Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (5): 981-988.doi: 10.12122/j.issn.1673-4254.2024.05.21
• Basic Research • Previous Articles Next Articles
Qinzhi WANG1( ), Bing SONG2, Shirui HAO3, Zhiyuan XIAO4, Lianhui JIN5, Tong ZHENG1, Fang CHAI1(
), Bing SONG2, Shirui HAO3, Zhiyuan XIAO4, Lianhui JIN5, Tong ZHENG1, Fang CHAI1( )
)
Received:2024-01-12
Online:2024-05-20
Published:2024-06-06
Contact:
Fang CHAI
E-mail:1986045014@qq.com;lyfsdyyy@163.com
Qinzhi WANG, Bing SONG, Shirui HAO, Zhiyuan XIAO, Lianhui JIN, Tong ZHENG, Fang CHAI. Bioinformatic analysis of CCND2 expression in papillary thyroid carcinoma and its impact on immune infiltration[J]. Journal of Southern Medical University, 2024, 44(5): 981-988.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.05.21
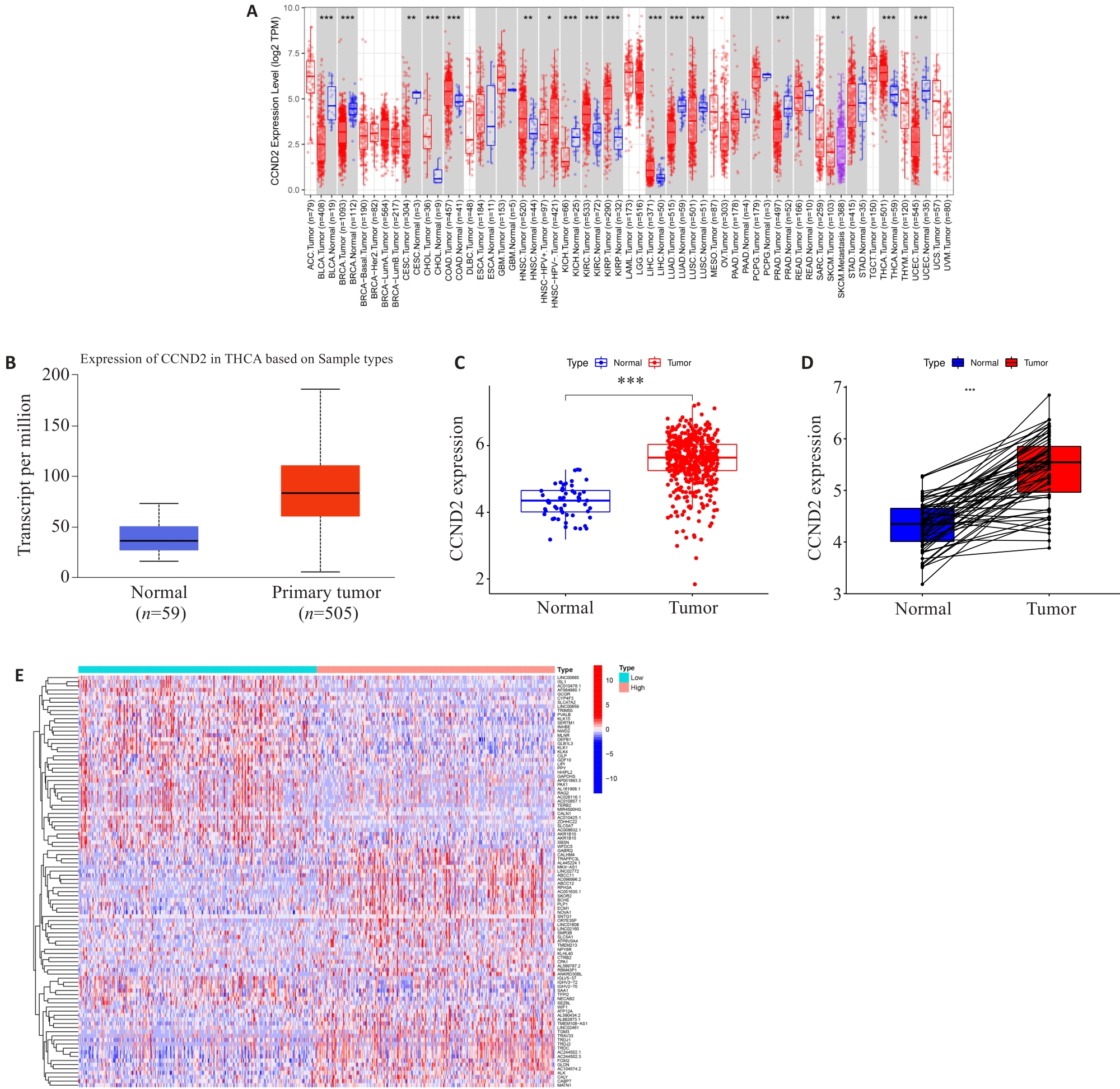
Fig.1 Differential expressions of CCND2 and related genes in thyroid cancer. A: CCND2expression in different types of cancer investigated with the Tumor Immune Estimation Resource (TIMER) database. B: CCND2 expression in THCA was examined by using the UALCAN database; C: Analysis of CCND2 expression in THCA and adjacent normal tissues in the TCGA database; D: Analysis of CCND2 expression in THCA and paired adjacent normal tissues in TCGA database; E: Expression heatmap of differential gene related to CCND2. ***P<0.001
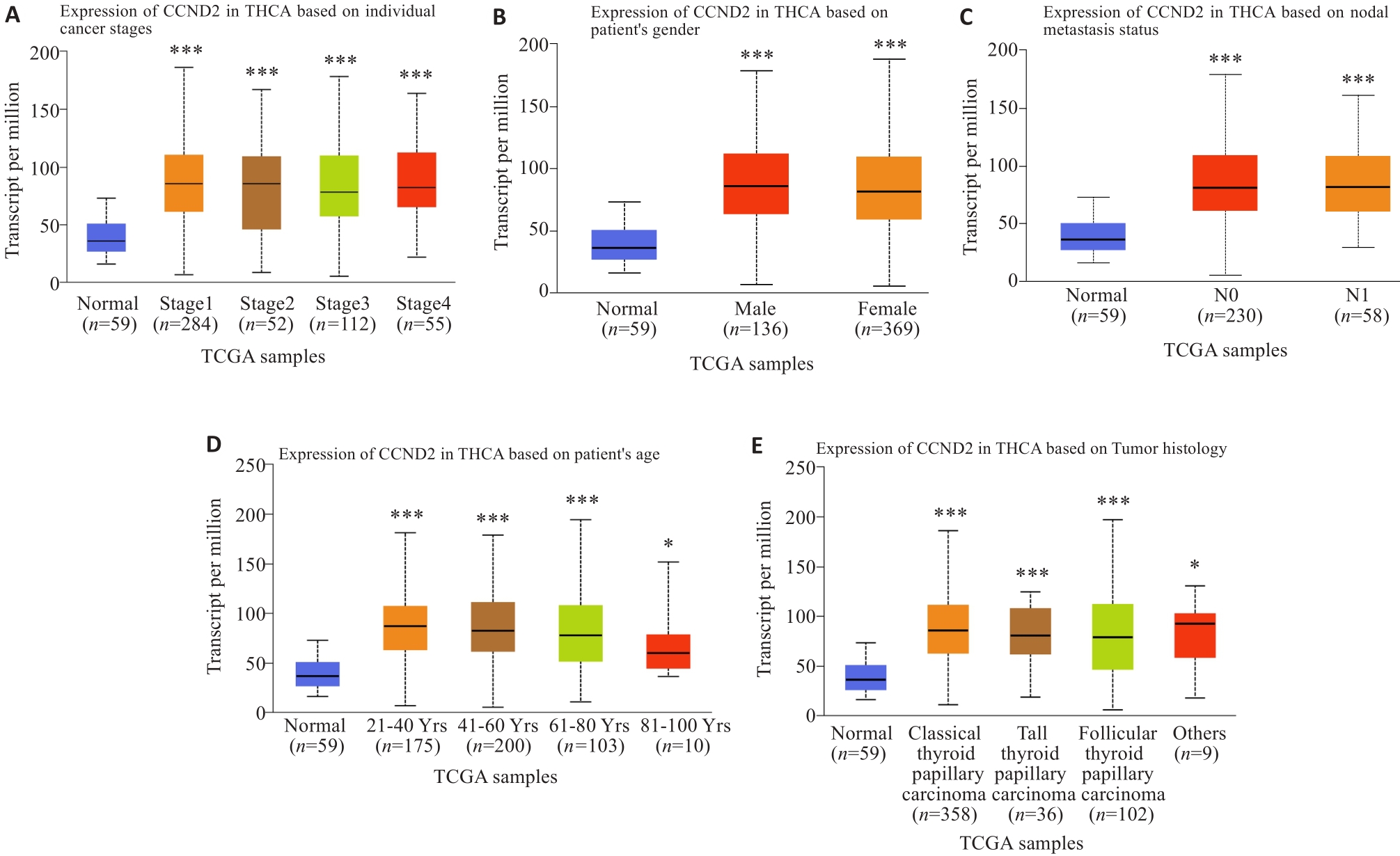
Fig.2 Correlation analysis of CCND2 expression with tumor stage (A), gender (B), lymph node metastasis (C), age (D) and tumor histology (E). *P<0.05, ***P<0.001 vs Normal group

Fig.5 Relationship between expression levels of CCND2 and TIICs. A: Correlation analyses of CCND2 with the infiltration of different immune cells using the TIMER database; B: Difference in infiltration fractions of 22 immune cell subsets in high- and low-risk groups. Red, the high-risk group and blue, low-risk group. *P<0.05, **P<0.01 vs groups.
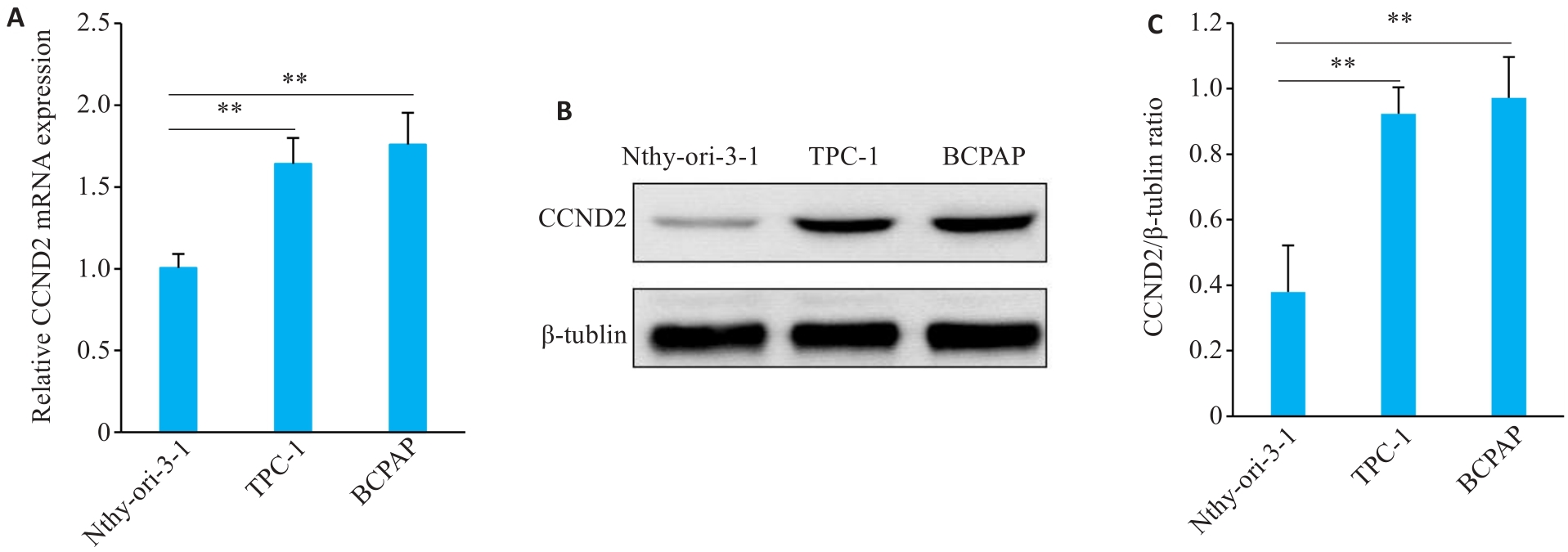
Fig.6 Expression of CCND2 in Nthy-ori-3-1, TPC-1, and BCPAP cells. A: RT-qPCR for detecting mRNA expression of CCNDD2. B: Western blot of CCND2 and β-tubulin (loading control). C: Relative expression level of CCND2/β-tubulin. Data are expressed as Mean±SD. **P<0.01.
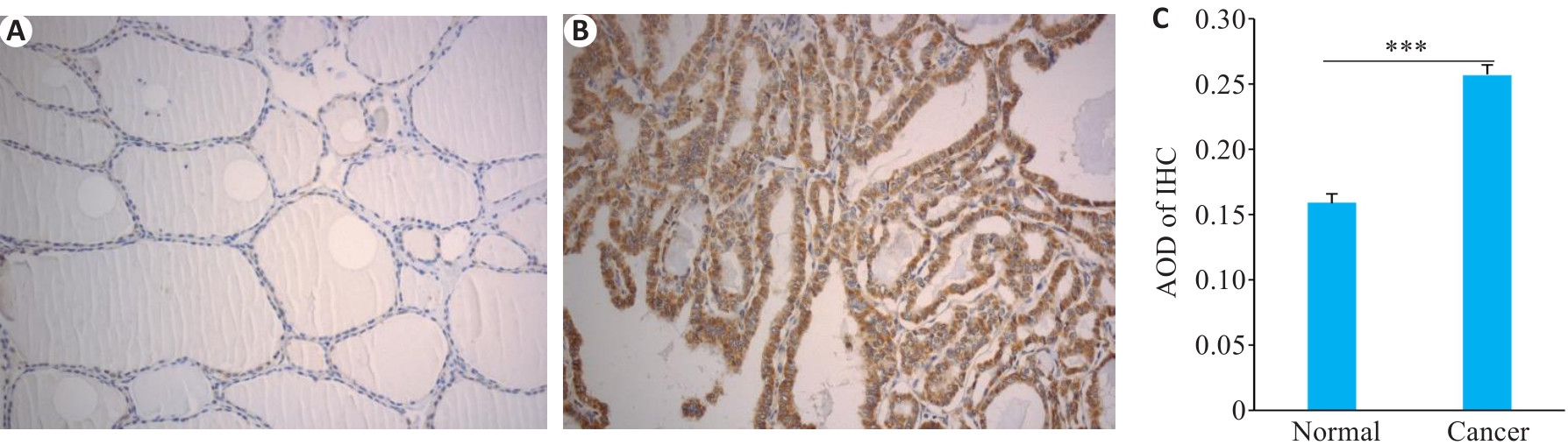
Fig.7 Expression of CCND2 in PTC tissues and adjacent tissues detected by immunohistochemistry (Original magnification:×200). A: Normal tissue. B: PTC tissue. C: Expression levels calculated with average optical density (AOD). ***P<0.001.
| Clinicopathological parameters | n | CCND2 positive | χ2 | P | |
|---|---|---|---|---|---|
| Gender | Male | 32 | 22 (68.75) | 0.4622 | 0.4966 |
| Female | 84 | 63 (75.00) | |||
| Age (year) | <55 | 79 | 55 (69.62) | 1.9627 | 0.1612 |
| ≥55 | 37 | 30 (81.08) | |||
| Tumor size (cm) | <1 | 76 | 50 (65.79) | 4.6911 | 0.0303 |
| ≥1 | 40 | 35 (75.00) | |||
| Multifocal | Yes | 40 | 26 (65.00) | 2.1353 | 0.1439 |
| No | 76 | 59 (77.63) | |||
| Extraglandular invasion | Yes | 13 | 11 (84.62) | 0.9614 | 0.3268 |
| No | 103 | 74 (71.84) | |||
| Lymphatic metastasis | Yes | 43 | 38 (88.37) | 7.9520 | 0.0048 |
| No | 73 | 47 (64.38) | |||
| TNM staging | I-II | 100 | 70 (70.00) | 12.1309 | 0.0005 |
| III-IV | 16 | 15 (93.75) |
Tab.1 Relationship between CCND2 expression and clinicopathological characteristics of PTC patients [n (%)]
| Clinicopathological parameters | n | CCND2 positive | χ2 | P | |
|---|---|---|---|---|---|
| Gender | Male | 32 | 22 (68.75) | 0.4622 | 0.4966 |
| Female | 84 | 63 (75.00) | |||
| Age (year) | <55 | 79 | 55 (69.62) | 1.9627 | 0.1612 |
| ≥55 | 37 | 30 (81.08) | |||
| Tumor size (cm) | <1 | 76 | 50 (65.79) | 4.6911 | 0.0303 |
| ≥1 | 40 | 35 (75.00) | |||
| Multifocal | Yes | 40 | 26 (65.00) | 2.1353 | 0.1439 |
| No | 76 | 59 (77.63) | |||
| Extraglandular invasion | Yes | 13 | 11 (84.62) | 0.9614 | 0.3268 |
| No | 103 | 74 (71.84) | |||
| Lymphatic metastasis | Yes | 43 | 38 (88.37) | 7.9520 | 0.0048 |
| No | 73 | 47 (64.38) | |||
| TNM staging | I-II | 100 | 70 (70.00) | 12.1309 | 0.0005 |
| III-IV | 16 | 15 (93.75) |
| 1 | Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68(6): 394-424. DOI: 10.3322/caac.21492 |
| 2 | 任 丽, 邹明远, 朱行春, 等. 姜黄素通过Keap1-Nrf2通路抑制甲状腺乳头状癌B-CPAP细胞的增殖、迁移及侵袭[J]. 南方医科大学学报, 2023, 43(8): 1356-62. |
| 3 | Mao JX, Zhang QH, Zhang HY, et al. Risk factors for lymph node metastasis in papillary thyroid carcinoma: a systematic review and meta-analysis[J]. Front Endocrinol, 2020, 11: 265. DOI: 10.3389/fendo.2020.00265 |
| 4 | Dotinga M, Vriens D, van Velden FHP, et al. Reinducing radioiodine-sensitivity in radioiodine-refractory thyroid cancer using lenvatinib (RESET): study protocol for a single-center, open label phase II trial[J]. Diagnostics: Basel, 2022, 12(12): 3154. DOI: 10.3390/diagnostics12123154 |
| 5 | Zhang XW, Xiong H, Duan JL, et al. The prognostic significance of the BIN1 and CCND2 gene in adult patients with acute myeloid leukemia[J]. Indian J Hematol Blood Transfus, 2022, 38(3): 481-91. DOI: 10.1007/s12288-021-01479-w |
| 6 | Conway JW, Braden J, Wilmott JS, et al. The effect of organ-specific tumor microenvironments on response patterns to immunotherapy[J]. Front Immunol, 2022, 13: 1030147. DOI: 10.3389/fimmu.2022.1030147 |
| 7 | Zhong L, Li YS, Xiong L, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives[J]. Signal Transduct Target Ther, 2021, 6: 201. DOI: 10.1038/s41392-021-00572-w |
| 8 | Li X, Fang J, Wei G, et al. CircMMP9 accelerates the progression of hepatocellular carcinoma through the miR-149/CCND2 axis[J]. J Gastrointest Oncol, 2022, 13(4): 1875-88. DOI: 10.21037/jgo-22-677 |
| 9 | Wang D, Zhang Y, Che YQ. CCND2 mRNA expression is correlated with R-CHOP treatment efficacy and prognosis in patients with ABC-DLBCL[J]. Front Oncol, 2020, 10: 1180. DOI: 10.3389/fonc.2020.01180 |
| 10 | Yue Q, Xu Y, Deng X, et al. Circ-PITX1 promotes the progression of non-small cell lung cancer through regulating the miR-1248/CCND2 axis[J]. Onco Targets Ther, 2021, 14: 1807-19. DOI: 10.2147/ott.s286820 |
| 11 | Richardson TE, Walker JM. CCND2 amplification is an independent adverse prognostic factor in IDH-mutant lower-grade astrocytoma[J]. Clin Neuropathol, 2021, 40(4): 209-14. DOI: 10.5414/np301354 |
| 12 | Dai W, Shi Y, Hu W, et al. Long noncoding RNA FAM225B facilitates proliferation and metastasis of nasopharyngeal carcinoma cells by regulating miR-613/CCND2 axis[J]. Bosn J Basic Med Sci, 2022, 22(1): 77-86. |
| 13 | Chen N, Yin D, Lun B, et al. LncRNA GAS8-AS1 suppresses papillary thyroid carcinoma cell growth through the miR-135b-5p/CCND2 axis[J]. Biosci Rep, 2019, 39(1): BSR20181440. DOI: 10.1042/bsr20181440 |
| 14 | Leone V, D’Angelo D, Rubio I, et al. MiR-1 is a tumor suppressor in thyroid carcinogenesis targeting CCND2, CXCR4, and SDF-1α[J]. J Clin Endocrinol Metab, 2011, 96(9): E1388-98. DOI: 10.1210/jc.2011-0345 |
| 15 | Yuan S, Liu Z, Yu S, et al. CCND2 and miR-206 as potential biomarkers in the clinical diagnosis of thyroid carcinoma by fine-needle aspiration cytology[J]. World J Surg Oncol, 2023, 21(1): 22. DOI: 10.1186/s12957-023-02899-w |
| 16 | Abbott M, Ustoyev Y. Cancer and the immune system: the history and background of immunotherapy[J]. Semin Oncol Nurs, 2019, 35(5): 150923. DOI: 10.1016/j.soncn.2019.08.002 |
| 17 | Kołat D, Kałuzińska Ż, Bednarek AK, et al. Determination of WWOX function in modulating cellular pathways activated by AP-2α and AP-2γ transcription factors in bladder cancer[J]. Cells, 2022, 11(9): 1382. DOI: 10.3390/cells11091382 |
| 18 | Varricchi G, Loffredo S, Marone G, et al. The immune landscape of thyroid cancer in the context of immune checkpoint inhibition[J]. Int J Mol Sci, 2019, 20(16): E3934. DOI: 10.3390/ijms20163934 |
| 19 | Zhang J, Gao J, Cui J, et al. Tumor-associated macrophages in tumor progression and the role of traditional Chinese medicine in regulating TAMs to enhance antitumor effects[J]. Front Immunol, 2022, 13: 1026898. DOI: 10.3389/fimmu.2022.1026898 |
| 20 | Morana O, Wood W, Gregory CD. The apoptosis paradox in cancer[J]. Int J Mol Sci, 2022, 23(3): 1328. DOI: 10.3390/ijms23031328 |
| [1] | Wei ZHOU, Jun NIE, Jia HU, Yizhi JIANG, Dafa ZHANG. Differential expressions of endoplasmic reticulum stress-associated genes in aortic dissection and their correlation with immune cell infiltration [J]. Journal of Southern Medical University, 2024, 44(5): 859-866. |
| [2] | Bei PEI, Yi ZHANG, Siyuan WEI, Yu MEI, Biao SONG, Gang DONG, Ziang WEN, Xuejun LI. Identification of potential pathogenic genes of intestinal metaplasia based on transcriptomic sequencing and bioinformatics analysis [J]. Journal of Southern Medical University, 2024, 44(5): 941-949. |
| [3] | LIANG Yihao, LAI Yingjun, YUAN Yanwen, YUAN Wei, ZHANG Xibo, ZHANG Bashan, LU Zhifeng. Screening of differentially expressed genes in gastric cancer based on GEO database and function and pathway enrichment analysis [J]. Journal of Southern Medical University, 2024, 44(3): 605-616. |
| [4] | CHENG Jiacong, LI Zhihui, LIU Yao, LI Cheng, HUANG Xin, TIAN Yinxin, SHEN Fubing. Bioinformatics analysis and validation of the interaction between PML protein and TAB1 protein [J]. Journal of Southern Medical University, 2024, 44(1): 179-186. |
| [5] | WANG Qiusheng, ZHANG Zhen, WANG Lian, WANG Yu, YAO Xinyu, WANG Yueyue, ZHANG Xiaofeng, GE Sitang, ZUO Lugen. High expression of death-associated protein 5 promotes glucose metabolism in gastric cancer cells and correlates with poor survival outcomes [J]. Journal of Southern Medical University, 2023, 43(7): 1063-1070. |
| [6] | ZHANG Weijian, ZOU Zhuoyue, ZHU Yongna, WANG Min, MA Caiyun, WU Junjie, SHI Xin, LIU Xi. Expression of interleukin-34 in tongue squamous cell carcinoma and its clinical implications [J]. Journal of Southern Medical University, 2023, 43(12): 2111-2117. |
| [7] | XIN Baoshan, LIU Gang, ZHANG Chengxiong, WANG Bing, SHI Lianghui. LncRNA LINC00342 regulates gastric cancer cell proliferation, migration and invasion by targeting miR-596 [J]. Journal of Southern Medical University, 2023, 43(10): 1761-1770. |
| [8] | ZHAO Haiyuan, LIU Gang, LI Yang, YANG Nianzhao, ZHAO Jun. High expression of ANKRD6 is an indicator for poor prognosis of colon adenocarcinoma [J]. Journal of Southern Medical University, 2023, 43(10): 1715-1724. |
| [9] | LIU Zhiyu, FANG Fengzhu, LI Jing, ZHAO Guangyue, ZANG Quanjin, ZHANG Feng, DIE Jun. RHPN2 is highly expressed in osteosarcoma cells to promote cell proliferation and migration and inhibit apoptosis [J]. Journal of Southern Medical University, 2022, 42(9): 1367-1373. |
| [10] | HONG Haining, ZHU Haonan, LI Chao, ZANG Chao, SANG Haiwei, CHEN Liwei, WANG Ansheng. FNDC1 is highly expressed in lung adenocarcinoma and closely related with poor prognosis [J]. Journal of Southern Medical University, 2022, 42(8): 1182-1190. |
| [11] | HUANG Ping, ZHU Jing, LI Hhua, WANG Yanzhao, TANG Yiming, LIU Qiang. Bioinformatic analysis of differentially expressed proteins in the dorsal raphe nucleus of rats after continuous treatment with olanzapine [J]. Journal of Southern Medical University, 2022, 42(8): 1221-1229. |
| [12] | WANG Kang, ZHANG Jun, DENG Muwen, JU Yongle, OUYANG Manzhao. METTL27 is a prognostic biomarker of colon cancer and associated with immune invasion [J]. Journal of Southern Medical University, 2022, 42(4): 486-497. |
| [13] | ZHANG Xiaoning, ZHANG Xiaoyu, LIU Peng, LIU Kuo, LI Wenwen, CHEN Qianqian, MA Wanshan. Prognostic implications and functional enrichment analysis of LTB4R in patients with acute myeloid leukemia [J]. Journal of Southern Medical University, 2022, 42(3): 309-320. |
| [14] | ZHU Yali, ZHANG Shujun, YANG Cheng, XUE Wei, ZHANG Jia, LI Jiajun, ZHAO Jinqiu, XU Jing, HUANG Wenxiang. Quantitative analysis of differential proteins in liver tissues of patients with non-alcoholic steatohepatitis using iTRAQ technology [J]. Journal of Southern Medical University, 2021, 41(9): 1381-1387. |
| [15] | . Expression of nicotinamide-N-methyltransferase in gastric cancer and its biological and clinicopathological significance [J]. Journal of Southern Medical University, 2021, 41(6): 828-838. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||