Journal of Southern Medical University ›› 2024, Vol. 44 ›› Issue (5): 867-875.doi: 10.12122/j.issn.1673-4254.2024.05.08
• Basic Research • Previous Articles Next Articles
Jinrui NIE1( ), Yahui WU1, Xuemei HAN2, Yaqi LI1, Haikuan WANG1(
), Yahui WU1, Xuemei HAN2, Yaqi LI1, Haikuan WANG1( ), Huitu ZHANG1(
), Huitu ZHANG1( )
)
Received:2023-11-10
Online:2024-05-20
Published:2024-06-04
Contact:
Haikuan WANG, Huitu ZHANG
E-mail:nie15128928292@163.com;hkwang@tust.edu.cn;hzhang@tust.edu.cn
Supported by:Jinrui NIE, Yahui WU, Xuemei HAN, Yaqi LI, Haikuan WANG, Huitu ZHANG. Preparation of Lactobacillus paracei TK1501 postbiotic and its inhibitory effect against Helicobacter pylori infection in mice[J]. Journal of Southern Medical University, 2024, 44(5): 867-875.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2024.05.08
| Elution time (min) | Flow rate (mL/min) | Mobile phase A% | Mobile phase B% |
|---|---|---|---|
| 0 | 0.8 | 95 | 5 |
| 5 | 0.5 | 80 | 20 |
| 15 | 0.5 | 80 | 20 |
| 20 | 0.8 | 95 | 5 |
Tab.1 Mobile phase elution gradients
| Elution time (min) | Flow rate (mL/min) | Mobile phase A% | Mobile phase B% |
|---|---|---|---|
| 0 | 0.8 | 95 | 5 |
| 5 | 0.5 | 80 | 20 |
| 15 | 0.5 | 80 | 20 |
| 20 | 0.8 | 95 | 5 |
| Protease types | Protease concentration (U/mL) | ||||
|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | 50 | |
| Pepsin | 25.1±0.3 | 21.3±0.4 | 19.3±0.1 | 16.7±0.3 | 15.8±0.2 |
| Trypsase | 25.3±0.5 | 19.6±0.3 | 16.8±0.1 | 15.3±0.3 | 13.7±0.3 |
| chymotrypsin | 24.8±0.1 | 21.5±0.5 | 14.7±0.4 | 10.8±0.6 | 10.7±0.4 |
| Proteinase K | 25.2±0.4 | 20.7±0.2 | 12.8±0.5 | 10.7±0.4 | 8.3±0.2 |
| Papain | 25.7±0.2 | 21.3±0.4 | 12.6±0.2 | 11.6±0.2 | 9.6±0.1 |
| Alkaline protease | 25.4±0.4 | 19.7±0.3 | 11.6±0.4 | 9.8±0.1 | 8.2±0.3 |
Tab.2 Sensitivity of L. paracasei TK1501 postbiotic to different proteases
| Protease types | Protease concentration (U/mL) | ||||
|---|---|---|---|---|---|
| 0 | 10 | 20 | 30 | 50 | |
| Pepsin | 25.1±0.3 | 21.3±0.4 | 19.3±0.1 | 16.7±0.3 | 15.8±0.2 |
| Trypsase | 25.3±0.5 | 19.6±0.3 | 16.8±0.1 | 15.3±0.3 | 13.7±0.3 |
| chymotrypsin | 24.8±0.1 | 21.5±0.5 | 14.7±0.4 | 10.8±0.6 | 10.7±0.4 |
| Proteinase K | 25.2±0.4 | 20.7±0.2 | 12.8±0.5 | 10.7±0.4 | 8.3±0.2 |
| Papain | 25.7±0.2 | 21.3±0.4 | 12.6±0.2 | 11.6±0.2 | 9.6±0.1 |
| Alkaline protease | 25.4±0.4 | 19.7±0.3 | 11.6±0.4 | 9.8±0.1 | 8.2±0.3 |
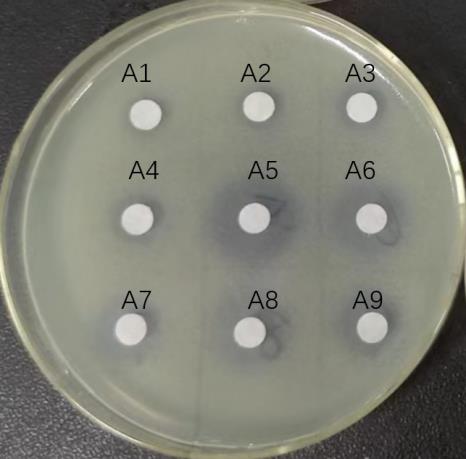
Fig.2 Bacterial ring for each gradient eluate of the effluent of macroporous adsorbent resin. A1-A3: Effluent of supernatant after large pore adsorption chromatography column; A4-A6: 10%, 20% and 40% ethanol eluent; A7-A9: 60%, 80%, and 100% ethanol eluent.
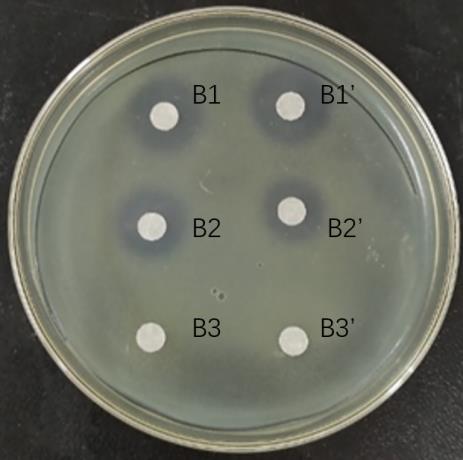
Fig.3 Bacterial inhibition by each gradient eluate of cation exchange columns. B1: Eluent of 20 mmol/L NaOAc+0.2 mol/L NaCl; B2: Eluent of 20 mmol/L NaOAc+0.5 mol/L NaCl; B3: Eluent of 20 mmol/L NaOAc +1.0 mol/L NaCl.

Fig.4 HPLC analysis of the ethanol eluents of the macroporous adsorbent resin and cation exchange resin. A: 20% ethanol elution chromatogram. B: 20 mmol/L sodium acetate+0.2 mol/L sodium chloride elution chromatogram of cation exchange resin.
| Indicator bacteria | Average number of colonies in the test group (CFU/mL) | Average colony number in control group (CFU/mL) | Sterilizing rate (%) |
|---|---|---|---|
| S. aureus | <10 | 4.5×104 | >99.98* |
| Hp | 2.2×104 | 6.5×104 | 66.10 |
| Salmonella | 2.4×104 | 5.5×104 | 56.40 |
| C. jejuni | 1.6×104 | 2.9×104 | 44.80** |
| C. perfringen | 2.2×104 | 5.8×104 | 63.80** |
| B. subtilis | 5.5×104 | 5.7×104 | 3.50 |
| Bifidobacterium | 4.4×104 | 4.7×104 | 6.40 |
| L. Beijerinck | 5.6×104 | 6.5×104 | 13.80 |
| E. coli | 2.8×104 | 3.1×104 | 9.60 |
| Bacteroides | 3.8×104 | 4.3×104 | 24.30 |
| C. Prazmowski | 4.6×104 | 4.7×104 | 2.00 |
| Proteus | 3.5×104 | 4.1×104 | 14.60 |
| Staphylococcus | 2.1×104 | 4.9×104 | 57.10* |
| Streptococcus | 4.3×104 | 4.8×104 | 10.40 |
Tab.3 Analysis of antibacterial spectrum of L. paracasei TK1501 postbiotic
| Indicator bacteria | Average number of colonies in the test group (CFU/mL) | Average colony number in control group (CFU/mL) | Sterilizing rate (%) |
|---|---|---|---|
| S. aureus | <10 | 4.5×104 | >99.98* |
| Hp | 2.2×104 | 6.5×104 | 66.10 |
| Salmonella | 2.4×104 | 5.5×104 | 56.40 |
| C. jejuni | 1.6×104 | 2.9×104 | 44.80** |
| C. perfringen | 2.2×104 | 5.8×104 | 63.80** |
| B. subtilis | 5.5×104 | 5.7×104 | 3.50 |
| Bifidobacterium | 4.4×104 | 4.7×104 | 6.40 |
| L. Beijerinck | 5.6×104 | 6.5×104 | 13.80 |
| E. coli | 2.8×104 | 3.1×104 | 9.60 |
| Bacteroides | 3.8×104 | 4.3×104 | 24.30 |
| C. Prazmowski | 4.6×104 | 4.7×104 | 2.00 |
| Proteus | 3.5×104 | 4.1×104 | 14.60 |
| Staphylococcus | 2.1×104 | 4.9×104 | 57.10* |
| Streptococcus | 4.3×104 | 4.8×104 | 10.40 |
| Groups | Feeding time (day) | |||
|---|---|---|---|---|
| 0 | 14 | 21 | 28 | |
| Blank group | 47.07±2.02** | 47.92±2.21** | 50.09±2.09* | 53.53±2.62* |
| Model group | 39.76±1.71 | 42.12±2.26 | 46.23±1.75 | 49.11±1.56 |
| Experiment group 1 | 41.05±1.97 | 43.99±2.35 | 46.95±2.62 | 49.28±2.71 |
| Experiment group 2 | 42.80±2.19 ## | 45.68±2.40 ## | 48.16±3.30 | 49.09±2.79 |
| Experiment group 3 | 42.85±1.80 ## | 45.14±2.50 ## | 47.29±3.45 | 48.08±2.65 |
Tab.4 Changes in body weight of the mice in different groups
| Groups | Feeding time (day) | |||
|---|---|---|---|---|
| 0 | 14 | 21 | 28 | |
| Blank group | 47.07±2.02** | 47.92±2.21** | 50.09±2.09* | 53.53±2.62* |
| Model group | 39.76±1.71 | 42.12±2.26 | 46.23±1.75 | 49.11±1.56 |
| Experiment group 1 | 41.05±1.97 | 43.99±2.35 | 46.95±2.62 | 49.28±2.71 |
| Experiment group 2 | 42.80±2.19 ## | 45.68±2.40 ## | 48.16±3.30 | 49.09±2.79 |
| Experiment group 3 | 42.85±1.80 ## | 45.14±2.50 ## | 47.29±3.45 | 48.08±2.65 |
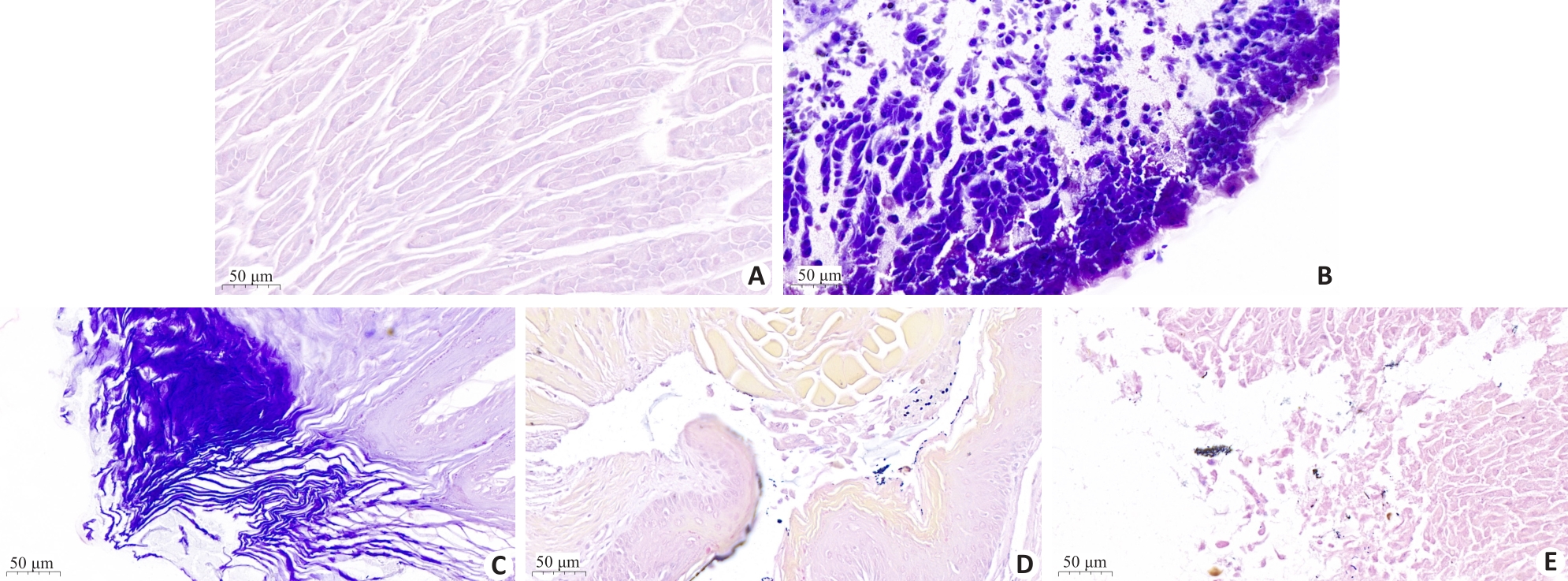
Fig. 8 HE staining of gastric tissues in different groups of mice. A: blank group; B: Hp infection model group; C: L. paracasei TK1501 postbiotic control group; D: L. paracasei TK1501 postbiotic low dose group; E: L. paracasei TK1501 postbiotic high dose group.
| Groups | Clump counts (CFU/mL) | Urease reagent detection |
|---|---|---|
| Blank | 3.40×10-1 | -- |
| Model | 2.57×10-3** | ++ |
| Experimental control | 2.39×10-3 | ++ |
| Low dose | 1.38×10-2 ## | - |
| High dose | 3.60×10-1## | - - |
Tab.5 Quantitative analysis of Hp in gastric mucosa of the mice in different groups
| Groups | Clump counts (CFU/mL) | Urease reagent detection |
|---|---|---|
| Blank | 3.40×10-1 | -- |
| Model | 2.57×10-3** | ++ |
| Experimental control | 2.39×10-3 | ++ |
| Low dose | 1.38×10-2 ## | - |
| High dose | 3.60×10-1## | - - |
| 1 | Lim SM. Anti-Helicobacter pylori activity of antimicrobial substances produced by lactic acid bacteria isolated from Baikkimchi[J]. J Korean Soc Appl Biol Chem, 2014, 57(5): 621-30. DOI: 10.1007/s13765-014-4198-6 |
| 2 | Li YL, Li XY, Tan ZJ. An overview of traditional Chinese medicine therapy for Helicobacter pylori-related gastritis[J]. Helicobacter, 2021, 26(3): e12799. DOI: 10.1111/hel.12799 |
| 3 | Shi YY, Yang ZW, Zhang T, et al. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer[J]. Cell Death Dis, 2019, 10(9): 625-33. DOI: 10.1038/s41419-019-1859-8 |
| 4 | Han TT, Jing XH, Bao JY, et al. H. pylori infection alters repair of DNA double-strand breaks via SNHG17[J]. J Clin Invest, 2020, 130(7): 3901-18. DOI: 10.1172/jci125581 |
| 5 | Feige MH, Vieth M, Sokolova O, et al. Helicobacter pylori induces direct activation of the lymphotoxin beta receptor and non-canonical nuclear factor-kappa B signaling[J]. Biochim Biophys Acta Mol Cell Res, 2018, 1865(4): 545-50. DOI: 10.1016/j.bbamcr.2018.01.006 |
| 6 | Aiba, Umeda K, Rahman S, et al. Synergistic effect of anti-Helicobacter pylori urease immunoglobulin Y from egg yolk of immunized hens and Lactobacillus johnsonii No.1088 to inhibit the growth of Helicobacter pylori in vitro and in vivo [J]. Vaccine, 2019, 37(23): 3106-12. DOI: 10.1016/j.vaccine.2019.04.045 |
| 7 | Watanabe Y, Oikawa R, Kodaka Y, et al. Cancer-related genetic variants of Helicobacter pylori strains determined using gastric wash-based whole-genome analysis with single-molecule real-time technology[J]. Int J Cancer, 2021, 148(1): 178-92. DOI: 10.1002/ijc.33257 |
| 8 | 黎文鸿, 李紫薇, 汪 娜, 等. 中国儿童幽门螺杆菌感染现状及其影响因素的Meta分析[J]. 中国全科医学, 2022, 25(28): 3569-78. |
| 9 | Resende C, Thiel A, Machado JC, et al. Gastric cancer: basic aspects[J]. Helicobacter, 2011, 16(): 38-44. DOI: 10.1111/j.1523-5378.2011.00879.x |
| 10 | 董奇灵, 赵慧慧, 李雪莹, 等. 中医药抗幽门螺杆菌感染的研究进展[J]. 中医药学报, 2023, 51(4): 108-11. |
| 11 | Ren S, Cai PP, Liu YQ, et al. Prevalence of Helicobacter pylori infection in China: a systematic review and meta-analysis[J]. J Gastroenterol Hepatol, 2022, 37(3): 464-70. DOI: 10.1111/jgh.15751 |
| 12 | 鞠俊杰. 含铋剂四联疗法与传统三联疗法治疗幽门螺杆菌(H.pylori)阳性小儿胃炎的临床疗效[J]. 中西医结合心血管病电子杂志, 2018, 6(9): 193-8. |
| 13 | Hu Y, Zhang M, Lu B, et al. Helicobacter pylori and antibiotic resistance, A continuing and intractable problem[J]. Helicobacter, 2016, 21(5): 349-63. DOI: 10.1111/hel.12299 |
| 14 | Gough EK. The impact of mass drug administration of antibiotics on the gut microbiota of target populations[J]. Infect Dis Poverty, 2022, 11(1): 76-83. DOI: 10.1186/s40249-022-00999-5 |
| 15 | Alava J, Altuğ G, Davies J, et al. Antibiotics as CECs: an overview of the hazards posed by antibiotics and antibiotic resistance[J]. Frontiers Marine Sci, 2016,3:102. DOI: 10.3389/fmars.2016.00024 |
| 16 | Bendjeddou K, Fons M, Strocker P, et al. Characterization and purification of a bacteriocin from Lactobacillus paracasei subsp. paracasei BMK2005, an intestinal isolate active against multidrug-resistant pathogens[J]. World J Microbiol Biotechnol, 2012, 28(4): 1543-52. DOI: 10.1007/s11274-011-0958-1 |
| 17 | Kumar SB, Arnipalli SR, Ziouzenkova O. Antibiotics in food chain: the consequences for antibiotic resistance[J]. Antibiotics, 2020, 9(10): 688-96. DOI: 10.3390/antibiotics9100688 |
| 18 | Bengoa AA, Errea AJ, Rumbo M, et al. Modulatory properties of Lactobacillus paracasei fermented milks on gastric inflammatory conditions[J]. Int Dairy J, 2020, 111: 104839-45. DOI: 10.1016/j.idairyj.2020.104839 |
| 19 | Das D, Sarkar S, Borsingh Wann S, et al. Current perspectives on the anti-inflammatory potential of fermented soy foods[J]. Food Res Int, 2022, 152: 110922-9. DOI: 10.1016/j.foodres.2021.110922 |
| 20 | Ali Mousavi Jam S, Talebi M, Alipour B, et al. The therapeutic effect of potentially probiotic Lactobacillus paracasei on dimethylhydrazine induced colorectal cancer in rats[J]. Food Biosci, 2021, 41: 101097. DOI: 10.1016/j.fbio.2021.101097 |
| 21 | Lv XC, Chen M, Huang ZR, et al. Potential mechanisms underlying the ameliorative effect of Lactobacillus paracasei FZU103 on the lipid metabolism in hyperlipidemic mice fed a high-fat diet[J]. Food Res Int, 2021, 139: 109956-63. DOI: 10.1016/j.foodres.2020.109956 |
| 22 | Xie Y, Guo QS, Wang GS. Preparative separation and purification of the total flavonoids in Scorzonera austriaca with macroporous resins[J]. Molecules, 2016, 21(6): 768-83. DOI: 10.3390/molecules21060768 |
| 23 | Friedrich V, Gerhard M. Vaccination against Helicobacter pylori-An approach for cancer prevention?[J]. Mol Aspects Med, 2023, 92: 101183. DOI: 10.1016/j.mam.2023.101183 |
| 24 | Berger H, Marques MS, Zietlow R, et al. Gastric cancer pathogenesis[J]. Helicobacter, 2016, 21(): 34-8. DOI: 10.1111/hel.12338 |
| 25 | 张 乐, 丁一珍, 潘媛娜, 等. 产细菌素乳酸菌的筛选及其益生性能评价[J]. 食品与发酵工业, 2023, 49(22): 19-26. |
| 26 | Cruz POD, Matos CJ, Nascimento YM, et al. Efficacy of potentially probiotic fruit-derived Lactobacillus fermentum, L. paracasei and L. plantarum to remove aflatoxin M1 in vitro [J]. Toxins, 2020, 13(1): 4-12. DOI: 10.3390/toxins13010004 |
| 27 | Lv XR, Ma HH, Sun MT, et al. A novel bacteriocin DY4-2 produced by Lactobacillus plantarum from cutlassfish and its application as bio-preservative for the control of Pseudomonas fluorescens in fresh turbot (Scophthalmus maximus) fillets[J]. Food Contr, 2018, 89: 22-31. DOI: 10.1016/j.foodcont.2018.02.002 |
| 28 | Miao JY, Liao WW, Pan ZY, et al. Isolation and identification of iron-chelating peptides from casein hydrolysates[J]. Food Funct, 2019, 10(5): 2372-81. DOI: 10.1039/c8fo02414f |
| 29 | Gomaa EZ. Synergistic antibacterial efficiency of bacteriocin and silver nanoparticles produced by probiotic Lactobacillus paracasei against multidrug resistant bacteria[J]. Int J Pept Res Ther, 2019, 25(3): 1113-25. DOI: 10.1007/s10989-018-9759-9 |
| 30 | 韩雪冰, 元香南, 方 俊, 等. 乳酸菌维持动物肠道健康的研究进展[J]. 中国科学: 生命科学, 2023, 53(4): 464-79. |
| 31 | 单春乔, 刘艳, 刘秋晨, 等. 嗜酸乳杆菌产细菌素生物学特性与临床应用的研究[C]// 第八届全国畜牧兽医青年科技工作者学术研讨会论文集. 绍兴, 2016: 440. |
| 32 | 柳 青. 乳酸杆菌产细菌素对金黄色葡萄球菌和肺炎链球菌临床株的体外抑菌效果研究[J]. 中国实用医药, 2022, 17(20): 192-4. |
| 33 | Techo S, Visessanguan W, Vilaichone RK, et al. Characterization and antibacterial activity against Helicobacter pylori of lactic acid bacteria isolated from Thai fermented rice noodle[J]. Probiotics Antimicrob Proteins, 2019, 11(1): 92-102. DOI: 10.1007/s12602-018-9385-z |
| 34 | Pei JJ, Yuan YH, Yue YL. Primary characterization of bacteriocin paracin C-a novel bacteriocin produced by Lactobacillus paracasei .[J]. Food Control, 2013, 34(1): 168-76. DOI: 10.1016/j.foodcont.2013.03.040 |
| 35 | Mendiola AS, Cardona AE. The IL-1β phenomena in neuroinflammatory diseases[J]. J Neural Transm, 2018, 125(5): 781-95. DOI: 10.1007/s00702-017-1732-9 |
| 36 | Wang YJ, Che MX, Xin JG, et al. The role of IL-1β and TNF‑α in intervertebral disc degeneration[J]. Biomedecine Pharmacother, 2020, 131: 110660. DOI: 10.1016/j.biopha.2020.110660 |
| 37 | Sato Y, Okamoto K, Kida Y, et al. Overview of chemotherapy for gastric cancer[J]. J Clin Med, 2023, 12(4): 1336-45. DOI: 10.3390/jcm12041336 |
| 38 | Shi YY, Wang P, Guo YL, et al. Helicobacter pylori-induced DNA damage is a potential driver for human gastric cancer AGS cells[J]. DNA Cell Biol, 2019, 38(3): 272-80. DOI: 10.1089/dna.2018.4487 |
| 39 | Ren ZJ, Li JY, Du XH, et al. Helicobacter pylori-induced progranulin promotes the progression of the gastric epithelial cell cycle by regulating CDK4[J]. J Microbiol Biotechnol, 2022, 32(7): 844-54. DOI: 10.4014/jmb.2203.03053 |
| [1] | . Extraction and identification of semen-derived exosomes using PEG6000 [J]. Journal of Southern Medical University, 2016, 36(11): 1531-. |
| [2] | . Optimization of the method for isolating and culturing rat mesenchymal stem cells [J]. Journal of Southern Medical University, 2014, 34(11): 1621-. |
| [3] | . Chemical constituents of the roots of Euphorbia pekinensis Rupr [J]. Journal of Southern Medical University, 2013, 33(12): 1748-. |
| [4] | . A method for isolating and identifying newborn rat sino-atrial node cells for patch-clamp study [J]. Journal of Southern Medical University, 2013, 33(03): 397-. |
| [5] | LU Dong-feng,WU Hao,HUANG Jing,LI Yan Department of Cardiology,Second Affiliated Hospital of Guangzhou Medical College,Guagnzhou 510260,China. Isolation,in vitro culture and identification of cardiac stem cells from neonatal SD rats [J]. Journal of Southern Medical University, 2006, 26(11): 1629-1632. |
| [6] | Lü Ben-qiang1, XING Xue-feng1, LUO Jia-bo1 1Key New Drug Research Lab, College of Traditional Chinese Medicine, Southern Medical University, Guangzhou 510515, China; 2Department of Pharmacy, 421 Hospital of PLA, Guangzhou 510310, China. Isolation of the chemical constituents from Shuanghuanglian injection and their structural identification [J]. Journal of Southern Medical University, 2006, 26(10): 1471-. |
| [7] | YUN Xue-xia, HU Jing, CHEN Qing Department of Epidemiology, Southern Medical University, Guangzhou 510515, China. Screening, isolation and identification of nisin resistance determinant gene in strains of Lactococcus lactis [J]. Journal of Southern Medical University, 2006, 26(06): 839-842. |
| [8] | CUI Jian-xiu1, ZHAO Guo-dong1, HUANG Wen-qi2. Analysis of the association between double-lumen endobronchial tube and inner diameter of the left main bronchus [J]. Journal of Southern Medical University, 2005, 25(07): 799-801. |
| [9] | LIU Zhi-lin1, ZHANG Xiang-nian2, ZHAO Shu-jin2, ZHOU Ri-xiao3. Extraction and purification of psoralen from Psoralea corylifolia L. [J]. Journal of Southern Medical University, 2005, 25(06): 751-752. |
| [10] | WEN Xiao-yun1, ZHU Zheng-guang1, LIN Bi-run2, XIE Shuang-da2, LEI Lin-sheng1, WU Shu-guang1. Isolation and structural analysis of a new polysaccharid, streptomyces polysaccharide [J]. Journal of Southern Medical University, 2005, 25(03): 251-254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||