Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (8): 1643-1653.doi: 10.12122/j.issn.1673-4254.2025.08.09
Zixian CHEN( ), Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN(
), Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN( )
)
Received:2025-04-06
Online:2025-08-20
Published:2025-09-05
Contact:
Mingkun CHEN
E-mail:czx961147842@163.com;chenmk1@smu.edu.cn
Supported by:Zixian CHEN, Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN. A risk prediction model for prognosis and immunotherapy response in prostate cancer patients based on immunosuppressive neutrophil Neu_2 subsets[J]. Journal of Southern Medical University, 2025, 45(8): 1643-1653.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.08.09
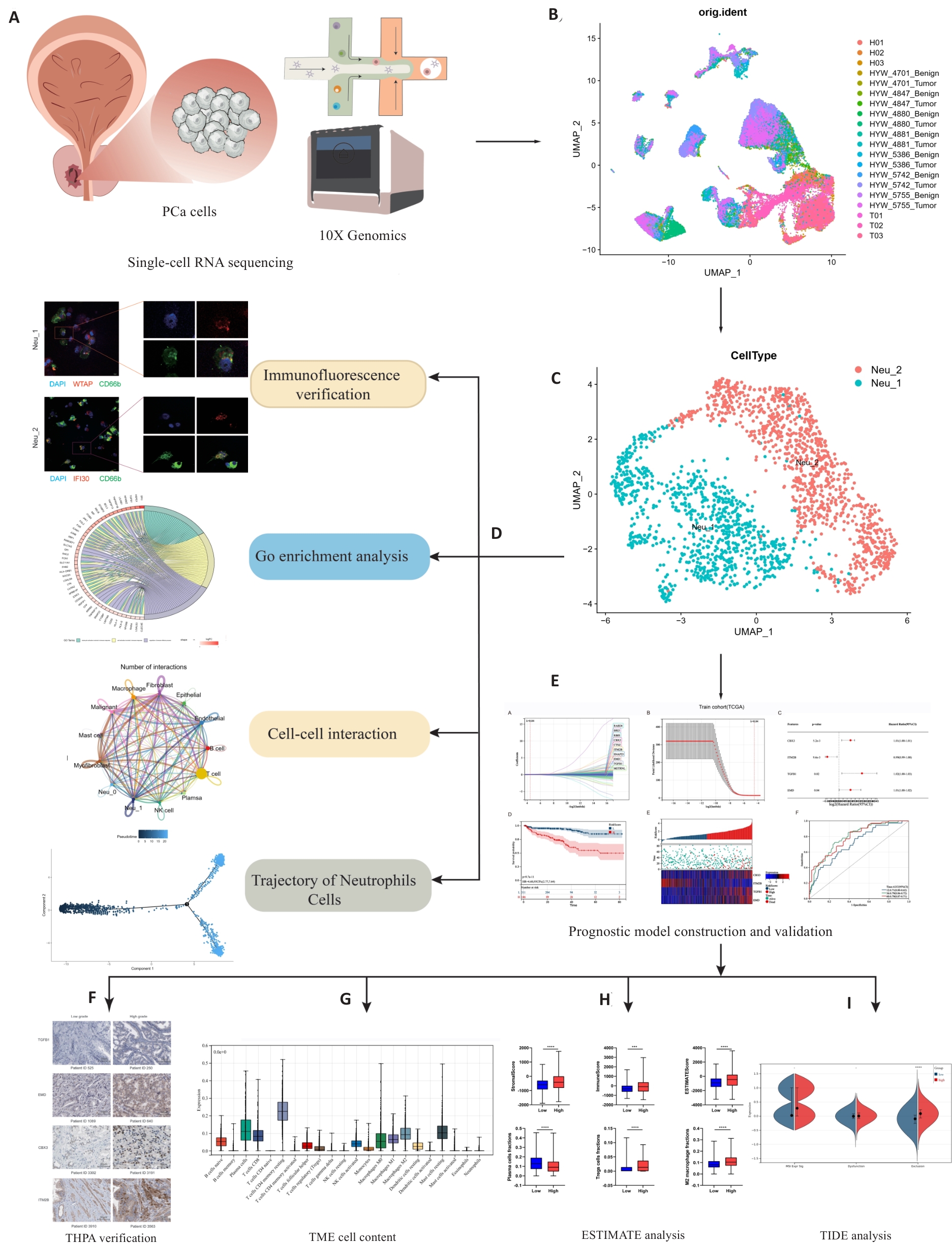
Fig.1 Workflow of this study. Single-cell sequencing data were obtained from principal components analysis (A), followed by annotation and clustering of the data (B), resulting in the identification of two types of neutrophils: Neu_1 and Neu_2 (C). Tissue immunofluorescence verification, Gene Ontology (GO) enrichment analysis, cell interaction studies, and counter-time sequence analysis of these two cell types were used to elucidate their roles within the tumor microenvironment (TME) (D). Combining our findings with data from the TCGA database, we identified 4 independent prognostic factors for constructing and validating the prognostic models (E, F). We performed an analysis of immunotherapy predictions by analyzing and scoring the proportion of immune cells within the TME (G, H). ***P<0.001, ****P<0.0001. The patients with high levels of infiltrating Neu_2 neutrophils are likely to have poor responses to immunotherapy, as indicated by TIDE analysis (I). THPA: The Human Protein Atlas; ESTIMATE: Estimation of stromal and immune cells in malignant tumor tissues using expression data; TIDE: Tumor immune dysfunction and exclusion.

Fig.2 Integration and clustering of PCa scRNA-Seq data. A: t-SNE of 15 PCa samples. B: t-SNE of 15 cell clusters. C: Identification of 12 cell types by marker genes. D: Cell types exist in different samples. E: Dot plot showed the expression differences of various genes across the 12 cell types. F: Expression differences of 12 cell types between the control and tumor groups. G: Heat map showing expressions of the characteristic genes across different cell subpopulations.

Fig.3 Cell map of neutrophil subtypes. A: t-SNE of the 20 cell clusters. B: t-SNE of control group and tumor group. C: Neu_1 and Neu_2 neutrophils identified by marker genes. D: Heat map showing differential expressions of the marker genes between Neu_1 and Neu_2 neutrophils. E: Expressions of the marker genes as signatures of the two cell types. F: GO enrichment analysis of signaling pathways associated with Neu_1 and Neu_2 neutrophils. G: Immunofluorescent staining of the marker genes in a subset of neutrophils (CD66b), specifically WTAP in Neu_1 and IIFI30 in Neu_2, within prostate cancer (PCa) tissue. H: GO enrichment analysis chord diagram Neu_1 signaling pathways involved in neutrophils. I: GO enrichment analysis chord diagram Neu_2 signaling pathways involved in neutrophils. J: KEGG analysis of signaling pathways involved in Neu_1 and Neu_2 neutrophils.

Fig.4 Analysis of intercellular communication related to neutrophils. A: Number of interactions in the intercellular communication network. B: Interaction weights/strengths in intercellular communication networks. C: Neu_1 interaction between neutrophils and other cells. D: Neu_2 interaction between neutrophils and other cells. E: Number and intensity of interactions between Neu_1 neutrophils and different cell types. F: Number and intensity of interactions between Neu_2 neutrophils and different cell types. G: Bubble diagram of ligand-receptor pair-mediated interactions between Neu_1 cells and Neu_2 neutrophils and other cells.

Fig.6 Construction and validation of a neutrophil prognostic risk model. A, B: Screening of prognostic-related core genes by lasso-cox regression in TCGA training group. C, G: Forest diagram in TCGA training group and GSE70770 validation set. D, H: Kaplan-Meier curve for overall survival between different ICPI risk groups in TCGA training group and GSE70770 validation set. E, I: Validation of centralized risk scores and expression heat maps of 4 genes in TCGA training group and GSE70770 validation set. F, J: Time-dependent ROC curve analysis in TCGA training group and GSE70770 validation set. K: Verification of expressions of the 4 key genes in PCa tissues by immunohistochemical staining from THPA database.
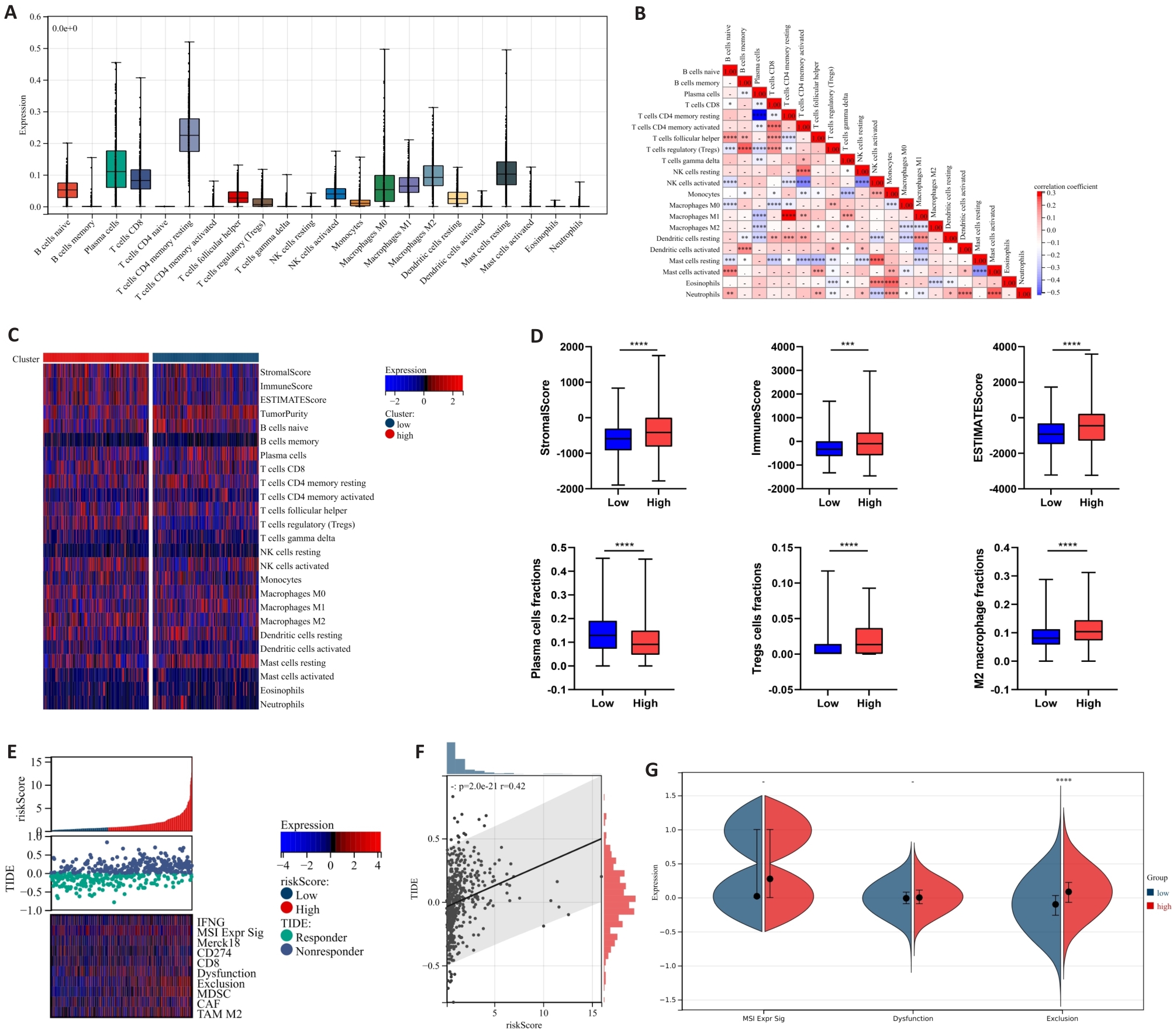
Fig.7 Correlation between neutrophil risk prognostic model and immune infiltration as well as immune response in PCa. A: Calculation of 22 immune cell infiltration ratios in PCa tissues based on CIBERSORT. B: Correlation analysis between immune cells in PCa tissues. C: Differences in immune cell infiltration expression between high-risk and low-risk groups. D: Differences in immune scores and infiltration ratios of some immune cells (plasma cells, Tregs cells, and M2-TAMs cells) between high-risk and low-risk groups. E, F: Correlation analysis between TIDE score expression and risk score. G: Differences in microsatellite instability, immune dysfunction and immune rejection scores between high-risk and low-risk groups. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
| [1] | Davidsson S, Fiorentino M, Andrén O, et al. Inflammation, focal atrophic lesions, and prostatic intraepithelial neoplasia with respect to risk of lethal prostate cancer[J]. Cancer Epidemiol Biomarkers Prev, 2011, 20(10): 2280-7. doi:10.1158/1055-9965.epi-11-0373 |
| [2] | Conti DV, Darst BF, Moss LC, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction[J]. Nat Genet, 2021, 53(1): 65-75. doi:10.1038/s41588-021-00786-2 |
| [3] | Nicholson LT, Fong L. Immune checkpoint inhibition in prostate cancer[J]. Trends Cancer, 2020, 6(3): 174-7. doi:10.1016/j.trecan.2020.01.003 |
| [4] | Pang K, Shi ZD, Wei LY, et al. Research progress of therapeutic effects and drug resistance of immunotherapy based on PD-1/PD-L1 blockade[J]. Drug Resist Updat, 2023, 66: 100907. doi:10.1016/j.drup.2022.100907 |
| [5] | Riley RS, June CH, Langer R, et al. Delivery technologies for cancer immunotherapy[J]. Nat Rev Drug Discov, 2019, 18(3): 175-96. doi:10.1038/s41573-018-0006-z |
| [6] | Iacovelli R, Ciccarese C, Brunelli M, et al. PD-L1 expression in de novo metastatic castration-sensitive prostate cancer[J]. J Immunother, 2019, 42(7): 269-73. doi:10.1097/cji.0000000000000287 |
| [7] | Cong LL, Zhao Q, Sun HY, et al. A novel long non-coding RNA SLNCR1 promotes proliferation, migration, and invasion of melanoma via transcriptionally regulating SOX5[J]. Cell Death Discov, 2024, 10(1): 160. doi:10.1038/s41420-024-01922-7 |
| [8] | Morimoto K, Yamada T, Takayama K. The landscape of immune therapy in vulnerable patients with advanced non-small cell lung cancer: a narrative review[J]. Transl Lung Cancer Res, 2023, 12(11): 2310-21. doi:10.21037/tlcr-23-581 |
| [9] | Bian XJ, Wang WF, Abudurexiti M, et al. Integration analysis of single-cell multi-omics reveals prostate cancer heterogeneity[J]. Adv Sci (Weinh), 2024, 11(18): e2305724. doi:10.1002/advs.202305724 |
| [10] | Bancaro N, Calì B, Troiani M, et al. Apolipoprotein E induces pathogenic senescent-like myeloid cells in prostate cancer[J]. Cancer Cell, 2023, 41(3): 602-19. e11. doi:10.1016/j.ccell.2023.02.004 |
| [11] | Angappulige DH, Mahajan NP, Mahajan K. Epigenetic underpinnings of tumor-immune dynamics in prostate cancer immune suppression[J]. Trends Cancer, 2024, 10(4): 369-81. doi:10.1016/j.trecan.2024.01.004 |
| [12] | Qin Y, Liu YL, Xiang XY, et al. Cuproptosis correlates with immunosuppressive tumor microenvironment based on pan-cancer multiomics and single-cell sequencing analysis[J]. Mol Cancer, 2023, 22(1): 59. doi:10.1186/s12943-023-01752-8 |
| [13] | Zhang YJ, Wang D, Peng M, et al. Single-cell RNA sequencing in cancer research[J]. J Exp Clin Cancer Res, 2021, 40(1): 81. doi:10.1186/s13046-021-01874-1 |
| [14] | Kock KH, Tan LM, Han KY, et al. Asian diversity in human immune cells[J]. Cell, 2025, 188(8): 2288-306. e24. doi:10.1016/j.cell.2025.02.017 |
| [15] | Hu JY, Chen ZH, Bao L, et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes[J]. Mol Ther, 2020, 28(7): 1658-72. doi:10.1016/j.ymthe.2020.04.023 |
| [16] | Ding SN, Qiao N, Zhu QC, et al. Single-cell atlas reveals a distinct immune profile fostered by T cell-B cell crosstalk in triple negative breast cancer[J]. Cancer Commun (Lond), 2023, 43(6): 661-84. doi:10.1002/cac2.12429 |
| [17] | Roma-Rodrigues C, Mendes R, Baptista PV, et al. Targeting tumor microenvironment for cancer therapy[J]. Int J Mol Sci, 2019, 20(4): 840. doi:10.3390/ijms20040840 |
| [18] | Brand A, Singer K, Koehl GE, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells[J]. Cell Metab, 2016, 24(5): 657-71. doi:10.1016/j.cmet.2016.08.011 |
| [19] | Maynard JP, Godwin TN, Lu JY, et al. Localization of macrophage subtypes and neutrophils in the prostate tumor microenvironment and their association with prostate cancer racial disparities[J]. Prostate, 2022, 82(16): 1505-19. doi:10.1002/pros.24424 |
| [20] | Lou Y, Fan ZG, Ren SC. The role of neutrophils and their immu-nosuppressive effects on prostate cancer[J]. Histol Histopathol, 2025: 18890. |
| [21] | Bailey-Whyte M, Minas TZ, Dorsey TH, et al. Systemic inflammation indices and association with prostate cancer survival in a diverse patient cohort[J]. Cancers (Basel), 2023, 15(6): 1869. doi:10.3390/cancers15061869 |
| [22] | Xiao ZP, Song SS, Chen D, et al. Proteolysis targeting Chimera (PROTAC) for macrophage migration inhibitory factor (MIF) has anti-proliferative activity in lung cancer cells[J]. Angew Chem Int Ed, 2021, 60(32): 17514-21. doi:10.1002/anie.202101864 |
| [23] | Zhu GQ, Tang YL, Geng N, et al. HIF-α/MIF and NF-κB/IL-6 axes contribute to the recruitment of CD11b+Gr-1+myeloid cells in hypoxic microenvironment of HNSCC[J]. Neoplasia, 2014, 16(2): 168-79. doi:10.1593/neo.132034 |
| [24] | Zhu GQ, Tang Z, Huang R, et al. CD36+ cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor[J]. Cell Discov, 2023, 9(1): 25. doi:10.1038/s41421-023-00529-z |
| [25] | Ding XC, Wang LL, Zhang XD, et al. The relationship between expression of PD-L1 and HIF-1α in glioma cells under hypoxia[J]. J Hematol Oncol, 2021, 14(1): 92. doi:10.1186/s13045-021-01102-5 |
| [26] | Xu XT, Lai C, Luo JW, et al. The predictive significance of chromobox family members in prostate cancer in humans[J]. Cell Oncol (Dordr), 2024, 47(4): 1315-31. doi:10.1007/s13402-024-00929-7 |
| [27] | Mališić E, Petrović N, Brengues M, et al. Association of polymorphisms in TGFB1 XRCC1 XRCC3 genes and CD8 T-lymphocyte apoptosis with adverse effect of radiotherapy for prostate cancer[J]. Sci Rep, 2022, 12(1): 21306. doi:10.1038/s41598-022-25328-6 |
| [28] | Xiao Y, Yu DH. Tumor microenvironment as a therapeutic target in cancer[J]. Pharmacol Ther, 2021, 221: 107753. doi:10.1016/j.pharmthera.2020.107753 |
| [29] | Duan QQ, Zhang HL, Zheng JN, et al. Turning cold into hot: firing up the tumor microenvironment[J]. Trends Cancer, 2020, 6(7): 605-18. doi:10.1016/j.trecan.2020.02.022 |
| [30] | Yang JF, Xing XD, Luo L, et al. Mitochondria-ER contact mediated by MFN2-SERCA2 interaction supports CD8+ T cell metabolic fitness and function in tumors[J]. Sci Immunol, 2023, 8(87): eabq2424. doi:10.1126/sciimmunol.abq2424 |
| [1] | Xiaojuan GUO, Ruijuan DU, Liping CHEN, Kelei GUO, Biao ZHOU, Hua BIAN, Li HAN. WW domain-containing ubiquitin E3 ligase 1 regulates immune infiltration in tumor microenvironment of ovarian cancer [J]. Journal of Southern Medical University, 2025, 45(5): 1063-1073. |
| [2] | Jinguang LUO, Huaixiang TAO, Zhiyuan WEN, Long CHEN, Hao HU, Han GUAN. Tumor-associated fibroblasts promotes proliferation and migration of prostate cancer cells by suppressing FBXL3 via upregulating hsa-miR-18b-5p [J]. Journal of Southern Medical University, 2024, 44(7): 1284-1296. |
| [3] | Bei PEI, Yi ZHANG, Siyuan WEI, Yu MEI, Biao SONG, Gang DONG, Ziang WEN, Xuejun LI. Identification of potential pathogenic genes of intestinal metaplasia based on transcriptomic sequencing and bioinformatics analysis [J]. Journal of Southern Medical University, 2024, 44(5): 941-949. |
| [4] | WANG Zining, YANG Ming, LI Shuanglei, CHI Haitao, WANG Junhui, XIAO Cangsong. A transcriptomic analysis of correlation between mitochondrial function and energy metabolism remodeling in mice with myocardial fibrosis following myocardial infarction [J]. Journal of Southern Medical University, 2024, 44(4): 666-674. |
| [5] | SHAO Shan, BAI Weichao, ZHOU Pengcheng, LUO Minna, ZHAO Xinhan, LEI Jianjun. Metformin suppresses hypoxia-inducible factor-1α expression in cancer-associated fibroblasts to block tumor-stromal cross-talk in breast cancer [J]. Journal of Southern Medical University, 2024, 44(3): 428-436. |
| [6] | Yanni WANG, Xia HUANG, Fuheng CHEN, Yuanyuan GAO, Xiangrong CUI, Qin YAN, Xuan JING. Association between serum BIN1 level and Killip class in patients with acute myocardial infraction [J]. Journal of Southern Medical University, 2024, 44(12): 2388-2395. |
| [7] | Chengling CUI, Yuzhen XU, Chaoqun TANG, Jiaying JIANG, Ying HU, Jie SHUANG. Molecular mechanism of high-altitude hypoxia-induced lipid metabolism disorder in mouse spleen tissue [J]. Journal of Southern Medical University, 2024, 44(10): 2024-2032. |
| [8] | WU Xiuhua, FAN Yingjing, YE Yongnong, LI Ping, ZHU Qing'an, CHEN Zesen, LI Bo, WANG Wen, ZHENG Lei. A transcriptomic study of osteoporosis induced by ketogenic diet in mice [J]. Journal of Southern Medical University, 2023, 43(8): 1440-1446. |
| [9] | DENG Ting, DU Boyu, XI Xueyan. Colorectal cancer cells induce the formation of cancer-associated fibroblasts by activating the ERK signaling pathway in fibroblasts [J]. Journal of Southern Medical University, 2023, 43(6): 943-951. |
| [10] | ZHANG Ziran, TAN Jiale, YU Zihang, LIU Chengdong, WANG Jian, WU Dehua, BAI Xue. FARSB stratifies prognosis and cold tumor microenvironment across different cancer types: an integrated single cell and bulk RNA sequencing analysis [J]. Journal of Southern Medical University, 2023, 43(5): 667-679. |
| [11] | MAO Jianying, YANG Wenjing, GUO He, DONG Ruili, REN Lifang, LI Shubin. A cervical cancer tissue-derived decellularized extracellular matrix scaffold for cervical cancer tissue reconstruction in vitro [J]. Journal of Southern Medical University, 2023, 43(2): 157-165. |
| [12] | ZHANG Weijian, ZOU Zhuoyue, ZHU Yongna, WANG Min, MA Caiyun, WU Junjie, SHI Xin, LIU Xi. Expression of interleukin-34 in tongue squamous cell carcinoma and its clinical implications [J]. Journal of Southern Medical University, 2023, 43(12): 2111-2117. |
| [13] | GUO Li, MA Yinling, LI Ting, LI Jinping. CD40LG is a novel immune- and stroma-related prognostic biomarker in the tumor microenvironment of invasive breast cancer [J]. Journal of Southern Medical University, 2022, 42(9): 1267-1278. |
| [14] | XIE Yan, LI Cheng, ZHANG Lulu, ZANG Shiming, YU Fei, WANG Shukui, WANG Feng. 68Ga-PSMA-I&T PET/CT for assessment of tumor burden in primary lesions of treatment-naïve prostate cancer [J]. Journal of Southern Medical University, 2022, 42(8): 1143-1148. |
| [15] | YANG Ming, ZHU Xudong, SHEN Yang, HE Qi, QIN Yuan, SHAO Yiqun, YUAN Lin, YE Hesong. High expression of MYBL2 promotes progression and predicts a poor survival outcome of prostate cancer [J]. Journal of Southern Medical University, 2022, 42(8): 1109-1118. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||