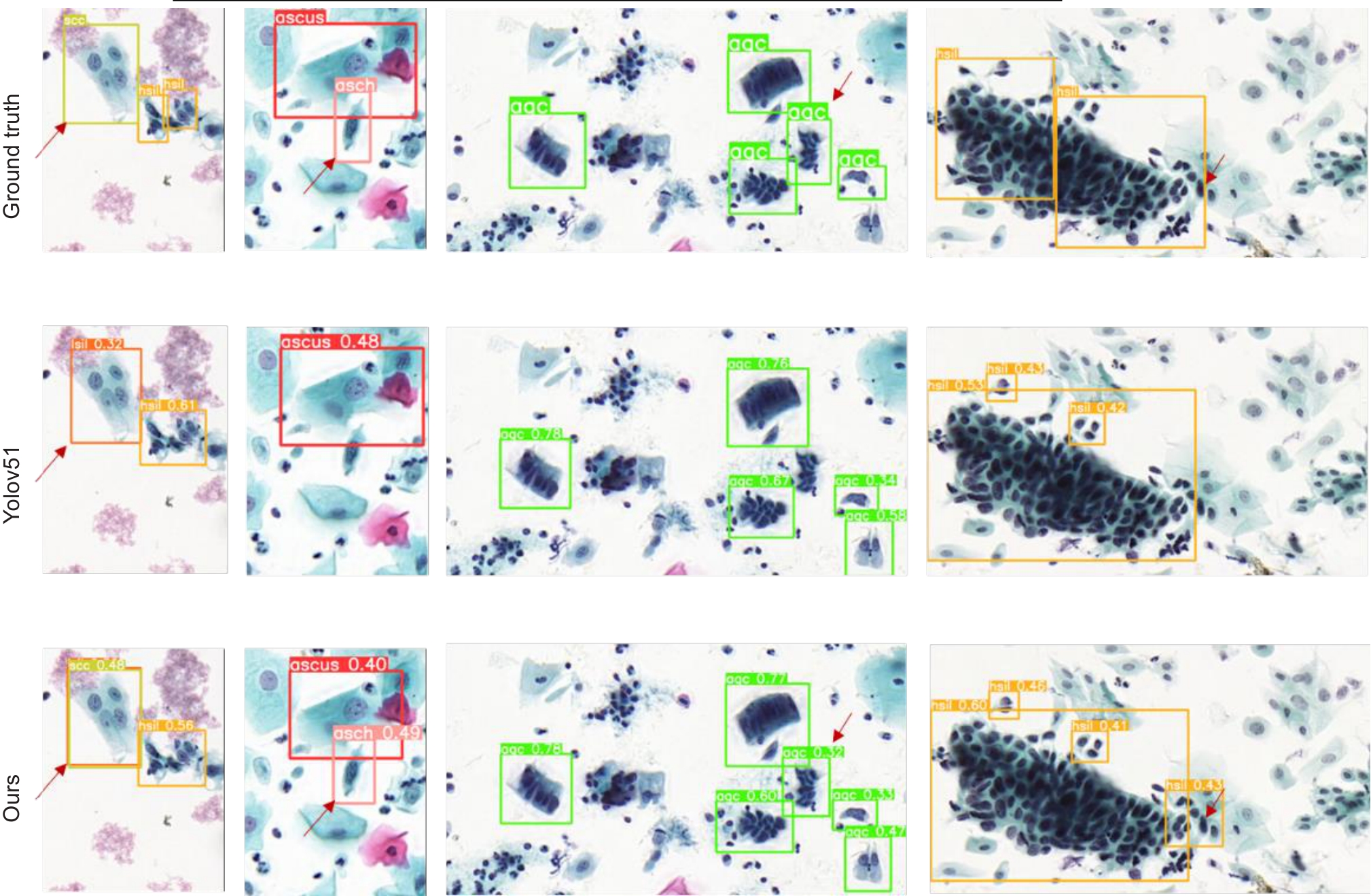
Fig.5 Comparison of the yolov5l model and Trans-YOLOv5 model for abnormal cell detection in cervical cytology images. Arrows highlight the areas where misdiagnosis or omission occurs with the original YOLOv5 model. The images in the first column show that the baseline model misdiagnosed scc as lsil, which is correctly identified by the proposed Trans-YOLOv5 model. The images in the second, third and fourth columns show that the Trans-YOLOv5 model model correctly diagnosed the abnormal cells that were missed by the baseline model (The images are cropped for convenience of presentation).
| Methods | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv5l | 47.0% | 25.0% | 59.2% | 53.6% | 43.4% | 65.8% | 48.6% | 50.2% | 79.1% | 78.7% | 61.1% | 55.6% | 49.4% |
| +AUG | 45.9% | 24.6% | 56.4% | 53.2% | 33.1% | 70.6% | 67.9% | 82.8% | 86.9% | 86.5% | 77.6% | 62.6% | 47.6% |
| +LS | 54.4% | 32.7% | 64.6% | 59.5% | 33.2% | 73.3% | 66.5% | 84.1% | 71.8% | 85.7% | 75.9% | 63.8% | 52.7% |
| +CBT3 | 52.8% | 32.9% | 60.5% | 60.0% | 37.2% | 73.7% | 68.7% | 87.0% | 79.8% | 90.4% | 81.2% | 65.8% | 52.9% |
| +ADH | 55.5% | 32.5% | 64.1% | 60.1% | 36.5% | 74.5% | 67.3% | 87.9% | 78.3% | 88.1% | 79.8% | 65.9% | 53.5% |
Tab.5 Ablation experiments of the YOLOv5L model after incorporation of data augmentation (AUG), label smoothing (LS), CBT3 and ADH modules
| Methods | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv5l | 47.0% | 25.0% | 59.2% | 53.6% | 43.4% | 65.8% | 48.6% | 50.2% | 79.1% | 78.7% | 61.1% | 55.6% | 49.4% |
| +AUG | 45.9% | 24.6% | 56.4% | 53.2% | 33.1% | 70.6% | 67.9% | 82.8% | 86.9% | 86.5% | 77.6% | 62.6% | 47.6% |
| +LS | 54.4% | 32.7% | 64.6% | 59.5% | 33.2% | 73.3% | 66.5% | 84.1% | 71.8% | 85.7% | 75.9% | 63.8% | 52.7% |
| +CBT3 | 52.8% | 32.9% | 60.5% | 60.0% | 37.2% | 73.7% | 68.7% | 87.0% | 79.8% | 90.4% | 81.2% | 65.8% | 52.9% |
| +ADH | 55.5% | 32.5% | 64.1% | 60.1% | 36.5% | 74.5% | 67.3% | 87.9% | 78.3% | 88.1% | 79.8% | 65.9% | 53.5% |
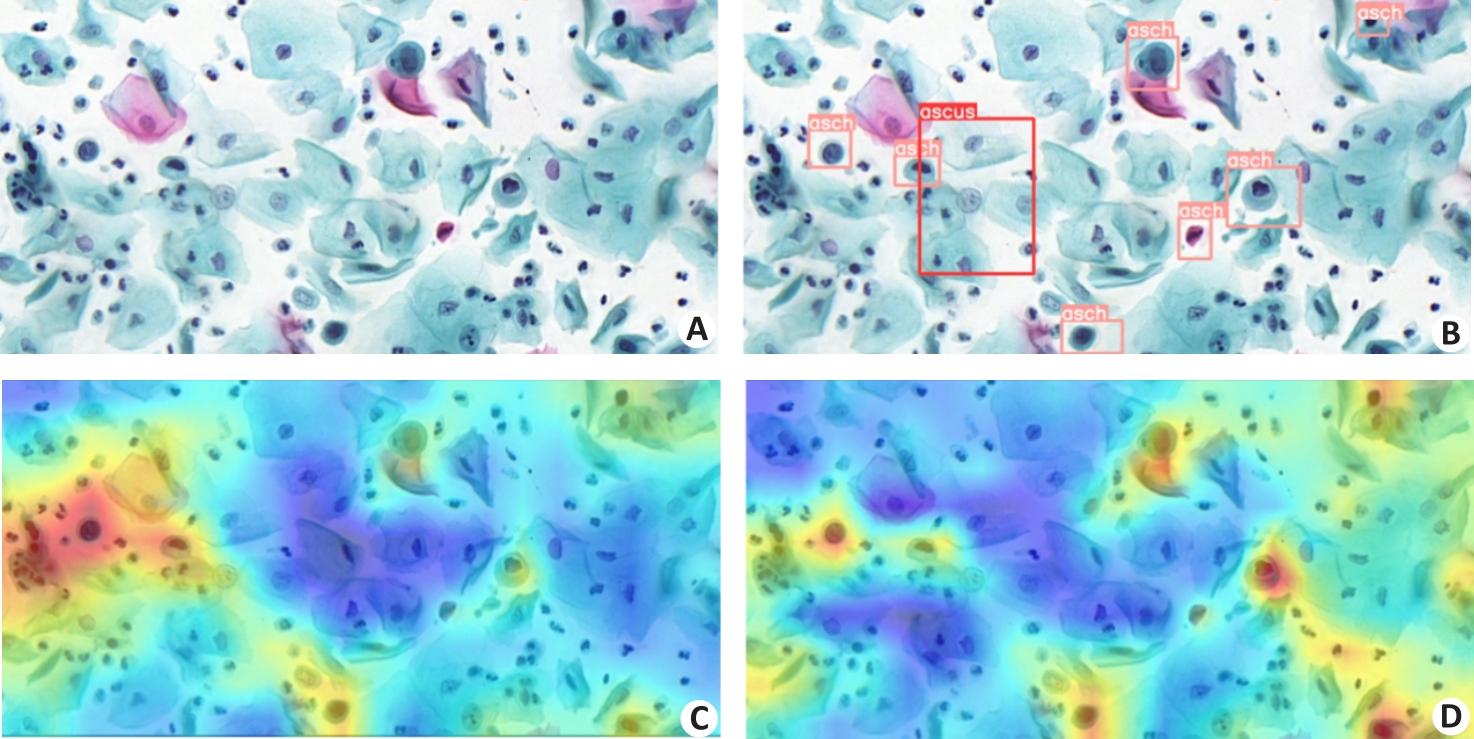
Fig.6 Visualization of model feature maps before and after addition of CBT3 to YOLOv5. A: Original image. B: Annotated image with category labels. C: Feature map of the original YOLOv5. D: Feature map of YOLOv5 after adding CBT3.
| Head | params | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv5l | 0.08 M | 54.4% | 32.7% | 64.6% | 59.5% | 33.2% | 73.3% | 66.5% | 84.1% | 71.8% | 85.7% | 75.9% | 63.8% | 52.7% |
| YOLOX[ | 7.58 M | 48.4% | 29.1% | 57.9% | 42.6% | 28.8% | 71.3% | 52.7% | 67.1% | 81.4% | 74.8% | 0.003% | 50.4% | 45.9% |
| TSCODE[ | 71.83 M | 48.6% | 32.1% | 62.4% | 57.6% | 35.1% | 73.1% | 36.0% | 71.7% | 77.2% | 88.8% | 75.6% | 62.4% | 52.1% |
| Efficient head | 4.04 M | 52.4% | 35.0% | 60.5% | 58.7% | 36.8% | 74.5% | 64.9% | 66.9% | 75.5% | 87.6% | 73.5% | 62.4% | 53.1% |
| ADH | 4.06 M | 51.8% | 31.6% | 63.4% | 58.6% | 34.8% | 73.7% | 68.2% | 89.1% | 77.5% | 84.3% | 77.8% | 64.6% | 52.4% |
Tab.6 Performance comparison of the state-of-the-art decoupling heads
| Head | params | ascus | asch | lsil | hsil | scc | agc | trich | cand | flora | herps | actin | mAP | mAP(*) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv5l | 0.08 M | 54.4% | 32.7% | 64.6% | 59.5% | 33.2% | 73.3% | 66.5% | 84.1% | 71.8% | 85.7% | 75.9% | 63.8% | 52.7% |
| YOLOX[ | 7.58 M | 48.4% | 29.1% | 57.9% | 42.6% | 28.8% | 71.3% | 52.7% | 67.1% | 81.4% | 74.8% | 0.003% | 50.4% | 45.9% |
| TSCODE[ | 71.83 M | 48.6% | 32.1% | 62.4% | 57.6% | 35.1% | 73.1% | 36.0% | 71.7% | 77.2% | 88.8% | 75.6% | 62.4% | 52.1% |
| Efficient head | 4.04 M | 52.4% | 35.0% | 60.5% | 58.7% | 36.8% | 74.5% | 64.9% | 66.9% | 75.5% | 87.6% | 73.5% | 62.4% | 53.1% |
| ADH | 4.06 M | 51.8% | 31.6% | 63.4% | 58.6% | 34.8% | 73.7% | 68.2% | 89.1% | 77.5% | 84.3% | 77.8% | 64.6% | 52.4% |
| 1 | Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-63. DOI: 10.3322/caac.21834 |
| 2 | Solomon D, Davey D, Kurman R, et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology [J]. JAMA, 2002, 287(16): 2114-9. |
| 3 | William W, Ware A, Basaza-Ejiri AH, et al. A pap-smear analysis tool (PAT) for detection of cervical cancer from pap-smear images [J]. Biomedical engineering online, 2019, 18: 1-22. DOI: 10.1186/s12938-019-0634-5 |
| 4 | 王嘉旭, 薛 鹏, 江 宇, 等. 人工智能在宫颈癌筛查中的应用研究进展[J]. 中国肿瘤临床, 2021, 48(09): 468-71. DOI: 10.3969/j.issn.1000-8179.2021.09.056 |
| 5 | Nayar R, Wilbur DC. The Bethesda system for reporting cervical cytology: definitions, criteria, and explanatory notes [M]. Springer, 2015. 103-285. DOI: 10.1007/978-3-319-11074-5 |
| 6 | Chankong T, Theera-Umpon N, Auephanwiriyakul S. Automatic cervical cell segmentation and classification in Pap smears [J]. Methods Programs Biomed, 2014, 113(2): 539-56. DOI: 10.1016/j.cmpb.2013.12.012 |
| 7 | Bora K, Chowdhury M, Mahanta L B, et al. Automated classification of Pap smear images to detect cervical dysplasia [J]. Comput Methods Programs Biomed, 2017, 138: 31-47. DOI: 10.1016/j.cmpb.2016.10.001 |
| 8 | Arya M, Mittal N, Singh G. Texture-based feature extraction of smear images for the detection of cervical cancer [J]. IET Comput Vision, 2018, 12(8): 1049-59. DOI: 10.1049/iet-cvi.2018.5349 |
| 9 | Wang P, Wang L, Li Y, et al. Automatic cell nuclei segmentation and classification of cervical Pap smear images [J]. Biomed Signal Process Control, 2019, 48: 93-103. DOI: 10.1016/j.bspc.2018.09.008 |
| 10 | Zhang L, Lu L, Nogues I, et al. DeepPap: deep convolutional networks for cervical cell classification [J]. IEEE J Biomed Health Inform, 2017, 21(6): 1633-43. DOI: 10.1109/jbhi.2017.2705583 |
| 11 | Taha B, Dias J, Werghi N. Classification of cervical-cancer using pap-smear images: a convolutional neural network approach [C]. Medical Image Understanding and Analysis: 21st Annual Conference, MIUA 2017, Edinburgh, UK, July 11-13, 2017, Proceedings 21. Springer International Publishing, 2017: 261-72. DOI: 10.1007/978-3-319-60964-5_23 |
| 12 | Wieslander H, Forslid G, Bengtsson E, et al. Deep convolutional neural networks for detecting cellular changes due to malignancy[C]. Proceedings of the IEEE International Conference on Computer Vision Workshops. 2017: 82-9. DOI: 10.1109/iccvw.2017.18 |
| 13 | Jiang P, Li X, Shen H, et al. A systematic review of deep learning-based cervical cytology screening: from cell identification to whole slide image analysis[J]. Artif Intell Rev, 2023, 56(): 2687-758. DOI: 10.1007/s10462-023-10588-z |
| 14 | Ren S, He K, Girshick R, et al. Faster R-CNN: Towards real-time object detection with region proposal networks[J]. IEEE Trans Pattern Anal Mach Intell, 2016, 39(6): 1137-49. DOI: 10.1109/tpami.2016.2577031 |
| 15 | Liang Y, Tang Z, Yan M, et al. Comparison detector for cervical cell/clumps detection in the limited data scenario[J]. Neurocomputing, 2021, 437: 195-205. DOI: 10.1016/j.neucom.2021.01.006 |
| 16 | Cao L, Yang J, Rong Z, et al. A novel attention-guided convolutional network for the detection of abnormal cervical cells in cervical cancer screening[J]. Med Image Anal, 2021, 73: 102197. DOI: 10.1016/j.media.2021.102197 |
| 17 | Chen T, Zheng W, Ying H, et al. A task decomposing and cell comparing method for cervical lesion cell detection[J]. IEEE Trans Med Imaging, 2022, 41(9): 2432-42. DOI: 10.1109/tmi.2022.3163171 |
| 18 | Liang Y, Feng S, Liu Q, et al. Exploring contextual relationships for cervical abnormal cell detection[J]. IEEE J Biomed Health Inform, 2023, 27(8): 4086-97. DOI: 10.1109/jbhi.2023.3276919 |
| 19 | Cai Z, Vasconcelos N. Cascade R-CNN: Delving into high quality object detection[C]//Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018: 6154-62. DOI: 10.1109/cvpr.2018.00644 |
| 20 | He K, Gkioxari G, Doll´ar P, et al. Mask R-CNN[C]. Proceedings of the IEEE International Conference on Computer Vision. 2017: 2961-9. DOI: 10.1109/iccv.2017.322 |
| 21 | Yi L, Lei Y, Fan Z, et al. Automatic detection of cervical cells using dense-cascade R-CNN[C]. Pattern Recognition and Computer Vision: Third Chinese Conference, PRCV 2020, 2020, Proceedings, Part II 3. Springer International Publishing, 2020: 602-13. DOI: 10.1007/978-3-030-60639-8_50 |
| 22 | Ma B, Zhang J, Cao F, et al. MACD R-CNN: an abnormal cell nucleus detection method[J]. IEEE Access, 2020, 8: 166658-69. DOI: 10.1109/access.2020.3020123 |
| 23 | Redmon J, Divvala S, Girshick R, et al. You only look once: unified, real-time object detection[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016: 779-88. DOI: 10.1109/cvpr.2016.91 |
| 24 | Xiang, Y, Sun W, Pan C, et al. A novel automation-assisted cervical cancer reading method based on convolutional neural network[J]. Biocybernet Biomed Engin, 2020, 40(2): 611-23. DOI: 10.1016/j.bbe.2020.01.016 |
| 25 | Liang Y, Pan C, Sun W, et al. Global context-aware cervical cell detection with soft scale anchor matching[J]. Comput Methods Programs Biomed, 2021, 204: 106061. DOI: 10.1016/j.cmpb.2021.106061 |
| 26 | Jia D, He Z, Zhang C, et al. Detection of cervical cancer cells in complex situation based on improved YOLOv3 network[C]. Multimed Tools Appl, 2022, 81(6): 8939-61. DOI: 10.1007/s11042-022-11954-9 |
| 27 | Fei M, Zhang X, Cao M, et al. Robust cervical abnormal cell detection via distillation from local-scale consistency refinement [C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2023: 652-61. DOI: 10.1007/978-3-031-43987-2_63 |
| 28 | Lin T Y, Goyal P, Girshick R, et al. Focal loss for dense object detection[C]//Proceedings of the IEEE International Conference on Computer Vision. 2017: 2980-2988. DOI: 10.1109/iccv.2017.324 |
| 29 | Srinivas A, Lin T Y, Parmar N, et al. Bottleneck transformers for visualrecognition[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 16519-16529. DOI: 10.1109/cvpr46437.2021.01625 |
| 30 | Woo S, Park J, Lee J Y, et al. Cbam: Convolutional block attention module[C]//Proceedings of the European Conference on Computer Vision (ECCV). 2018: 3-19. DOI: 10.1007/978-3-030-01234-2_1 |
| 31 | Duan S, Zhang M, Qiu S, et al. Tunnel lining crack detection model based on improved YOLOv5[J]. Tunnel Underground Space Technol, 2024, 147: 105713. DOI: 10.1016/j.tust.2024.105713 |
| 32 | Song C Y, Zhang F, Li J S, et al. Detection of maize tassels for UAV remote sensing image with an improved YOLOX model[J]. J Integr Agric, 2023, 22(6): 1671-83. DOI: 10.1016/j.jia.2022.09.021 |
| [1] | Yedong LIANG, Xiongfeng ZHU, Meiyan HUANG, Wencong ZHANG, Hanyu GUO, Qianjin FENG. CRAKUT:integrating contrastive regional attention and clinical prior knowledge in U-transformer for radiology report generation [J]. Journal of Southern Medical University, 2025, 45(6): 1343-1352. |
| [2] | Chen WANG, Mingqiang MENG, Mingqiang LI, Yongbo WANG, Dong ZENG, Zhaoying BIAN, Jianhua MA. Reconstruction from CT truncated data based on dual-domain transformer coupled feature learning [J]. Journal of Southern Medical University, 2024, 44(5): 950-959. |
| [3] | HUANG Pinyu, ZHONG Liming, ZHENG Kaiyi, CHEN Zeli, XIAO Ruolin, QUAN Xianyue, YANG Wei. Multi-phase CT synthesis-assisted segmentation of abdominal organs [J]. Journal of Southern Medical University, 2024, 44(1): 83-92. |
| [4] | ZHONG Youwen, CHE Wengang, GAO Shengxiang. A lightweight multiscale target object detection network for melanoma based on attention mechanism manipulation [J]. Journal of Southern Medical University, 2022, 42(11): 1662-1671. |
| [5] | . Temporal pattern of postmortem color changes in the pupil region of the cornea in rabbits [J]. Journal of Southern Medical University, 2018, 38(10): 1266-. |
| [6] | . Isolation and expression profiling of transformer 2 gene in Aedes aegypti [J]. Journal of Southern Medical University, 2013, 33(11): 1583-. |
| [7] | ZHU Xiao-xia, CHEN Yong-qing, CHEN Long-hua. Value of integrated positron-emission tomography and computed tomography in gross tumor volume delineation for radiotherapy for bone metastasis [J]. Journal of Southern Medical University, 2004, 24(06): 700-702,705. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
