Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (11): 2184-2191.doi: 10.12122/j.issn.1673-4254.2024.11.15
Previous Articles Next Articles
Qi QI1( ), Zheng ZHOU2, Jinzhao MA1, Bing YAO1, Li CHEN1(
), Zheng ZHOU2, Jinzhao MA1, Bing YAO1, Li CHEN1( )
)
Received:2024-06-12
Online:2024-11-20
Published:2024-11-29
Contact:
Li CHEN
E-mail:17720502549@163.com;chenli1978@nju.edu.cn
Qi QI, Zheng ZHOU, Jinzhao MA, Bing YAO, Li CHEN. Successful application of preimplantation genetic testing combined with third-generation sequencing for blocking hereditary spastic paraplegia[J]. Journal of Southern Medical University, 2024, 44(11): 2184-2191.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.11.15
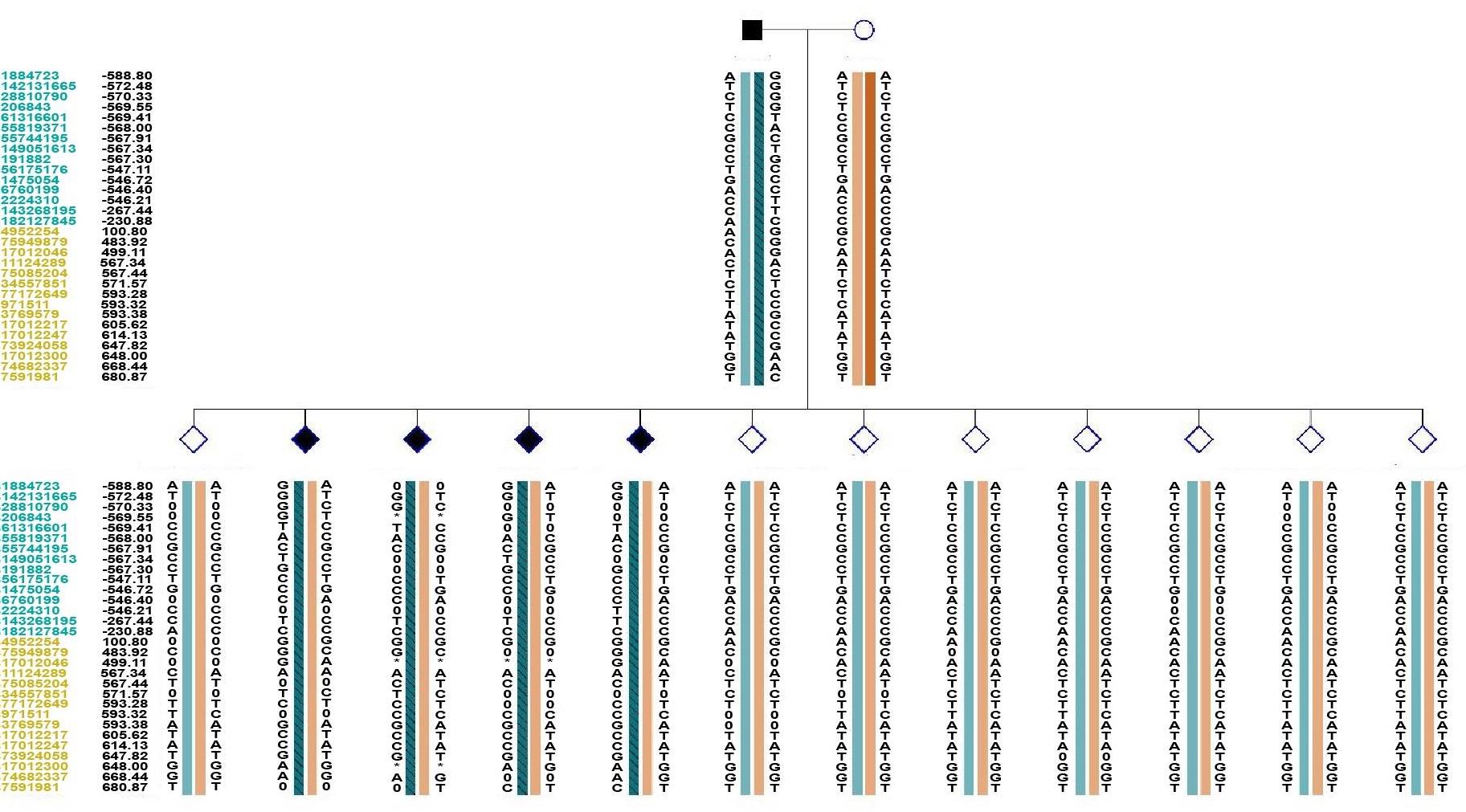
Fig.3 Single nucleotide polymorphisms (SNP)‑based genetic linkage map of the pedigree. ■: Affected male carrying pathogenic mutations; ○: Normal female not carrying pathogenic mutations; ◇: Unaffected, of unknown gender, not carrying pathogenic mutations; ◆: Affected, of unknown gender, carrying pathogenic mutations; *: ADO, allele drop out; 0: Missing data; Number: Distance (Kb) to the upstream or downstream of the gene; Blue: Location of SNP in the upstream of the chromosomal position of the target gene; Orange: Location of SNP in the downstream of the chromosomal position of the target gene; Left slash inside the haplotype: haplotype linked to maternal mutation; Right slash inside the haplotype: haplotype linked to paternal mutation.
| Probe ID | Chr | Pos | Male | Female | E-1 | E-2 | E-3 | E-4 | E-5 | E-6 | E-7 | E-8 | E-9 | E-10 | E-11 | E-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1884723 | 2 | 31477017 | A G | A A | A A | G A | 0 0 | G A | G A | A A | A A | A A | A A | A A | A A | A A |
| rs142131665 | 2 | 31493330 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | T T | T T | T T | T T |
| rs28810790 | 2 | 31495488 | C G | C C | 0 0 | G C | G C | 0 0 | 0 0 | C C | C C | C C | C C | C C | 0 0 | C C |
| rs206843 | 2 | 31496262 | T G | T T | 0 0 | G T | ** | G T | 0 0 | T T | T T | T T | T T | T T | 0 0 | T T |
| rs61316601 | 2 | 31496406 | C T | C C | C C | T C | T C | 0 0 | T C | C C | C C | C C | C C | C C | C C | C C |
| rs55819371 | 2 | 31497812 | C A | C C | C C | A C | A C | A C | A C | C C | C C | C C | C C | C C | C C | C C |
| rs55744195 | 2 | 31497904 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs149051613 | 2 | 31498476 | C T | C C | C C | T C | 0 0 | T C | 0 0 | C C | C C | C C | C C | C C | C C | C C |
| rs191882 | 2 | 31498514 | C G | C C | C C | G C | 0 0 | G C | G C | C C | C C | C C | C C | C C | C C | C C |
| rs56175176 | 2 | 31516496 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs1475054 | 2 | 31516895 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs6760199 | 2 | 31517208 | A C | A A | 0 0 | C A | C A | 0 0 | C A | A A | A A | A A | A A | 0 0 | A A | A A |
| rs2224310 | 2 | 31517397 | C T | C C | C C | 0 0 | 0 0 | 0 0 | T C | C C | C C | C C | C C | 0 0 | C C | C C |
| rs143268195 | 2 | 31796171 | C T | C C | C C | T C | T C | T C | T C | C C | C C | C C | C C | C C | C C | C C |
| rs182127845 | 2 | 31832726 | A C | C C | A C | C C | C C | C C | C C | A C | A C | A C | A C | A C | A C | A C |
| rs4952254 | 2 | 32258435 | A G | G G | 0 0 | G G | G G | G G | G G | A G | A G | A G | A G | A G | A G | A G |
| rs75949879 | 2 | 32641555 | C G | C C | C C | G C | G C | 0 0 | G C | C C | C C | 0 0 | C C | C C | C C | C C |
| rs17012046 | 2 | 32656752 | A G | A A | 0 0 | G A | ** | ** | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs11124289 | 2 | 32724978 | C A | A A | C A | A A | A A | A A | A A | C A | C A | C A | C A | C A | C A | C A |
| rs75085204 | 2 | 32725079 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs34557851 | 2 | 32729211 | C T | C C | 0 0 | T C | T C | 0 0 | 0 0 | C C | 0 0 | C C | C C | C C | C C | C C |
| rs77172649 | 2 | 32750915 | T C | T T | T T | C T | C T | 0 0 | C T | T T | T T | T T | T T | T T | T T | T T |
| rs971511 | 2 | 32750955 | T C | C C | T C | 0 0 | C C | C C | C C | 0 0 | T C | T C | T C | T C | T C | T C |
| rs3769579 | 2 | 32751020 | A G | A A | A A | G A | G A | G A | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs17012217 | 2 | 32763264 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs17012247 | 2 | 32771766 | A C | A A | A A | C A | C A | C A | C A | A A | A A | A A | A A | A A | A A | A A |
| rs73924058 | 2 | 32805458 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | 0 0 | T T | T T | T T |
| rs17012300 | 2 | 32805642 | G A | G G | G G | A G | ** | A G | A G | G G | G G | G G | G G | G G | G G | G G |
| rs74682337 | 2 | 32826076 | G A | G G | G G | A G | A G | 0 0 | A G | G G | G G | G G | G G | G G | G G | G G |
| rs7591981 | 2 | 32838505 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
Tab.1 Single nucleotide polymorphism haplotype analysis
| Probe ID | Chr | Pos | Male | Female | E-1 | E-2 | E-3 | E-4 | E-5 | E-6 | E-7 | E-8 | E-9 | E-10 | E-11 | E-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1884723 | 2 | 31477017 | A G | A A | A A | G A | 0 0 | G A | G A | A A | A A | A A | A A | A A | A A | A A |
| rs142131665 | 2 | 31493330 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | T T | T T | T T | T T |
| rs28810790 | 2 | 31495488 | C G | C C | 0 0 | G C | G C | 0 0 | 0 0 | C C | C C | C C | C C | C C | 0 0 | C C |
| rs206843 | 2 | 31496262 | T G | T T | 0 0 | G T | ** | G T | 0 0 | T T | T T | T T | T T | T T | 0 0 | T T |
| rs61316601 | 2 | 31496406 | C T | C C | C C | T C | T C | 0 0 | T C | C C | C C | C C | C C | C C | C C | C C |
| rs55819371 | 2 | 31497812 | C A | C C | C C | A C | A C | A C | A C | C C | C C | C C | C C | C C | C C | C C |
| rs55744195 | 2 | 31497904 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs149051613 | 2 | 31498476 | C T | C C | C C | T C | 0 0 | T C | 0 0 | C C | C C | C C | C C | C C | C C | C C |
| rs191882 | 2 | 31498514 | C G | C C | C C | G C | 0 0 | G C | G C | C C | C C | C C | C C | C C | C C | C C |
| rs56175176 | 2 | 31516496 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs1475054 | 2 | 31516895 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs6760199 | 2 | 31517208 | A C | A A | 0 0 | C A | C A | 0 0 | C A | A A | A A | A A | A A | 0 0 | A A | A A |
| rs2224310 | 2 | 31517397 | C T | C C | C C | 0 0 | 0 0 | 0 0 | T C | C C | C C | C C | C C | 0 0 | C C | C C |
| rs143268195 | 2 | 31796171 | C T | C C | C C | T C | T C | T C | T C | C C | C C | C C | C C | C C | C C | C C |
| rs182127845 | 2 | 31832726 | A C | C C | A C | C C | C C | C C | C C | A C | A C | A C | A C | A C | A C | A C |
| rs4952254 | 2 | 32258435 | A G | G G | 0 0 | G G | G G | G G | G G | A G | A G | A G | A G | A G | A G | A G |
| rs75949879 | 2 | 32641555 | C G | C C | C C | G C | G C | 0 0 | G C | C C | C C | 0 0 | C C | C C | C C | C C |
| rs17012046 | 2 | 32656752 | A G | A A | 0 0 | G A | ** | ** | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs11124289 | 2 | 32724978 | C A | A A | C A | A A | A A | A A | A A | C A | C A | C A | C A | C A | C A | C A |
| rs75085204 | 2 | 32725079 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs34557851 | 2 | 32729211 | C T | C C | 0 0 | T C | T C | 0 0 | 0 0 | C C | 0 0 | C C | C C | C C | C C | C C |
| rs77172649 | 2 | 32750915 | T C | T T | T T | C T | C T | 0 0 | C T | T T | T T | T T | T T | T T | T T | T T |
| rs971511 | 2 | 32750955 | T C | C C | T C | 0 0 | C C | C C | C C | 0 0 | T C | T C | T C | T C | T C | T C |
| rs3769579 | 2 | 32751020 | A G | A A | A A | G A | G A | G A | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs17012217 | 2 | 32763264 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs17012247 | 2 | 32771766 | A C | A A | A A | C A | C A | C A | C A | A A | A A | A A | A A | A A | A A | A A |
| rs73924058 | 2 | 32805458 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | 0 0 | T T | T T | T T |
| rs17012300 | 2 | 32805642 | G A | G G | G G | A G | ** | A G | A G | G G | G G | G G | G G | G G | G G | G G |
| rs74682337 | 2 | 32826076 | G A | G G | G G | A G | A G | 0 0 | A G | G G | G G | G G | G G | G G | G G | G G |
| rs7591981 | 2 | 32838505 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| Embryo No. | Embryo grade | Detection result | Gene mutation site detection |
|---|---|---|---|
| E-1 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-2 | 4AB (D5) | Euploid | Carrying paternal mutations |
| E-3 | 4BB (D5) | Euploid | Carrying paternal mutations |
| E-4 | 3BB (D5) | Euploid | Carrying paternal mutations |
| E-5 | 4BA (D5) | Euploid | Carrying paternal mutations |
| E-6 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-7 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-8 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-9 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-10 | 4BA (D6) | Euploid | Not carrying paternal mutations |
| E-11 | 4BC (D6) | Euploid | Not carrying paternal mutations |
| E-12 | 4BB (D6) | Euploid | Not carrying paternal mutations |
Tab.2 Detection results of the blastocysts
| Embryo No. | Embryo grade | Detection result | Gene mutation site detection |
|---|---|---|---|
| E-1 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-2 | 4AB (D5) | Euploid | Carrying paternal mutations |
| E-3 | 4BB (D5) | Euploid | Carrying paternal mutations |
| E-4 | 3BB (D5) | Euploid | Carrying paternal mutations |
| E-5 | 4BA (D5) | Euploid | Carrying paternal mutations |
| E-6 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-7 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-8 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-9 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-10 | 4BA (D6) | Euploid | Not carrying paternal mutations |
| E-11 | 4BC (D6) | Euploid | Not carrying paternal mutations |
| E-12 | 4BB (D6) | Euploid | Not carrying paternal mutations |
| 1 | de Souza PVS, de Rezende Pinto WBV, de Rezende Batistella GN, et al. Hereditary spastic paraplegia: clinical and genetic hallmarks[J]. Cerebellum, 2017, 16(2): 525-51. |
| 2 | Meyyazhagan A, Kuchi Bhotla H, Pappuswamy M, et al. The puzzle of hereditary spastic paraplegia: from epidemiology to treatment[J]. Int J Mol Sci, 2022, 23(14): 7665. |
| 3 | 郑备红, 孙 艳, 邱淑敏, 等. 全外显子测序技术鉴定一个常染色体显性遗传性痉挛性截瘫3A型家系ATL1基因突变[J]. 实用医学杂志, 2021, 37(13): 1761-4. |
| 4 | Ho NJ, Chen X, Lei Y, et al. Decoding hereditary spastic paraplegia pathogenicity through transcriptomic profiling[J]. Zool Res, 2023, 44(3): 650-62. |
| 5 | Zhu Z, Zhang C, Zhao G, et al. Novel mutations in the SPAST gene cause hereditary spastic paraplegia[J]. Parkinsonism Relat Disord. 2019, 69: 125-33. |
| 6 | Murala S, Nagarajan E, Bollu PC. Hereditary spastic paraplegia[J]. Neurol Sci, 2021, 42(3): 883-94. |
| 7 | Shribman S, Reid E, Crosby AH, et al. Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches[J]. Lancet Neurol, 2019, 18(12): 1136-46. |
| 8 | Elert-Dobkowska E, Stepniak I, Radziwonik-Fraczyk W, et al. SPAST intragenic CNVs lead to hereditary spastic paraplegia via a haploinsufficiency mechanism[J]. Int J Mol Sci, 2024, 25(9): 5008. |
| 9 | Chen R, Du SY, Yao YY, et al. A novel SPAST mutation results in spastin accumulation and defects in microtubule dynamics[J]. Mov Disord, 2022, 37(3): 598-607. |
| 10 | Parodi L, Fenu S, Barbier M, et al. Spastic paraplegia due to SPAST mutations is modified by the underlying mutation and sex[J]. Brain. 2018,141(12): 3331-42. |
| 11 | Mo A, Saffari A, Kellner M, et al. Early-onset and severe complex hereditary spastic paraplegia caused by de novo variants in SPAST[J]. Mov Disord, 2022, 37(12): 2440-6. |
| 12 | Cui F, Sun LQ, Qiao J, et al. Genetic mutation analysis of hereditary spastic paraplegia: a retrospective study[J]. Medicine, 2020, 99(23): e20193. |
| 13 | Panza E, Meyyazhagan A, Orlacchio A. Hereditary spastic paraplegia: genetic heterogeneity and common pathways[J]. Exp Neurol, 2022, 357: 114203. |
| 14 | Méreaux JL, Banneau G, Papin M, et al. Clinical and genetic spectra of 1550 index patients with hereditary spastic paraplegia[J]. Brain, 2022, 145(3): 1029-37. |
| 15 | Lallemant-Dudek P, Durr A. Clinical and genetic update of hereditary spastic paraparesis[J]. Rev Neurol, 2021, 177(5): 550-6. |
| 16 | Meyyazhagan A, Orlacchio A. Hereditary spastic paraplegia: an update[J]. Int J Mol Sci, 2022, 23(3): 1697. |
| 17 | 姜红芳, 余永林, 阮雯聪, 等. 儿童遗传性痉挛性截瘫临床与康复进展[J]. 中国实用儿科杂志, 2023, 38(1): 19-23. |
| 18 | 何秉涛, 黄美欢, 贠国俊, 等. 遗传性痉挛性截瘫基因分型、分子诊断和治疗的研究进展[J]. 中国优生与遗传杂志, 2022, 30(3): 516-9. |
| 19 | Bellofatto M, de Michele G, Iovino A, et al. Management of hereditary spastic paraplegia: a systematic review of the literature[J]. Front Neurol, 2019, 10: 3. |
| 20 | Di Ludovico A, Ciarelli F, la Bella S, et al. The therapeutic effects of physical treatment for patients with hereditary spastic paraplegia: a narrative review[J]. Front Neurol, 2023, 14: 1292527. |
| 21 | Parikh F, Athalye A, Madon P, et al. Genetic counseling for pre-implantation genetic testing of monogenic disorders (PGT-M)[J]. Front Reprod Health, 2023, 5: 1213546. |
| 22 | Madero JI, Manotas MC, García-Acero M, et al. Preimplantation genetic testing in assisted reproduction[J].Minerva Obstet Gynecol. 2023,75(3): 260-72. |
| 23 | Cornelisse S, Zagers M, Kostova E, et al. Preimplantation genetic testing for aneuploidies (abnormal number of chromosomes) in in vitro fertilisation[J]. Cochrane Database Syst Rev, 2020, 9(9): CD005291. |
| 24 | Yan L, Cao Y, Chen ZJ, et al. Chinese experts' consensus guideline on preimplantation genetic testing of monogenic disorders[J]. Hum Reprod, 2023, 38(): ii3-13. |
| 25 | Fan XY, Tang D, Liao YH, et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing[J]. PLoS Biol, 2020, 18(12): e3001017. |
| 26 | Cheng C, Fei ZJ, Xiao PF. Methods to improve the accuracy of next-generation sequencing[J]. Front Bioeng Biotechnol, 2023, 11: 982111. |
| 27 | Niu WB, Wang LL, Xu JW, et al. Improved clinical outcomes of preimplantation genetic testing for aneuploidy using MALBAC-NGS compared with MDA-SNP array[J]. BMC Pregnancy Childbirth, 2020, 20(1): 388. |
| 28 | Scarano C, Veneruso I, de Simone RR, et al. The third-generation sequencing challenge: novel insights for the omic sciences[J]. Biomolecules, 2024, 14(5): 568. |
| 29 | Searle B, Müller M, Carell T, et al. Third-generation sequencing of epigenetic DNA[J]. Angew Chem Int Ed, 2023, 62(14): e202215704. |
| 30 | Tan VJ, Liu TM, Arifin Z, et al. Third-generation single-molecule sequencing for preimplantation genetic testing of aneuploidy and segmental imbalances[J]. Clin Chem, 2023, 69(8): 881-9. |
| 31 | van Dijk EL, Naquin D, Gorrichon K, et al. Genomics in the long-read sequencing era[J]. Trends Genet, 2023, 39(9): 649-71. |
| 32 | de Rycke M, Berckmoes V. Preimplantation genetic testing for monogenic disorders[J]. Genes, 2020, 11(8): 871. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||