南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (8): 1643-1653.doi: 10.12122/j.issn.1673-4254.2025.08.09
• • 上一篇
陈子贤( ), 周家伟, 谭磊, 黄志鹏, 薛康颐, 陈明坤(
), 周家伟, 谭磊, 黄志鹏, 薛康颐, 陈明坤( )
)
收稿日期:2025-04-06
出版日期:2025-08-20
发布日期:2025-09-05
通讯作者:
陈明坤
E-mail:czx961147842@163.com;chenmk1@smu.edu.cn
作者简介:陈子贤,在读硕士研究生,E-mail: czx961147842@163.com
基金资助:
Zixian CHEN( ), Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN(
), Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN( )
)
Received:2025-04-06
Online:2025-08-20
Published:2025-09-05
Contact:
Mingkun CHEN
E-mail:czx961147842@163.com;chenmk1@smu.edu.cn
Supported by:摘要:
目的 鉴定前列腺癌(PCa)患者免疫抑制性中性粒细胞亚群并构建基于中性粒细胞亚群相关的免疫预后风险模型。 方法 从基因表达综合数据库及癌症基因组图谱数据库收集PCa患者单细胞、转录组数据,通过无监督聚类鉴定前列腺癌中性粒细胞亚群,通过功能富集、细胞互作、伪时序分析鉴定中性粒细胞亚群的生物学功能及对患者免疫调控的影响;通过LASSO-Cox回归构建免疫抑制性中性粒细胞亚群相关预后风险模型,通过生存分析、ROC曲线探讨高低风险组预后差异,采用CIBERSORT、TIDE评分分析预后风险模型与PCa免疫浸润及免疫应答的关系。 结果 和邻近正常组织相比,PCa组织内中性粒细胞浸润比例显著增加(P<0.05)。PCa相关中性粒细胞可聚类为2个独立细胞亚群:Neu_1和Neu_2,其中Neu_2细胞表现为高富集的免疫调节功能和分化成熟状态,并上调TGFB1、ITGB2、LGALS3等免疫抑制性细胞因子;基于Neu_2细胞亚群基因特征构建免疫相关预后风险模型、生存分析和免疫差异分析显示,高风险组患者具有更短的生化复发时间(P<0.05)和有更高比例的Tregs、M2-TAMs细胞浸润(P<0.05);TIDE分析显示,高风险组患者具有免疫排斥和更差的免疫应答评分。 结论 PCa相关中性粒细胞存在显著异质性,基于免疫抑制特征的Neu_2细胞群构建的相关预后风险模型可有效预测PCa患者生存预后及免疫应答反应。
陈子贤, 周家伟, 谭磊, 黄志鹏, 薛康颐, 陈明坤. 基于免疫抑制性Neu_2中性粒细胞亚群模型精准预测前列腺癌生存预后及免疫治疗应答[J]. 南方医科大学学报, 2025, 45(8): 1643-1653.
Zixian CHEN, Jiawei ZHOU, Lei TAN, Zhipeng HUANG, Kangyi XUE, Mingkun CHEN. A risk prediction model for prognosis and immunotherapy response in prostate cancer patients based on immunosuppressive neutrophil Neu_2 subsets[J]. Journal of Southern Medical University, 2025, 45(8): 1643-1653.
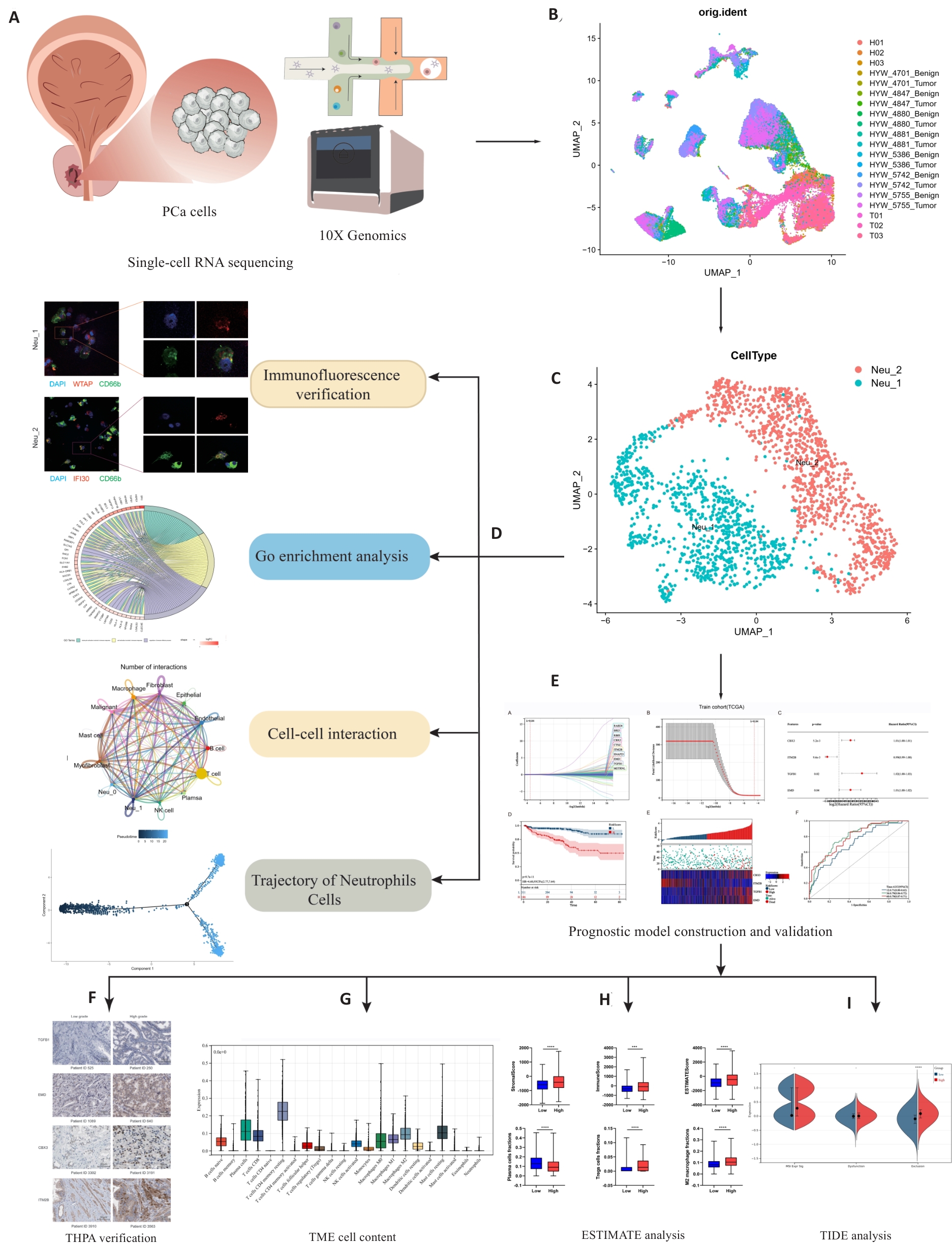
图1 本研究的工作流程
Fig.1 Workflow of this study. Single-cell sequencing data were obtained from principal components analysis (A), followed by annotation and clustering of the data (B), resulting in the identification of two types of neutrophils: Neu_1 and Neu_2 (C). Tissue immunofluorescence verification, Gene Ontology (GO) enrichment analysis, cell interaction studies, and counter-time sequence analysis of these two cell types were used to elucidate their roles within the tumor microenvironment (TME) (D). Combining our findings with data from the TCGA database, we identified 4 independent prognostic factors for constructing and validating the prognostic models (E, F). We performed an analysis of immunotherapy predictions by analyzing and scoring the proportion of immune cells within the TME (G, H). ***P<0.001, ****P<0.0001. The patients with high levels of infiltrating Neu_2 neutrophils are likely to have poor responses to immunotherapy, as indicated by TIDE analysis (I). THPA: The Human Protein Atlas; ESTIMATE: Estimation of stromal and immune cells in malignant tumor tissues using expression data; TIDE: Tumor immune dysfunction and exclusion.

图2 PCa scRNA-Seq数据的集成和聚类
Fig.2 Integration and clustering of PCa scRNA-Seq data. A: t-SNE of 15 PCa samples. B: t-SNE of 15 cell clusters. C: Identification of 12 cell types by marker genes. D: Cell types exist in different samples. E: Dot plot showed the expression differences of various genes across the 12 cell types. F: Expression differences of 12 cell types between the control and tumor groups. G: Heat map showing expressions of the characteristic genes across different cell subpopulations.

图3 中性粒细胞亚型的细胞图谱
Fig.3 Cell map of neutrophil subtypes. A: t-SNE of the 20 cell clusters. B: t-SNE of control group and tumor group. C: Neu_1 and Neu_2 neutrophils identified by marker genes. D: Heat map showing differential expressions of the marker genes between Neu_1 and Neu_2 neutrophils. E: Expressions of the marker genes as signatures of the two cell types. F: GO enrichment analysis of signaling pathways associated with Neu_1 and Neu_2 neutrophils. G: Immunofluorescent staining of the marker genes in a subset of neutrophils (CD66b), specifically WTAP in Neu_1 and IIFI30 in Neu_2, within prostate cancer (PCa) tissue. H: GO enrichment analysis chord diagram Neu_1 signaling pathways involved in neutrophils. I: GO enrichment analysis chord diagram Neu_2 signaling pathways involved in neutrophils. J: KEGG analysis of signaling pathways involved in Neu_1 and Neu_2 neutrophils.

图4 与中性粒细胞相关的细胞间通讯分析
Fig.4 Analysis of intercellular communication related to neutrophils. A: Number of interactions in the intercellular communication network. B: Interaction weights/strengths in intercellular communication networks. C: Neu_1 interaction between neutrophils and other cells. D: Neu_2 interaction between neutrophils and other cells. E: Number and intensity of interactions between Neu_1 neutrophils and different cell types. F: Number and intensity of interactions between Neu_2 neutrophils and different cell types. G: Bubble diagram of ligand-receptor pair-mediated interactions between Neu_1 cells and Neu_2 neutrophils and other cells.

图6 中性粒细胞预后风险模型的构建和验证
Fig.6 Construction and validation of a neutrophil prognostic risk model. A, B: Screening of prognostic-related core genes by lasso-cox regression in TCGA training group. C, G: Forest diagram in TCGA training group and GSE70770 validation set. D, H: Kaplan-Meier curve for overall survival between different ICPI risk groups in TCGA training group and GSE70770 validation set. E, I: Validation of centralized risk scores and expression heat maps of 4 genes in TCGA training group and GSE70770 validation set. F, J: Time-dependent ROC curve analysis in TCGA training group and GSE70770 validation set. K: Verification of expressions of the 4 key genes in PCa tissues by immunohistochemical staining from THPA database.
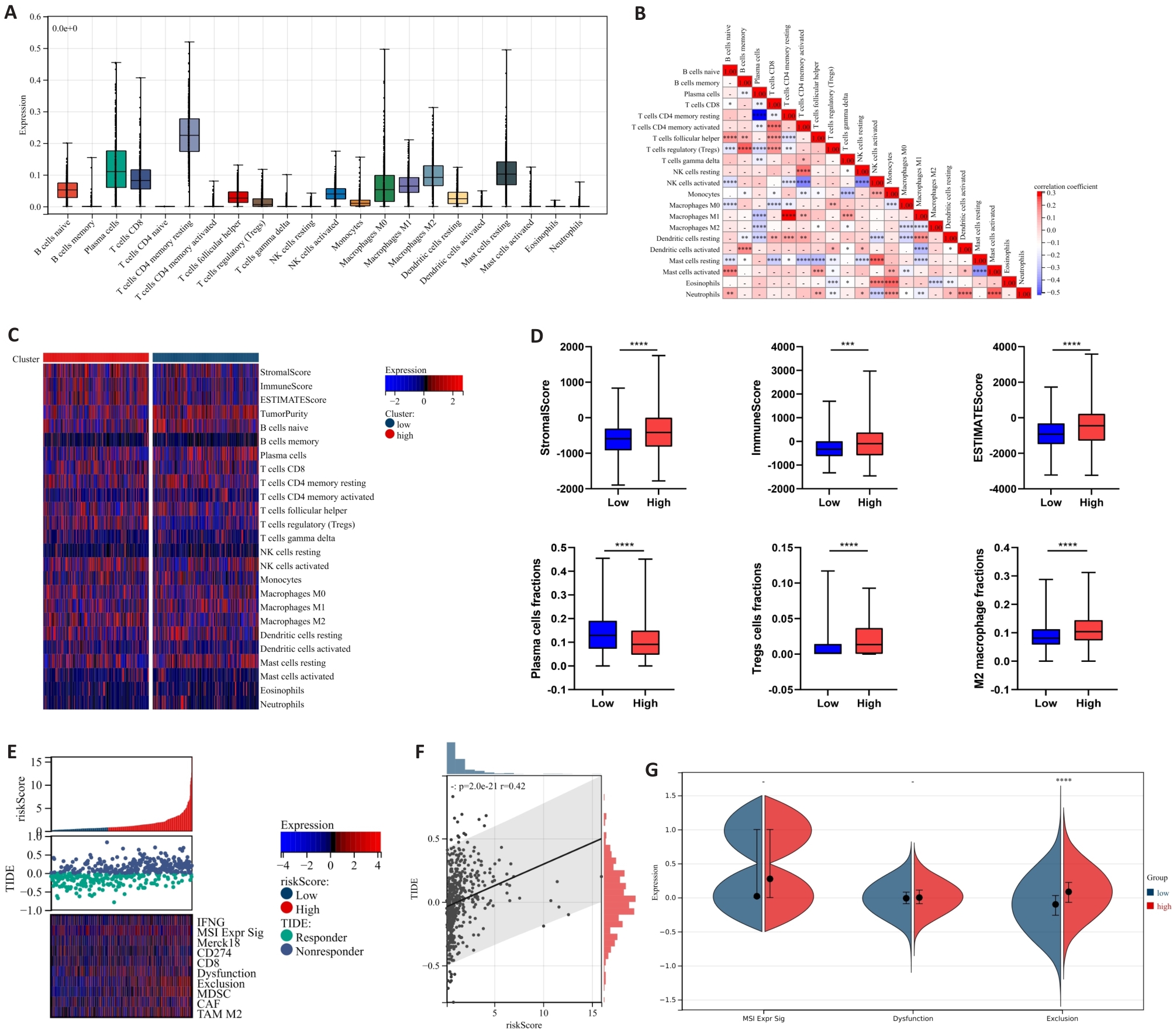
图7 中性粒细胞风险预后模型与PCa免疫浸润及免疫应答的相关性
Fig.7 Correlation between neutrophil risk prognostic model and immune infiltration as well as immune response in PCa. A: Calculation of 22 immune cell infiltration ratios in PCa tissues based on CIBERSORT. B: Correlation analysis between immune cells in PCa tissues. C: Differences in immune cell infiltration expression between high-risk and low-risk groups. D: Differences in immune scores and infiltration ratios of some immune cells (plasma cells, Tregs cells, and M2-TAMs cells) between high-risk and low-risk groups. E, F: Correlation analysis between TIDE score expression and risk score. G: Differences in microsatellite instability, immune dysfunction and immune rejection scores between high-risk and low-risk groups. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
| [1] | Davidsson S, Fiorentino M, Andrén O, et al. Inflammation, focal atrophic lesions, and prostatic intraepithelial neoplasia with respect to risk of lethal prostate cancer[J]. Cancer Epidemiol Biomarkers Prev, 2011, 20(10): 2280-7. doi:10.1158/1055-9965.epi-11-0373 |
| [2] | Conti DV, Darst BF, Moss LC, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction[J]. Nat Genet, 2021, 53(1): 65-75. doi:10.1038/s41588-021-00786-2 |
| [3] | Nicholson LT, Fong L. Immune checkpoint inhibition in prostate cancer[J]. Trends Cancer, 2020, 6(3): 174-7. doi:10.1016/j.trecan.2020.01.003 |
| [4] | Pang K, Shi ZD, Wei LY, et al. Research progress of therapeutic effects and drug resistance of immunotherapy based on PD-1/PD-L1 blockade[J]. Drug Resist Updat, 2023, 66: 100907. doi:10.1016/j.drup.2022.100907 |
| [5] | Riley RS, June CH, Langer R, et al. Delivery technologies for cancer immunotherapy[J]. Nat Rev Drug Discov, 2019, 18(3): 175-96. doi:10.1038/s41573-018-0006-z |
| [6] | Iacovelli R, Ciccarese C, Brunelli M, et al. PD-L1 expression in de novo metastatic castration-sensitive prostate cancer[J]. J Immunother, 2019, 42(7): 269-73. doi:10.1097/cji.0000000000000287 |
| [7] | Cong LL, Zhao Q, Sun HY, et al. A novel long non-coding RNA SLNCR1 promotes proliferation, migration, and invasion of melanoma via transcriptionally regulating SOX5[J]. Cell Death Discov, 2024, 10(1): 160. doi:10.1038/s41420-024-01922-7 |
| [8] | Morimoto K, Yamada T, Takayama K. The landscape of immune therapy in vulnerable patients with advanced non-small cell lung cancer: a narrative review[J]. Transl Lung Cancer Res, 2023, 12(11): 2310-21. doi:10.21037/tlcr-23-581 |
| [9] | Bian XJ, Wang WF, Abudurexiti M, et al. Integration analysis of single-cell multi-omics reveals prostate cancer heterogeneity[J]. Adv Sci (Weinh), 2024, 11(18): e2305724. doi:10.1002/advs.202305724 |
| [10] | Bancaro N, Calì B, Troiani M, et al. Apolipoprotein E induces pathogenic senescent-like myeloid cells in prostate cancer[J]. Cancer Cell, 2023, 41(3): 602-19. e11. doi:10.1016/j.ccell.2023.02.004 |
| [11] | Angappulige DH, Mahajan NP, Mahajan K. Epigenetic underpinnings of tumor-immune dynamics in prostate cancer immune suppression[J]. Trends Cancer, 2024, 10(4): 369-81. doi:10.1016/j.trecan.2024.01.004 |
| [12] | Qin Y, Liu YL, Xiang XY, et al. Cuproptosis correlates with immunosuppressive tumor microenvironment based on pan-cancer multiomics and single-cell sequencing analysis[J]. Mol Cancer, 2023, 22(1): 59. doi:10.1186/s12943-023-01752-8 |
| [13] | Zhang YJ, Wang D, Peng M, et al. Single-cell RNA sequencing in cancer research[J]. J Exp Clin Cancer Res, 2021, 40(1): 81. doi:10.1186/s13046-021-01874-1 |
| [14] | Kock KH, Tan LM, Han KY, et al. Asian diversity in human immune cells[J]. Cell, 2025, 188(8): 2288-306. e24. doi:10.1016/j.cell.2025.02.017 |
| [15] | Hu JY, Chen ZH, Bao L, et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes[J]. Mol Ther, 2020, 28(7): 1658-72. doi:10.1016/j.ymthe.2020.04.023 |
| [16] | Ding SN, Qiao N, Zhu QC, et al. Single-cell atlas reveals a distinct immune profile fostered by T cell-B cell crosstalk in triple negative breast cancer[J]. Cancer Commun (Lond), 2023, 43(6): 661-84. doi:10.1002/cac2.12429 |
| [17] | Roma-Rodrigues C, Mendes R, Baptista PV, et al. Targeting tumor microenvironment for cancer therapy[J]. Int J Mol Sci, 2019, 20(4): 840. doi:10.3390/ijms20040840 |
| [18] | Brand A, Singer K, Koehl GE, et al. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells[J]. Cell Metab, 2016, 24(5): 657-71. doi:10.1016/j.cmet.2016.08.011 |
| [19] | Maynard JP, Godwin TN, Lu JY, et al. Localization of macrophage subtypes and neutrophils in the prostate tumor microenvironment and their association with prostate cancer racial disparities[J]. Prostate, 2022, 82(16): 1505-19. doi:10.1002/pros.24424 |
| [20] | Lou Y, Fan ZG, Ren SC. The role of neutrophils and their immu-nosuppressive effects on prostate cancer[J]. Histol Histopathol, 2025: 18890. |
| [21] | Bailey-Whyte M, Minas TZ, Dorsey TH, et al. Systemic inflammation indices and association with prostate cancer survival in a diverse patient cohort[J]. Cancers (Basel), 2023, 15(6): 1869. doi:10.3390/cancers15061869 |
| [22] | Xiao ZP, Song SS, Chen D, et al. Proteolysis targeting Chimera (PROTAC) for macrophage migration inhibitory factor (MIF) has anti-proliferative activity in lung cancer cells[J]. Angew Chem Int Ed, 2021, 60(32): 17514-21. doi:10.1002/anie.202101864 |
| [23] | Zhu GQ, Tang YL, Geng N, et al. HIF-α/MIF and NF-κB/IL-6 axes contribute to the recruitment of CD11b+Gr-1+myeloid cells in hypoxic microenvironment of HNSCC[J]. Neoplasia, 2014, 16(2): 168-79. doi:10.1593/neo.132034 |
| [24] | Zhu GQ, Tang Z, Huang R, et al. CD36+ cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor[J]. Cell Discov, 2023, 9(1): 25. doi:10.1038/s41421-023-00529-z |
| [25] | Ding XC, Wang LL, Zhang XD, et al. The relationship between expression of PD-L1 and HIF-1α in glioma cells under hypoxia[J]. J Hematol Oncol, 2021, 14(1): 92. doi:10.1186/s13045-021-01102-5 |
| [26] | Xu XT, Lai C, Luo JW, et al. The predictive significance of chromobox family members in prostate cancer in humans[J]. Cell Oncol (Dordr), 2024, 47(4): 1315-31. doi:10.1007/s13402-024-00929-7 |
| [27] | Mališić E, Petrović N, Brengues M, et al. Association of polymorphisms in TGFB1 XRCC1 XRCC3 genes and CD8 T-lymphocyte apoptosis with adverse effect of radiotherapy for prostate cancer[J]. Sci Rep, 2022, 12(1): 21306. doi:10.1038/s41598-022-25328-6 |
| [28] | Xiao Y, Yu DH. Tumor microenvironment as a therapeutic target in cancer[J]. Pharmacol Ther, 2021, 221: 107753. doi:10.1016/j.pharmthera.2020.107753 |
| [29] | Duan QQ, Zhang HL, Zheng JN, et al. Turning cold into hot: firing up the tumor microenvironment[J]. Trends Cancer, 2020, 6(7): 605-18. doi:10.1016/j.trecan.2020.02.022 |
| [30] | Yang JF, Xing XD, Luo L, et al. Mitochondria-ER contact mediated by MFN2-SERCA2 interaction supports CD8+ T cell metabolic fitness and function in tumors[J]. Sci Immunol, 2023, 8(87): eabq2424. doi:10.1126/sciimmunol.abq2424 |
| [1] | 郭晓娟, 杜瑞娟, 陈丽平, 郭克磊, 周彪, 卞华, 韩立. WW结构域E3泛素连接酶1调控卵巢癌肿瘤微环境中的免疫浸润[J]. 南方医科大学学报, 2025, 45(5): 1063-1073. |
| [2] | 杨锐, 舒翊秦, 文晖杰, 蔡熹, 王震, 张棽, 向阳, 吴昊. 枫杨总黄酮通过抑制中性粒细胞胞外陷阱网的释放减轻大鼠类风湿关节炎[J]. 南方医科大学学报, 2024, 44(9): 1645-1652. |
| [3] | 骆金光, 陶怀祥, 闻志远, 陈龙, 胡昊, 关翰. 肿瘤相关成纤维细胞上调hsa-miR-18b-5p靶向FBXL3促进前列腺癌的增殖及转移[J]. 南方医科大学学报, 2024, 44(7): 1284-1296. |
| [4] | 裴蓓, 张艺, 魏思源, 梅语, 宋标, 董港, 温子昂, 李学军. 基于转录组学测序及生物信息学方法鉴定肠上皮化生的潜在致病基因[J]. 南方医科大学学报, 2024, 44(5): 941-949. |
| [5] | 王梓凝, 杨 明, 李双磊, 迟海涛, 王军惠, 肖苍松. 心肌梗死后心肌纤维化小鼠心肌线粒体功能和能量代谢重塑相关性的转录组学分析[J]. 南方医科大学学报, 2024, 44(4): 666-674. |
| [6] | 邵 珊, 白薇超, 邹鹏程, 罗敏娜, 赵新汉, 雷建军. 二甲双胍阻断乳腺癌细胞-间质细胞的交互作用:基于抑制肿瘤相关成纤维细胞缺氧诱导因子-1α的表达[J]. 南方医科大学学报, 2024, 44(3): 428-436. |
| [7] | 王燕妮, 黄霞, 陈福恒, 高圆圆, 崔向荣, 严琴, 井宣. 急性心肌梗死患者血清中 BIN1 水平与 Killip 分级之间的关系[J]. 南方医科大学学报, 2024, 44(12): 2388-2395. |
| [8] | 崔成玲, 许玉珍, 唐超群, 蒋佳颖, 胡英, 双杰. 高原低氧暴露导致小鼠脾脏组织脂代谢发生紊乱的分子机制[J]. 南方医科大学学报, 2024, 44(10): 2024-2032. |
| [9] | 吴秀华, 范应静, 叶永浓, 李 萍, 朱青安, 陈泽森, 李 博, 王 文, 郑 磊. 生酮饮食导致小鼠骨质疏松的转录组学[J]. 南方医科大学学报, 2023, 43(8): 1440-1446. |
| [10] | 邓 婷, 杜伯雨, 郗雪艳. 结直肠癌细胞通过激活成纤维细胞的ERK通路诱导癌症相关成纤维细胞的形成[J]. 南方医科大学学报, 2023, 43(6): 943-951. |
| [11] | 张自然, 谭家乐, 于子航, 刘承栋, 王 剑, 吴德华, 白 雪. FARSB对泛肿瘤的预后和冷肿瘤微环境的分层作用:基于整合单细胞和大块组织的DNA测序[J]. 南方医科大学学报, 2023, 43(5): 667-679. |
| [12] | 毛建英, 杨文静, 郭 和, 董瑞丽, 任丽芳, 李树斌. 体外重建宫颈癌的三维培养模型:基于人宫颈癌组织的去细胞支架[J]. 南方医科大学学报, 2023, 43(2): 157-165. |
| [13] | 张伟健, 邹琸玥, 朱永娜, 王 敏, 马彩云, 武峻捷, 石 昕, 刘 茜. IL-34在舌鳞状细胞癌中的表达及意义[J]. 南方医科大学学报, 2023, 43(12): 2111-2117. |
| [14] | 唐 娟, 陈 娟, 林国新, 张 浩, 桂 明, 李楠楠, 谷依红, 罗林娟, 孙 剑. 中性粒细胞/淋巴细胞、血小板/淋巴细胞比值是评价托法替尼治疗后类风湿关节炎活动性的有效指标[J]. 南方医科大学学报, 2023, 43(10): 1651-1656. |
| [15] | 郭 丽, 马银玲, 李 婷, 李金平. CD40LG是与浸润性乳腺癌肿瘤微环境中免疫和基质相关的预后标志物[J]. 南方医科大学学报, 2022, 42(9): 1267-1278. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||