南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (9): 1919-1926.doi: 10.12122/j.issn.1673-4254.2025.09.11
• • 上一篇
冯志惠1( ), 李文月2, 张铭修1, 王培培3, 帅阳阳3, 张宏1(
), 李文月2, 张铭修1, 王培培3, 帅阳阳3, 张宏1( )
)
收稿日期:2025-02-05
出版日期:2025-09-20
发布日期:2025-09-28
通讯作者:
张宏
E-mail:feng18740536649@163.com;zhanghong@chnu.edu.cn
作者简介:冯志惠,硕士,E-mail: feng18740536649@163.com
基金资助:
Zhihui FENG1( ), Wenyue LI2, Mingxiu ZHANG1, Peipei WANG3, Yangyang SHUAI3, Hong ZHANG1(
), Wenyue LI2, Mingxiu ZHANG1, Peipei WANG3, Yangyang SHUAI3, Hong ZHANG1( )
)
Received:2025-02-05
Online:2025-09-20
Published:2025-09-28
Contact:
Hong ZHANG
E-mail:feng18740536649@163.com;zhanghong@chnu.edu.cn
Supported by:摘要:
目的 观察长链非编码RNA ENSG00000271218.1(lncRNA HClnc1)对肝癌细胞增殖、侵袭和迁移的影响,探索其作用机制。 方法 基于TCGA数据库分析HClnc1在临床肝癌组织中的表达水平;BrdU掺入、平板克隆及Transwell方法检测干扰/过表达HClnc1及干扰ODC1对肝癌细胞增殖、侵袭和迁移的影响;BrdU参入和Transwell方法检测同时过表达HClnc1和干扰ODC1对肝癌细胞增殖、侵袭和迁移的影响;qRT-PCR和免疫印迹分析干扰HClnc1对肝癌细胞ODC1蛋白和mRNA表达的影响;双荧光素酶报告基因分析ODC1启动子活性;Pull-down、质谱分析和Co-IP鉴定与HClnc1结合蛋白及其相互作用;RNA干扰及ChIP分析等方法研究与ODC1启动子区域结合的蛋白及结合效率。 结果 HClnc1在肝癌组织中明显升高(P<0.001);干扰HClnc1后增殖、侵袭和迁移的肝癌细胞数量明显减少(P<0.001);过表达HClnc1则增加了增殖、侵袭和迁移的细胞数量(P<0.001);干扰ODC1后减少了增殖、侵袭和迁移细胞数量,且抵消了过表达HClnc1的促进作用(P<0.01);干扰HClnc1后ODC1启动子活性明显降低(P<0.001);HClnc1均能与RBBP5和KAT2B蛋白结合,干扰HClnc1后则阻止了RBBP5和KAT2B相互结合;过表达HClnc1上调ODC1蛋白表达,干扰RBBP5或KAT2B则下调ODC1蛋白表达,并且减少HClnc1诱导的ODC1蛋白上调;RBBP5和KAT2B能直接结合在ODC1的启动子区域,干扰KAT2B或RBBP5则降低结合效率(P<0.001),而干扰HClnc1则会减少RBBP5和KAT2B与ODC1启动子区域的结合(P<0.001)。 结论 lncRNA HClnc1通过靶向RBBP5/KAT2B表观遗传修饰复合物,增加ODC1启动子活性,提高ODC1转录水平,从而促进肝癌细胞的生长和转移。
冯志惠, 李文月, 张铭修, 王培培, 帅阳阳, 张宏. lncRNA HClnc1通过靶向RBBP5/KAT2B复合物增强ODC1转录促进肝癌细胞生长和转移[J]. 南方医科大学学报, 2025, 45(9): 1919-1926.
Zhihui FENG, Wenyue LI, Mingxiu ZHANG, Peipei WANG, Yangyang SHUAI, Hong ZHANG. Long noncoding RNA HClnc1 promotes proliferation and migration of liver cancer cells by targeting RBBP5/KAT2B complex to enhance ODC1 transcription[J]. Journal of Southern Medical University, 2025, 45(9): 1919-1926.
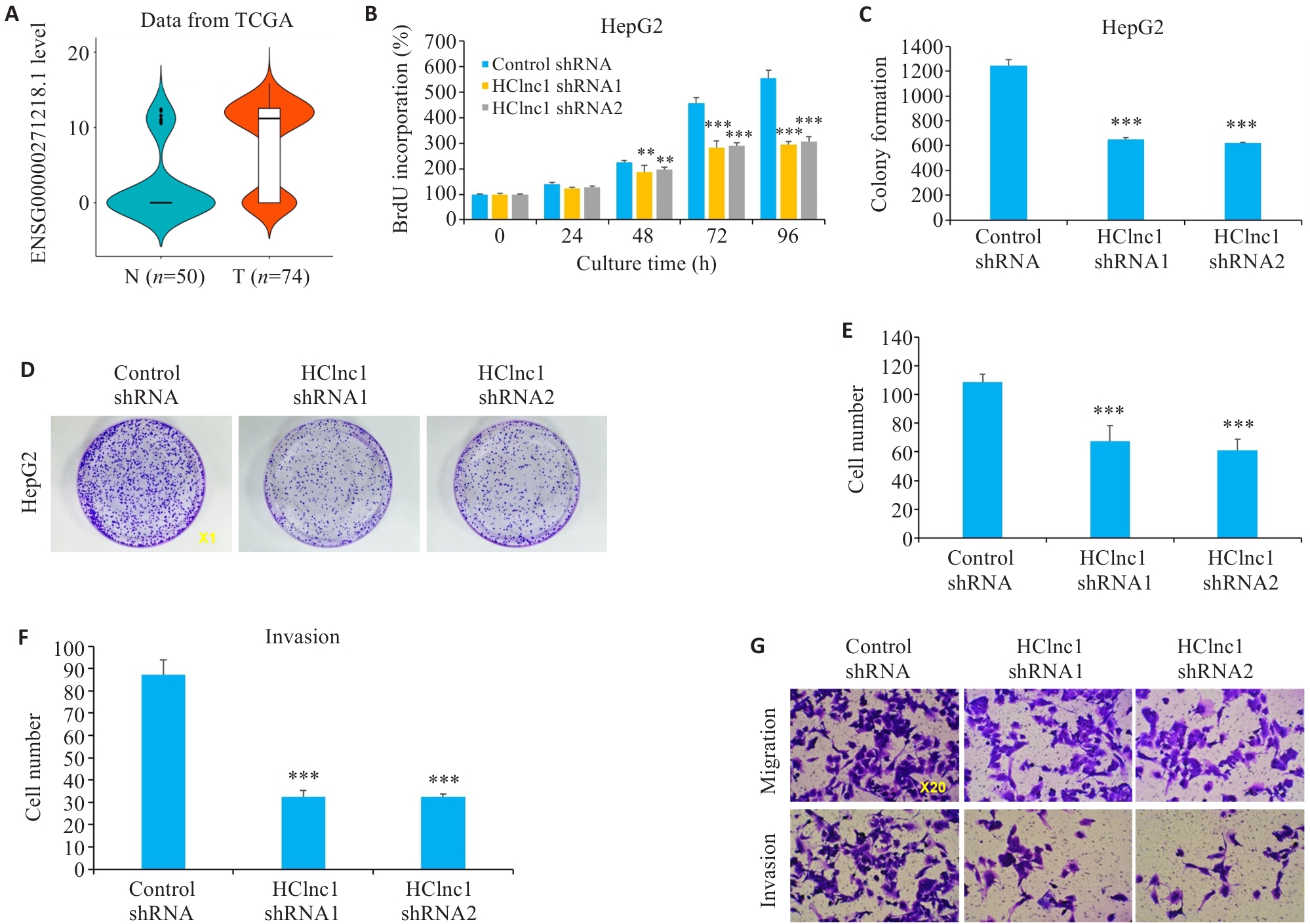
图1 干扰HClnc1对肝癌细胞增殖、侵袭和迁移的影响
Fig.1 Effects of HClnc1 silencing on proliferation, invasion, and migration of liver cancer cells. A: Expression of HClnc1 in HCC (T) and adjacent regular tissues (N) from TCGA database. B: Proliferation of HepG2 cells assessed using BrdUrd assay. C, D: Colony formation of HepG2 cells (Original magnification:×1). E-G: Migration and invasion of HepG2 cells assessed using Transwell assay. **P<0.01, ***P<0.001 vs control group.
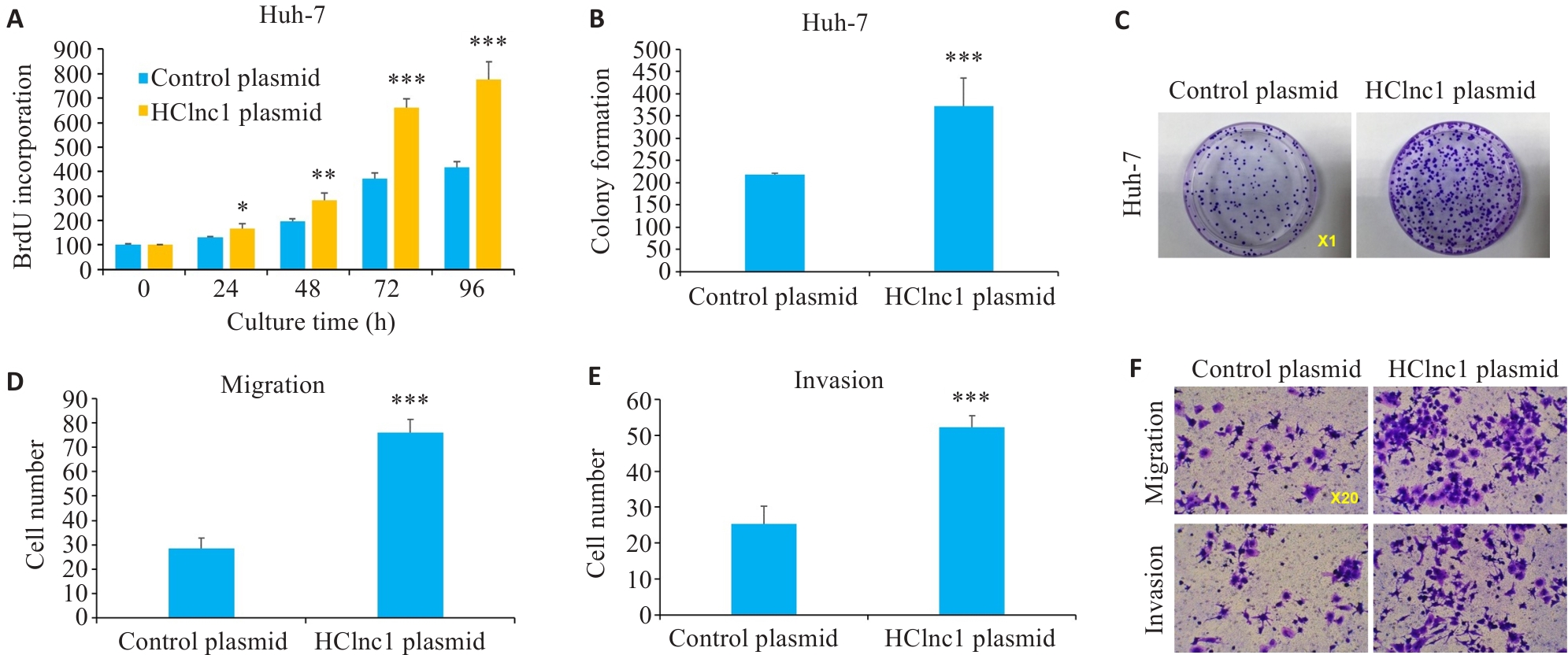
图2 过表达HClnc1对肝癌细胞增殖、侵袭和迁移的影响
Fig.2 Effects of HClnc1 overexpression on proliferation, invasion, and migration of liver cancer cells. A: Proliferation of Huh7 cells assessed using BrdUrd assay. B, C: Colony formation of Huh7 cells. D-F: Migration and invasion of Huh7 cells assessed using Transwell assay. *P<0.05, **P<0.01, ***P<0.001 vs control group.
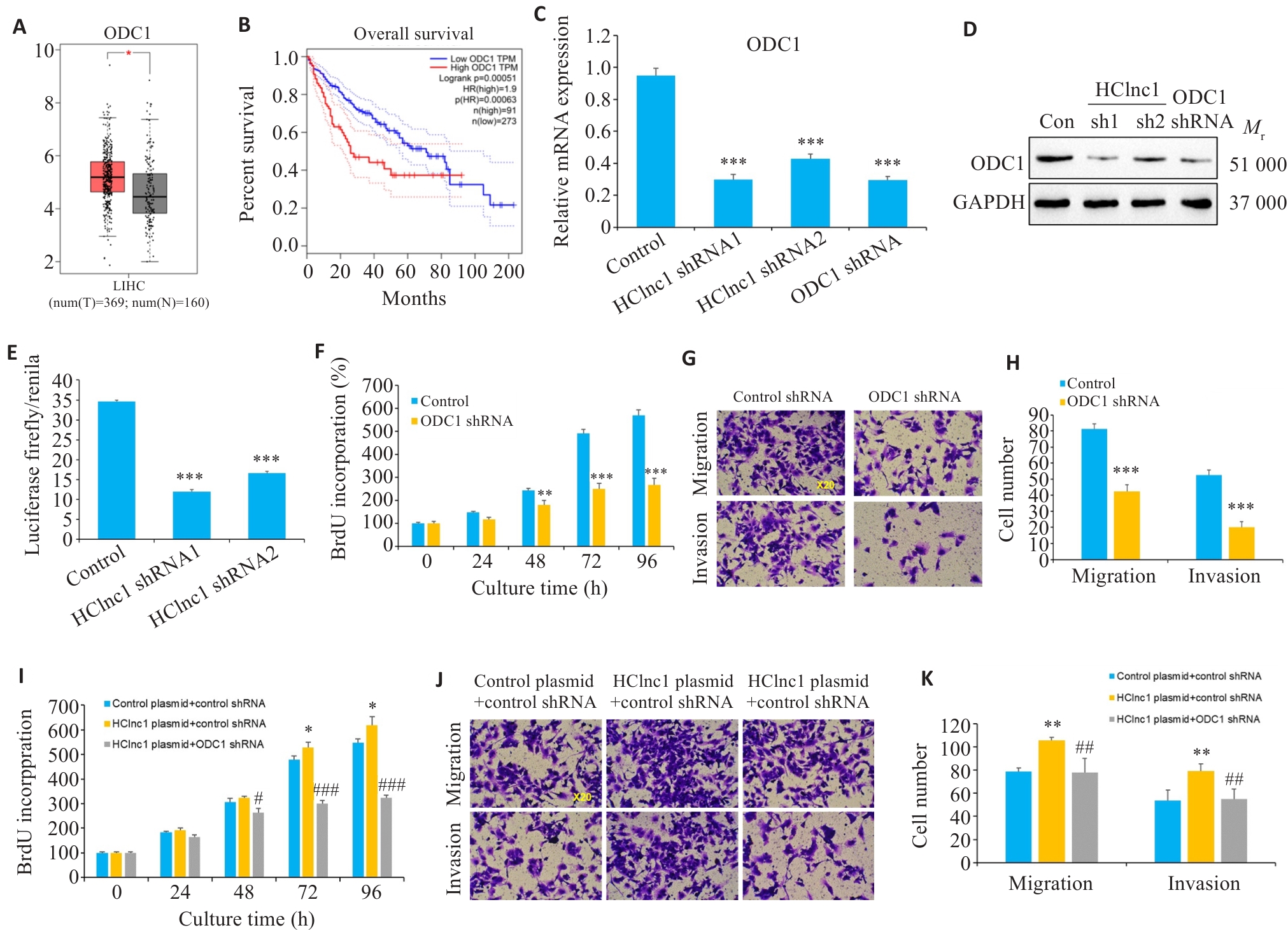
图3 干扰ODC1逆转HClnc1诱导的肝癌细胞增殖、侵袭和迁移
Fig.3 ODC1 knockdown reverses HClnc1-induced proliferation, invasion, and migration of hepatocellular carcinoma (HCC) cells. A, B: Gene expression levels of ODC1 in HCC and normal tissues from TCGA clinical samples and their correlation with survival. C, D: Relative mRNA and protein expression levels of ODC1 in HepG2 cells transfected with control, HClnc1 shRNA1/2, or ODC1 shRNA. E: ODC1 promoter activity assessed by luciferase activity assay. F: Proliferation of HepG2 cells assessed using BrdUrd assay. G-H: Migration and invasion of HepG2 cells assessed using Transwell assay (×20). I-K: HepG2 cells were co-transfected with control or HClnc1 plasmid plus control or ODC1 shRNA. I: Proliferation of HepG2 cells assessed using BrdUrd assay. J-K: Migration and invasion of HepG2 cells assessed using Transwell assay (×20). *P<0.05, **P<0.01, ***P<0.001 vs control or control plasmid +control shRNA. #P<0.05, ##P<0.01, ###P<0.001 vs HClnc1 plasmid+control shRNA.
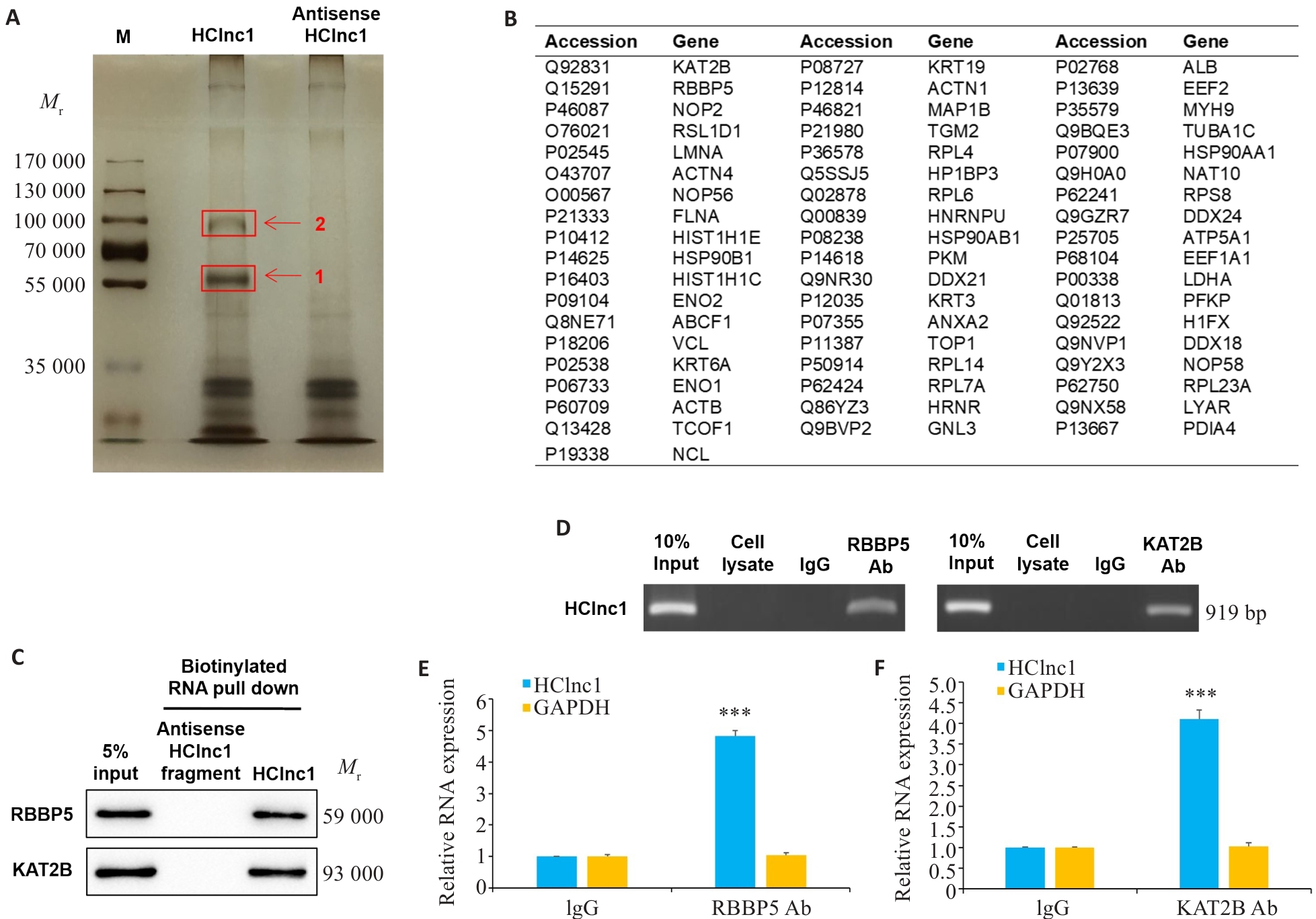
图4 HClin1与RBBP5和KAT2B蛋白结合
Fig.4 HClin1 binds to RBBP5 and KAT2B proteins. A: Silver staining of biotinylated HClnc1-associated proteins. The proteins were excised from the two HClnc1-specific bands (red arrows) for mass spectrometry analysis. B: 55 identified proteins. C: Western blotting of proteins from antisense HClnc1 and HClnc1 pull-down assay. D: RNA immunoprecipitation with anti-RBBP5 or anti-KAT2B antibody. E, F: Relative RNA levels of HClnc1 and GAPDH detected using real-time PCR assay. The nuclear fraction of HepG2 cells was immunoprecipitated with anti-RBBP5, anti-KAT2B, or IgG control antibodies. ***P<0.001 vs IgG.
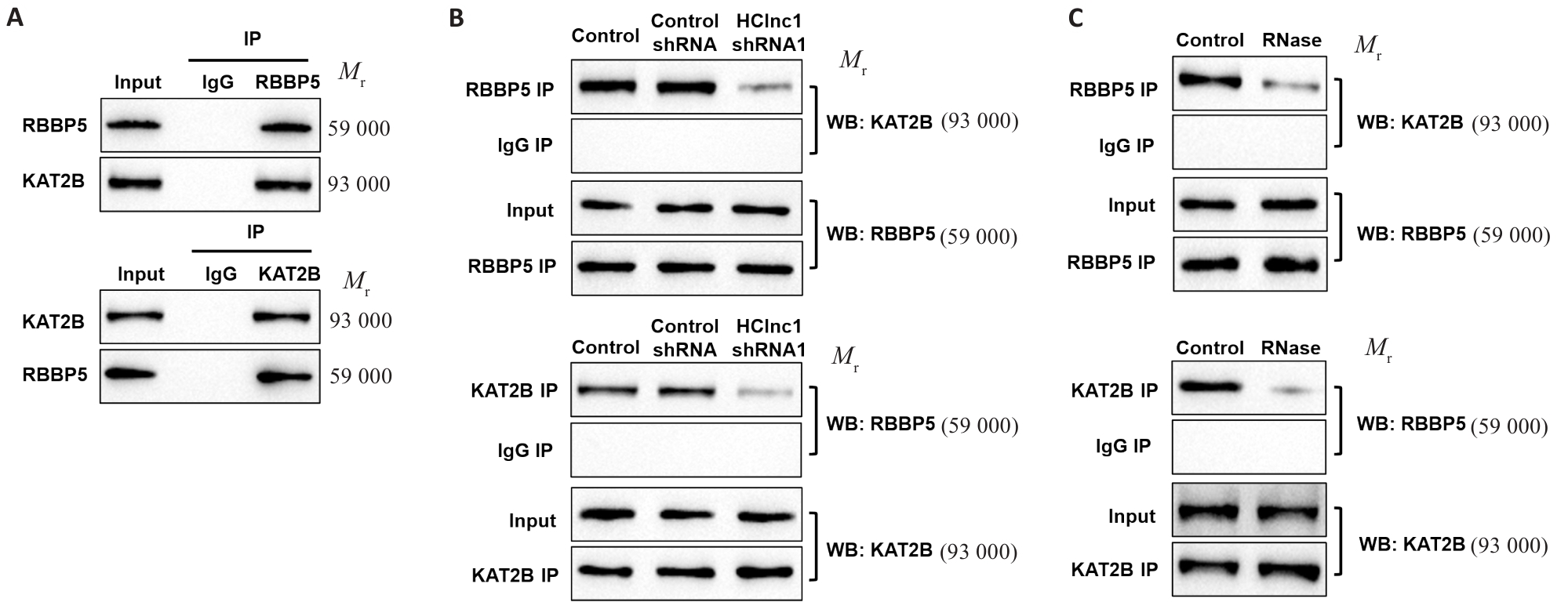
图5 干扰HClnc1阻止RBBP5和KAT2B蛋白相互结合
Fig.5 Knockdown of HClnc1 disrupts the protein-protein interaction between RBBP5 and KAT2B. A: Co-immunoprecipitation of RBBP5 and KAT2B in HepG2 cells. B, C: Interaction of RBBP5 and KAT2B after HClnc1 shRNA transfection (B) or RNase treatment (C).
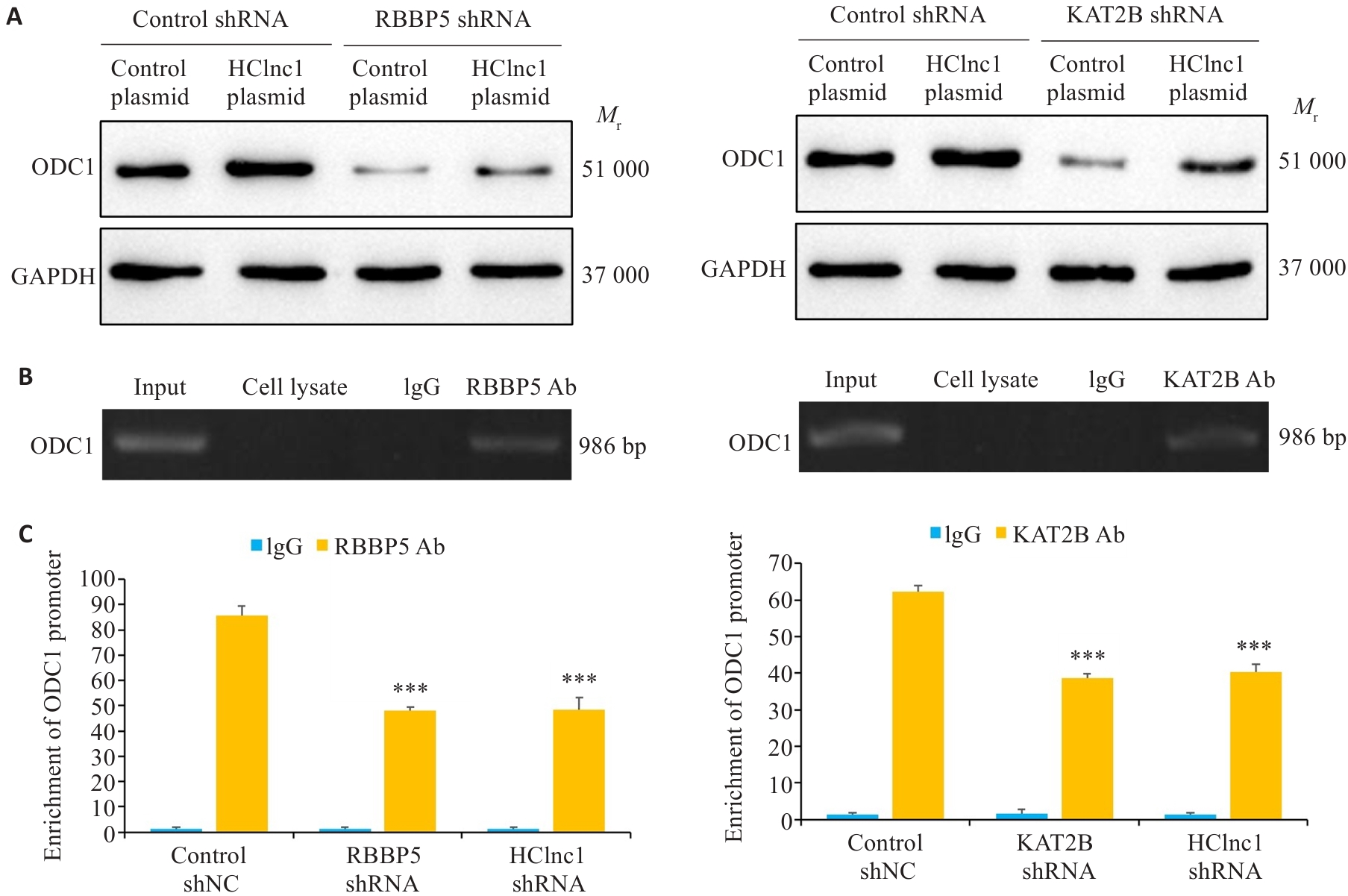
图6 HClnc1通过链接RBBP5/KAT2B调控ODC1转录
Fig.6 HClnc1 regulates the transcription of ODC1 by connecting RBBP5 and KAT2B. A: ODC1 protein levels in HepG2 cells transfected with control shRNA or RBBP5/KAT2B shRNA detected using Western blotting (with GAPDH as the internal control). B: Detection of ODC1 DNA in anti-RBBP5 or anti-KAT2B antibody-immunoprecipitated chromatin of HepG2 cells. C: ODC1 gene promoter levels in chromatin immunoprecipitation samples detected using real-time PCR analysis. ***P<0.001 vs control shNC or control plasmid.
| [1] | Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2024, 74(3): 229-63. doi:10.3322/caac.21834 |
| [2] | Han BF, Zheng RS, Zeng HM, et al. Cancer incidence and mortality in China, 2022[J]. J Natl Cancer Cent, 2024, 4(1): 47-53. doi:10.1016/j.jncc.2024.01.006 |
| [3] | Wen NY, Cai YL, Li FY, et al. The clinical management of hepatocellular carcinoma worldwide: a concise review and comparison of current guidelines: 2022 update[J]. Biosci Trends, 2022, 16(1): 20-30. doi:10.5582/bst.2022.01061 |
| [4] | Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma[J]. Nat Rev Dis Primers, 2021, 7: 6. doi:10.1038/s41572-020-00240-3 |
| [5] | Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome[J]. Nat Genet, 2015, 47(3): 199-208. doi:10.1038/ng.3192 |
| [6] | Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways[J]. Cancer Cell, 2016, 29(4): 452-63. doi:10.1016/j.ccell.2016.03.010 |
| [7] | Kim MY. Long non-coding RNAs in cancer[J]. Non Coding RNA Res, 2019, 4(2): 45. doi:10.1016/j.ncrna.2019.02.003 |
| [8] | Coan M, Haefliger S, Ounzain S, et al. Targeting and engineering long non-coding RNAs for cancer therapy[J]. Nat Rev Genet, 2024, 25(8): 578-95. doi:10.1038/s41576-024-00693-2 |
| [9] | Huang Z, Zhou JK, Peng Y, et al. The role of long noncoding RNAs in hepatocellular carcinoma[J]. Mol Cancer, 2020, 19(1): 77. doi:10.1186/s12943-020-01188-4 |
| [10] | Peng L, Yuan XQ, Zhang CY, et al. The emergence of long non-coding RNAs in hepatocellular carcinoma: an update[J]. J Cancer, 2018, 9(14): 2549-58. doi:10.7150/jca.24560 |
| [11] | Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets[J]. J Hepatol, 2017, 67(3): 603-18. doi:10.1016/j.jhep.2017.04.009 |
| [12] | 李菲凡, 向俊馨, 刘佳慧, 等. LncRNA FEZF1-AS1通过miR-130a-5p/CCND1轴促进非小细胞肺癌发展的分子机制研究[J]. 南方医科大学学报, 2024, 44(5): 841-50. |
| [13] | 冯 雯, 赖跃兴, 王 静, 等. 长链非编码RNA ABHD11-AS1促进胃癌细胞糖酵解并加速肿瘤恶性进展[J]. 南方医科大学学报, 2023, 43(9): 1485-92. |
| [14] | Deng YQ, Gao M, Lu D, et al. Compound-composed Chinese medicine of Huachansu triggers apoptosis of gastric cancer cells through increase of reactive oxygen species levels and suppression of proteasome activities[J]. Phytomedicine, 2024, 123: 155169. doi:10.1016/j.phymed.2023.155169 |
| [15] | Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern[J]. Cancer Discov, 2016, 6(7): 784-801. doi:10.1158/2159-8290.cd-15-0921 |
| [16] | Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway[J]. Biomed Pharm-acother, 2016, 83: 450-5. doi:10.1016/j.biopha.2016.06.056 |
| [17] | Chen ZL, Yu C, Zhan L, et al. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384[J]. Am J Cancer Res, 2016, 6(10): 2299-309. |
| [18] | Huang RY, Wang XC, Zhang WJ, et al. Down-regulation of LncRNA DGCR5 correlates with poor prognosis in hepatocellular carcinoma[J]. Cell Physiol Biochem, 2016, 40(3/4): 707-15. doi:10.1159/000452582 |
| [19] | Xiao CH, Wang C, Cheng SJ, et al. The significance of low levels of LINC RP1130-1 expression in human hepatocellular carcinoma[J]. Biosci Trends, 2016, 10(5): 378-85. doi:10.5582/bst.2016.01123 |
| [20] | Fei MM, Li XY, Liang SH, et al. LncRNA PWRN1 inhibits the progression of hepatocellular carcinoma by activating PKM2 activity[J]. Cancer Lett, 2024, 584: 216620. doi:10.1016/j.canlet.2024.216620 |
| [21] | Zhang X, Li ZL, Nie HZ, et al. The IGF2BP2-lncRNA TRPC7-AS1 axis promotes hepatocellular carcinoma cell proliferation and invasion[J]. Cell Signal, 2024, 117: 111078. doi:10.1016/j.cellsig.2024.111078 |
| [22] | Cha H, Kim M, Ahn N, et al. Role of UPF1 in lncRNA-HEIH regulation for hepatocellular carcinoma therapy[J]. Exp Mol Med, 2024, 56(2): 344-54. doi:10.1038/s12276-024-01158-6 |
| [23] | Ma K, Chu JH, Liu YF, et al. Hepatocellular carcinoma LINC01116 outcompetes T cells for linoleic acid and accelerates tumor progression[J]. Adv Sci (Weinh), 2024, 11(21): e2400676. doi:10.1002/advs.202400676 |
| [24] | Li XY, Liang SH, Fei MM, et al. LncRNA CRNDE drives the progression of hepatocellular carcinoma by inducing the immunosuppressive niche[J]. Int J Biol Sci, 2024, 20(2): 718-32. doi:10.7150/ijbs.85471 |
| [25] | Casero RA, Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities[J]. Nat Rev Cancer, 2018, 18(11): 681-95. doi:10.1038/s41568-018-0050-3 |
| [26] | Zhang XP, Zou WB, Li ZQ, et al. The heterogeneity of cellular metabolism in the tumour microenvironment of hepatocellular carcinoma with portal vein tumour thrombus[J]. Cell Prolif, 2025, 58(1): e13738. doi:10.1111/cpr.13738 |
| [27] | Ji GY, Liu J, Zhao ZQ, et al. Polyamine anabolism promotes chemotherapy-induced breast cancer stem cell enrichment[J]. Adv Sci (Weinh), 2024, 11(40): e2404853. doi:10.1002/advs.202470240 |
| [28] | Li YP, Huang ZJ, He QK, et al. Pirin promotes the progression of non-small-cell lung cancer by increasing ODC1 to suppress autophagy[J]. J Proteome Res, 2024, 23(5): 1713-24. doi:10.1021/acs.jproteome.3c00871 |
| [29] | Ye Z, Zeng ZR, Shen YY, et al. ODC1 promotes proliferation and mobility via the AKT/GSK3β/β-catenin pathway and modulation of acidotic microenvironment in human hepatocellular carcinoma[J]. Onco Targets Ther, 2019, 12: 4081-92. doi:10.2147/ott.s198341 |
| [30] | Meng XQ, Peng JX, Xie XS, et al. Roles of lncRNA LVBU in regulating urea cycle/polyamine synthesis axis to promote colorectal carcinoma progression[J]. Oncogene, 2022, 41(36): 4231-43. doi:10.1038/s41388-022-02413-8 |
| [31] | Choi Y, Oh ST, Won MA, et al. Targeting ODC1 inhibits tumor growth through reduction of lipid metabolism in human hepatocellular carcinoma[J]. Biochem Biophys Res Commun, 2016, 478(4): 1674-81. doi:10.1016/j.bbrc.2016.09.002 |
| [32] | Zhang HX, Li XR, Liu ZY, et al. Elevated expression of HIGD1A drives hepatocellular carcinoma progression by regulating polyamine metabolism through c-Myc-ODC1 nexus[J]. Cancer Metab, 2024, 12(1): 7. doi:10.1186/s40170-024-00334-6 |
| [33] | Zhang GX, Lan YJ, Xie AM, et al. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements[J]. J Biol Chem, 2019, 294(43): 15613-22. doi:10.1074/jbc.ra119.008732 |
| [34] | Zhang P, Chaturvedi CP, Tremblay V, et al. A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation[J]. Genes Dev, 2015, 29(2): 123-8. doi:10.1101/gad.254870.114 |
| [35] | Fournier M, Orpinell M, Grauffel C, et al. KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification[J]. Nat Commun, 2016, 7: 13227. doi:10.1038/ncomms13227 |
| [36] | He BY, Pan HL, Zheng FQ, et al. Long noncoding RNA LINC00930 promotes PFKFB3-mediated tumor glycolysis and cell proliferation in nasopharyngeal carcinoma[J]. J Exp Clin Cancer Res, 2022, 41(1): 77. doi:10.1186/s13046-022-02282-9 |
| [37] | Fang M, Zhang MJ, Wang YQ, et al. Long noncoding RNA AFAP1-AS1 is a critical regulator of nasopharyngeal carcinoma tumo-rigenicity[J]. Front Oncol, 2020, 10: 601055. doi:10.3389/fonc.2020.601055 |
| [1] | 张兆君, 吴琼, 谢苗苗, 叶洳吟, 耿晨晨, 石纪雯, 杨清玲, 王文锐, 石玉荣. 层状双氢氧化物负载si-NEAT1通过miR-133b/PD-L1轴调控乳腺癌紫杉醇耐药及巨噬细胞极化[J]. 南方医科大学学报, 2025, 45(8): 1718-1731. |
| [2] | 于滢, 涂丽, 刘洋, 宋雪翼, 邵倩倩, 唐小龙. TGF-β通过miR-23a-3p/IRF1轴下调主要组织相容性复合体I类表达促进肝癌免疫逃逸[J]. 南方医科大学学报, 2025, 45(7): 1397-1408. |
| [3] | 龚秀莹, 侯顺福, 赵苗苗, 王晓娜, 张致涵, 刘清华, 尹崇高, 李洪利. LncRNA SNHG15通过miR-30b-3p调控COX6B1轴促进肺腺癌细胞增殖、迁移和侵袭的分子机制[J]. 南方医科大学学报, 2025, 45(7): 1498-1505. |
| [4] | 宋添力, 王一民, 孙童, 刘绪, 黄胜, 冉云. 正肝方对二乙基亚硝胺诱导的肝癌大鼠的抗癌作用及机制:基于激活Hippo/YAP通路[J]. 南方医科大学学报, 2025, 45(4): 799-809. |
| [5] | 邹金华, 王惠, 张冬艳. SLC1A5通过促进M2型巨噬细胞极化促进肝癌进展[J]. 南方医科大学学报, 2025, 45(2): 269-284. |
| [6] | 徐朦, 陈丽娜, 吴金玉, 刘丽丽, 施美, 周灏, 张国梁. “白花蛇舌草-半枝莲”治疗原发性肝癌的机制研究:基于网络药理学、分子对接及体外实验验证[J]. 南方医科大学学报, 2025, 45(1): 80-89. |
| [7] | 张力苹, 刘喜娟, 胡潇, 王嘉丽, 余锡贺, 栗国梁, 游海敏, 张启周, 张海波. 经动脉化疗栓塞续贯肝动脉灌注化疗联合TKI和PD-1单抗在晚期肝癌一线治疗中的疗效观察[J]. 南方医科大学学报, 2024, 44(9): 1831-1838. |
| [8] | 何华星, 刘璐琳, 刘颖茵, 陈纳川, 孙素霞. 丁酸钠与索拉非尼可能通过YAP诱导铁死亡协同抑制肝癌细胞增殖[J]. 南方医科大学学报, 2024, 44(7): 1425-1430. |
| [9] | 陈 浩, 李振汉, 王明婷, 卢林明, 唐乾利, 罗良平. 高表达UBE2S通过增加癌细胞干性促进肝癌的进程机制[J]. 南方医科大学学报, 2024, 44(3): 455-464. |
| [10] | 赵培培, 周志刚, 杨媛媛, 黄树升, 涂逸轩, 涂剑. 铁死亡诱导剂Erastin下调ACSL4抑制肝癌细胞体外增殖[J]. 南方医科大学学报, 2024, 44(11): 2131-2136. |
| [11] | 黄萃园, 孙运平, 李文强, 刘丽, 王伟, 张静. Nlrp6过表达通过调控AMPK-Srebp1c轴抑制脂质合成抑制肝癌细胞的增殖[J]. 南方医科大学学报, 2024, 44(10): 1910-1917. |
| [12] | 范喜瑞, 戚之琳, 邓园洁, 杨子晗, 孙丽, 李国豪, 梁娟娟, 吴菲, 袁力文. LncRNA MAGI2-AS3通过靶向调控miR-1269a/PTEN/AKT通路增强非小细胞肺癌对顺铂化疗的敏感性[J]. 南方医科大学学报, 2024, 44(10): 2033-2043. |
| [13] | 孔 祥, 张 腾, 张 妍, 高灵犀, 汪 文, 汪梦燕, 王国栋, 吕 坤. 过表达lncRNA HEM2M改善非酒精性脂肪肝病小鼠的肝脏损伤[J]. 南方医科大学学报, 2024, 44(1): 1-8. |
| [14] | 罗 瑞, 田龙海, 杨永曜. 高良姜素通过下调lncRNA H19的表达抑制ox-LDL诱导的人源正常主动脉内皮细胞血管生成活性[J]. 南方医科大学学报, 2024, 44(1): 52-59. |
| [15] | 辛 辰, 王笑影, 李 响, 陈 宇, 王 雪, 宁佳曦, 杨 适, 王忠琼. LncRNA SOX2OT靶向SIRT1/自噬通路增强胆管癌细胞5-FU耐药[J]. 南方医科大学学报, 2024, 44(1): 187-193. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||