南方医科大学学报 ›› 2025, Vol. 45 ›› Issue (11): 2466-2474.doi: 10.12122/j.issn.1673-4254.2025.11.19
高志强( ), 林洁, 洪鹏, 胡再宏, 崔孔孔, 王语, 董军君, 石秦林, 田小毛(
), 林洁, 洪鹏, 胡再宏, 崔孔孔, 王语, 董军君, 石秦林, 田小毛( ), 魏光辉
), 魏光辉
收稿日期:2024-05-19
出版日期:2025-11-20
发布日期:2025-11-28
通讯作者:
田小毛
E-mail:1815234622@qq.com;xiao-mao@ hospital.cqmu.edu.cn
作者简介:高志强,硕士,E-mail:1815234622@qq.com
基金资助:
Zhiqiang GAO( ), Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN(
), Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN( ), Guanghui WEI
), Guanghui WEI
Received:2024-05-19
Online:2025-11-20
Published:2025-11-28
Contact:
Xiaomao TIAN
E-mail:1815234622@qq.com;xiao-mao@ hospital.cqmu.edu.cn
Supported by:摘要:
目的 探究环状RNA表达谱及其在肾母细胞瘤(WT)预后和进展中的潜在作用。 方法 搜集4对配对WT组织送环状RNA测序,筛选差异表达环状RNA。使用qPCR对表达水平最高的前6个候选环状RNA进行临床样本表达验证。选择有代表性的环状RNA在较大临床队列中分析其与临床病理特征和预后的相关性。最后,使用Sanger测序和RNase R酶消化实验验证该分子的成环位点和结构稳定性。 结果 基于高通量环状RNA测序在WT中鉴定到23,978个环状RNA分子,其中10,884个环状RNA已获得注释。根据筛选参数(|log2FC|>2和P值<0.05),去除表达丰度较低的样本,鉴定出614个差异表达的环状RNA分子,其中包括269个上调和345个下调。基于临床样本表达验证hsa_circ_0001900表达丰度最高与临床来源的肿瘤组织中和测序结果表达趋势一致,诊断ROC曲线显示该分子具有良好的鉴别诊断能力(曲线下面积为0.72)。临床相关性分析提示其表达水平与WT体积呈正相关(R=0.35,P=0.040),同时高表达组患儿的无复发生存率(RFS)具有明显降低的趋势(4年无复发生存率分别为81.25% vs 85.11%,P=0.496)。最后,本研究设计针对“反向剪接”位点的发散引物,该分子的成环剪接位点序列得到Sanger测序的验证,Rnase R酶消化实验进一步证实该分子的稳定共价结构。 结论 本研究首次展示WT中环状RNA的全面表达谱,其中hsa_circ_0001900的表达水平与WT肿瘤大小和患儿预后有关,可能是WT进展的关键驱动基因。
高志强, 林洁, 洪鹏, 胡再宏, 崔孔孔, 王语, 董军君, 石秦林, 田小毛, 魏光辉. 高通量环状RNA测序揭示hsa_circ_0001900在肾母细胞瘤中特异性高表达且与不良预后相关[J]. 南方医科大学学报, 2025, 45(11): 2466-2474.
Zhiqiang GAO, Jie LIN, Peng HONG, Zaihong HU, Kongkong CUI, Yu WANG, Junjun DONG, Qinlin SHI, Xiaomao TIAN, Guanghui WEI. High-throughput circular RNA sequencing reveals tumor-specific high expression of hsa_circ_0001900 in Wilms tumor in association with poor prognosis[J]. Journal of Southern Medical University, 2025, 45(11): 2466-2474.
| Samples | Output of data (Gb) | Q30 (%) |
|---|---|---|
| Tumor tissue-1 | 15.060 | 88.84 |
| Tumor tissue-2 | 13.910 | 88.83 |
| Tumor tissue-3 | 14.581 | 89.52 |
| Tumor tissue-4 | 13.873 | 89.72 |
| Adjacent control-1 | 13.325 | 90.70 |
| Adjacent control-2 | 14.272 | 90.45 |
| Adjacent control-3 | 15.289 | 90.44 |
| Adjacent control-4 | 11.826 | 90.67 |
表1 测序结果质控表
Tab.1 Quality control for sequencing results
| Samples | Output of data (Gb) | Q30 (%) |
|---|---|---|
| Tumor tissue-1 | 15.060 | 88.84 |
| Tumor tissue-2 | 13.910 | 88.83 |
| Tumor tissue-3 | 14.581 | 89.52 |
| Tumor tissue-4 | 13.873 | 89.72 |
| Adjacent control-1 | 13.325 | 90.70 |
| Adjacent control-2 | 14.272 | 90.45 |
| Adjacent control-3 | 15.289 | 90.44 |
| Adjacent control-4 | 11.826 | 90.67 |
| Genes | Pre-primer (5'→3') | Post-primer (3'→5') |
|---|---|---|
| hsa_circ_0001900 | CGTTCAGTGCCTCGAAAGAAC | CTGGTCCCCTTTCAGGATGAG |
| hsa_circ_0004425 | AAGTGTGGACCCTGCAAATA | CCATTTGATCCACAGACAGGA |
| hsa_circ_0005230 | CTCTTTGTTTTGCACACTAGGGA | ACCAGGTGAGCAGTCAAGAA |
| hsa_circ_0075828 | GGAAGTGGCTAATGGATCTG | ATGGAGAACAGCCATCCATG |
| hsa_circ_0008122 | TGTCTCAAAGCCAGACCTGAT | ATTCTGTTGTTCGGTGTCCAG |
| hsa_circ_0006151 | GGCACCAGCATAGCCAGTAGT | GATGACCAGGCGCTTTTCACA |
| GAPDH | CCTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGTG |
表2 基因q-PCR引物表
Tab.2 Primer sequences for q-PCR
| Genes | Pre-primer (5'→3') | Post-primer (3'→5') |
|---|---|---|
| hsa_circ_0001900 | CGTTCAGTGCCTCGAAAGAAC | CTGGTCCCCTTTCAGGATGAG |
| hsa_circ_0004425 | AAGTGTGGACCCTGCAAATA | CCATTTGATCCACAGACAGGA |
| hsa_circ_0005230 | CTCTTTGTTTTGCACACTAGGGA | ACCAGGTGAGCAGTCAAGAA |
| hsa_circ_0075828 | GGAAGTGGCTAATGGATCTG | ATGGAGAACAGCCATCCATG |
| hsa_circ_0008122 | TGTCTCAAAGCCAGACCTGAT | ATTCTGTTGTTCGGTGTCCAG |
| hsa_circ_0006151 | GGCACCAGCATAGCCAGTAGT | GATGACCAGGCGCTTTTCACA |
| GAPDH | CCTTCCTGGGCATGGAGTC | TGATCTTCATTGTGCTGGGTG |
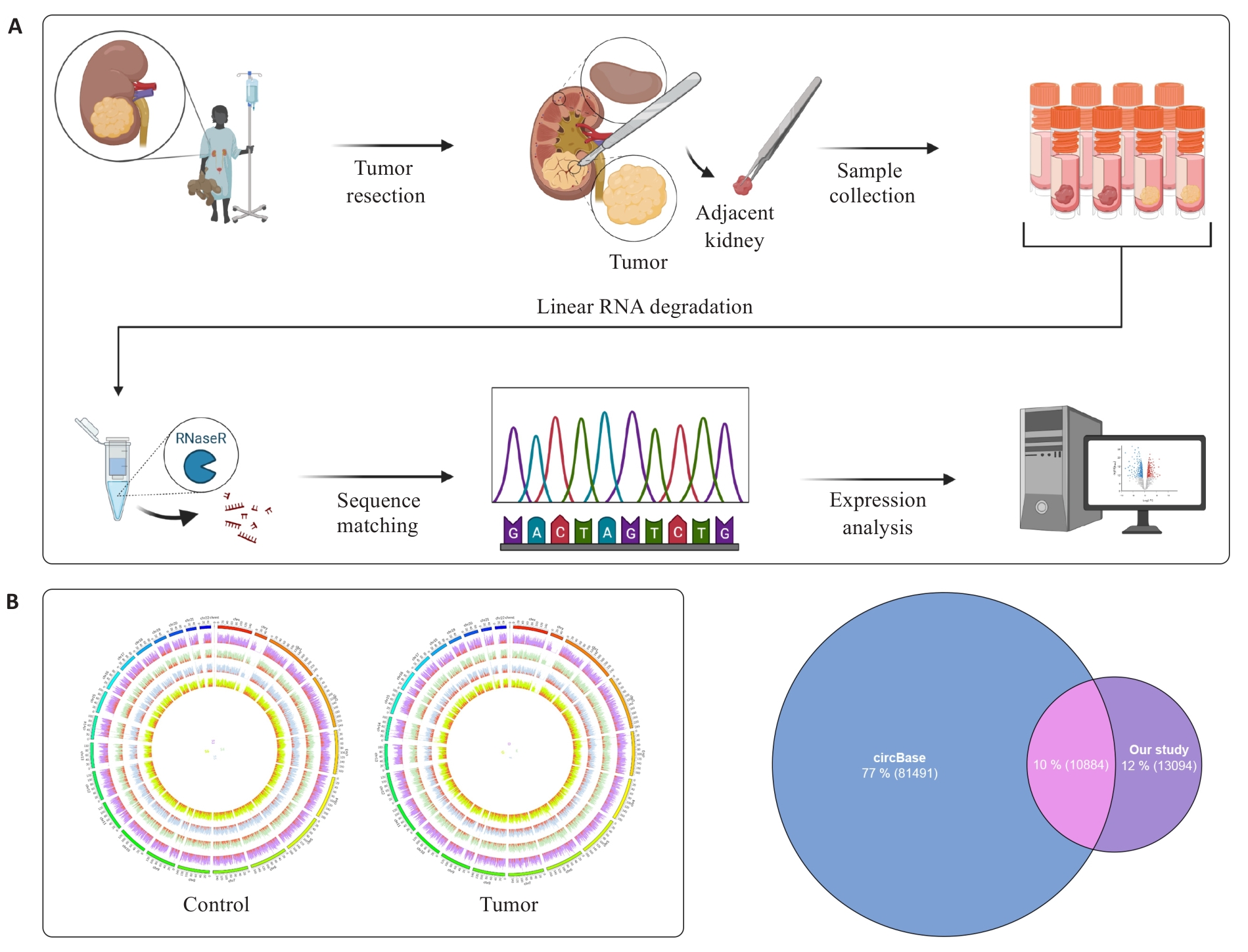
图1 环状RNA的鉴定和注释
Fig.1 Identification and annotation of circRNAs A: Schematic diagram of Wilms tumor (WT) circular RNA sequencing analysis. B: Circos showing the full view of the circRNAs, with the outer circles representing chromosomes, the inner circles representing samples, and the red part of each sample representing "spliced" readings of the circRNAs. C: Venn diagram of circRNA sequencing data annotated with circBase database.

图2 环状RNA的分类和差异表达模式
Fig.2 Classification and differential expression patterns of circRNAs. A: Circular RNA classification. B: Principal component analysis (PCA). C: Volcano plot of the differentially expressed circRNAs. Red dots indicate upregulation and blue dots indicate downregulation. The horizontal dashed line represents the truncated P value of 0.05, and the vertical dashed line represents the 2-fold logFC value.
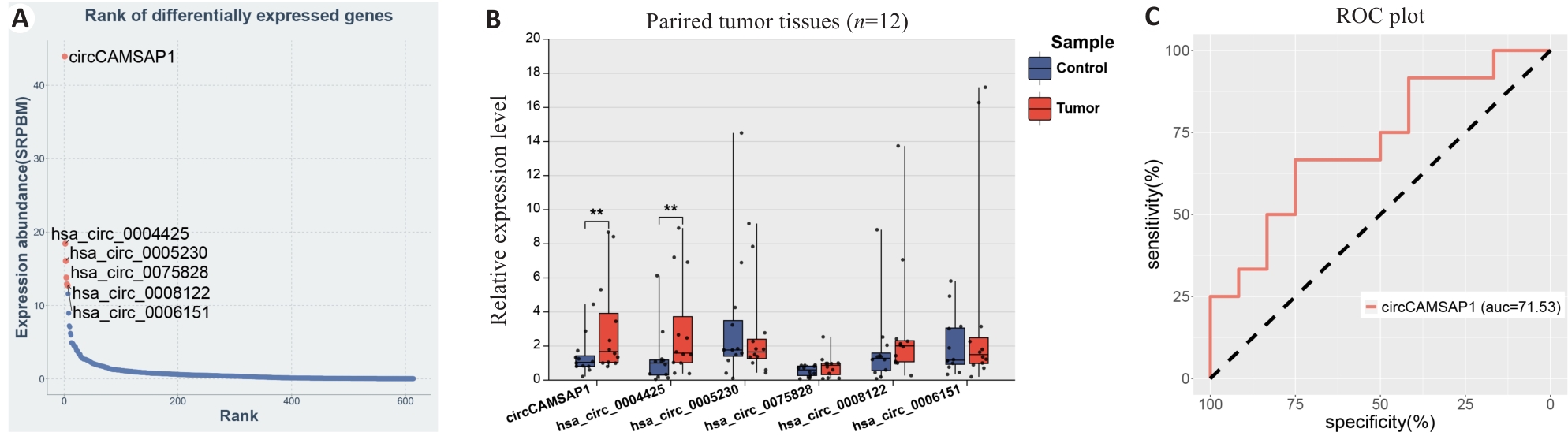
图3 临床样本验证提示hsa_circ_0001900在WT中呈现出肿瘤特异性表达模式
Fig.3 Validation of tumor-specific expression pattern of hsa_circ_0001900 in clinical samples of WT A: Ranking map of circRNA expression abundance. B: Expression validation of the top 6 circRNAs in clinical samples. C: ROC diagnostic curve of hsa_circ_0001900. *P<0.05, **P<0.01 vs Tumor group.
| circRNA | Chromosomal loci | Direction of ring formation | Genome length | Splice sequence length | Gene ID | Gene of parent | Trend of expression |
|---|---|---|---|---|---|---|---|
| hsa_circ_0008122 | chr19:23134080-23136043 | + | 1964 | 223 | ENSG00000183850 | ZNF730 | up |
| hsa_circ_0001900 | chr9:135881633-135883078 | - | 1446 | 425 | ENSG00000130559 | CAMSAP1 | up |
| hsa_circ_0006151 | chr10:68468123-68470163 | - | 2041 | 367 | ENSG00000138346 | DNA2 | up |
| hsa_circ_0004425 | chr9:100498765-100516771 | + | 18007 | 364 | ENSG00000241697 | TMEFF1 | up |
| hsa_circ_0075828 | Chr6:22020339-22020542 | + | 204 | 204 | ENSG00000272168 | CASC15 | up |
| hsa_circ_0005230 | Chr1:172140480-172144437 | - | 3958 | 3958 | ENSG00000230630 | DNM3OS | up |
表3 临床样本表达验证的环状RNA分子的基本特征
Tab.3 Characteristics of circRNA molecules for expression validation in clinical samples
| circRNA | Chromosomal loci | Direction of ring formation | Genome length | Splice sequence length | Gene ID | Gene of parent | Trend of expression |
|---|---|---|---|---|---|---|---|
| hsa_circ_0008122 | chr19:23134080-23136043 | + | 1964 | 223 | ENSG00000183850 | ZNF730 | up |
| hsa_circ_0001900 | chr9:135881633-135883078 | - | 1446 | 425 | ENSG00000130559 | CAMSAP1 | up |
| hsa_circ_0006151 | chr10:68468123-68470163 | - | 2041 | 367 | ENSG00000138346 | DNA2 | up |
| hsa_circ_0004425 | chr9:100498765-100516771 | + | 18007 | 364 | ENSG00000241697 | TMEFF1 | up |
| hsa_circ_0075828 | Chr6:22020339-22020542 | + | 204 | 204 | ENSG00000272168 | CASC15 | up |
| hsa_circ_0005230 | Chr1:172140480-172144437 | - | 3958 | 3958 | ENSG00000230630 | DNM3OS | up |

图4 临床相关性分析提示WT中hsa_circ_0001900的高表达与较大的肿瘤体积和较差的预后有关
Fig.4 Correlation analysis of hsa_circ_0001900 expression level in WT with tumor volume and prognosis. A: Expression levels of hsa_circ_0001900 in different cancers. B: Verification of differential expression of hsa_circ_0001900 in a patient cohort. C: The expression level of hsa_circ_0001900 is positively correlated with tumor volume. D: Children with high expression of hsa_circ_0001900 had shorter survival time. *P<0.05, **P<0.01 vs Tumor group.

图5 环状RNA分子hsa_circ_0001900环状结构的注释和验证
Fig.5 Annotation and verification of circular structure of hsa_circ_0001900. A: Sanger sequencing based on PCR products that verifies the reverse splicing sequence of hsa_circ_0001900. B: Agarose gel electrophoresis verifies the RNase R enzyme tolerance of hsa_circ_0001900 from a qualitative perspective. C: qPCR verifies the structural stability of hsa_circ_0001900 from a quantitative perspective. ****P<0.0001.
| [1] | Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression[J]. Mol Cancer, 2018, 17(1): 79. doi:10.1186/s12943-018-0827-8 |
| [2] | 田小毛, 向 彬, 刘 丰, et al. 环状RNA在儿童肾母细胞瘤中的表达谱、生物学功能和临床意义 [J]. 生物化学与生物物理进展, 2023, 50(3): 463-72. |
| [3] | Zhao Y, Li J, Li J, et al. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100[J]. Eur Rev Med Pharmacol Sci, 2020, 24(8): 4420-9. |
| [4] | Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer[J]. Nat Rev Cancer, 2018, 18(1): 5-18. doi:10.1038/nrc.2017.99 |
| [5] | Hare A, Zeng M, Rehemutula A, et al. Hsa-circ_0000064 accelerates the malignant progression of gastric cancer via sponging microRNA-621[J]. Kaohsiung J Med Sci, 2021, 37(10): 841-50. doi:10.1002/kjm2.12419 |
| [6] | Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis[J]. Cancer Res, 2017, 77(9): 2339-50. doi:10.1158/0008-5472.can-16-1883 |
| [7] | Yin WB, Yan MG, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection[J]. Clin Chim Acta, 2018, 487: 363-8. doi:10.1016/j.cca.2017.10.011 |
| [8] | Qiu M, Xia W, Chen R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma[J]. Cancer Res, 2018, 78(11): 2839-51. doi:10.1158/0008-5472.can-17-2808 |
| [9] | Zhang Y, Jiang J, Zhang J, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability[J]. Mol Cancer, 2021, 20(1): 101. doi:10.1186/s12943-021-01390-y |
| [10] | Hou L, Zhang J, Zhao F. Full-length circular RNA profiling by nanopore sequencing with CIRI-long[J]. Nat Protoc, 2023, 18(6): 1795-813. doi:10.1038/s41596-023-00815-w |
| [11] | López-Jiménez E, Rojas AM, Andrés-León E. RNA sequencing and prediction tools for circular RNAs analysis[J]. Adv Exp Med Biol, 2018, 1087: 17-33. doi:10.1007/978-981-13-1426-1_2 |
| [12] | Zhang J, Zhao F. Circular RNA discovery with emerging sequencing and deep learning technologies[J]. Nat Genet, 2025, 57(5): 1089-102. doi:10.1038/s41588-025-02157-7 |
| [13] | Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance[J]. Trends Cancer, 2020, 6(4): 319-36. doi:10.1016/j.trecan.2020.01.012 |
| [14] | Zhang J, Quan Y, Su X, et al. Circ-PRMT5 stimulates the proliferative ability in Wilms' tumor through the miR-7-5p/KLF4 axis[J]. Cell Mol Biol: Noisy-le-grand, 2023, 69(8): 232-6. doi:10.14715/cmb/2023.69.8.36 |
| [15] | 田小毛, 石秦林, 陆 鹏, 等. 生物标志物对Wilms瘤预后判断及治疗指导价值的研究进展 [J]. 现代医药卫生, 2020, 36(6): 855-9. |
| [16] | Xiang B, Chen ML, Gao ZQ, et al. CCNB1 is a novel prognostic biomarker and promotes proliferation, migration and invasion in Wilms tumor[J]. BMC Med Genomics, 2023, 16(1): 189. doi:10.1186/s12920-023-01627-3 |
| [17] | 高志强, 林 洁, 洪 鹏, 等. 基于高通量RNA测序分析Wilms瘤中关键基因对预后及免疫应答的影响 [J]. 南方医科大学学报, 2024, 44(4): 727-38. |
| [18] | Chen S, Zhou Y, Chen Y, et al. Fastp: an ultra-fast all-in-one FASTQ preprocessor[J]. Bioinformatics, 2018, 34(17): i884-90. doi:10.1093/bioinformatics/bty560 |
| [19] | Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol, 2015, 16: 4. doi:10.1186/s13059-014-0571-3 |
| [20] | Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs[J]. RNA, 2014, 20(11): 1666-70. doi:10.1261/rna.043687.113 |
| [21] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics[J]. Genome Res, 2009, 19(9): 1639-45. doi:10.1101/gr.092759.109 |
| [22] | Robic A, Kühn C. Beyond back splicing, a still poorly explored world: non-canonical circular RNAs[J]. Genes: Basel, 2020, 11(9): E1111. doi:10.3390/genes11091111 |
| [23] | Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis[J]. Cell Res, 2015, 25(8): 981-4. doi:10.1038/cr.2015.82 |
| [24] | Dome JS, Graf N, Geller JI, et al. Advances in wilms tumor treatment and biology: progress through international collaboration[J]. J Clin Oncol, 2015, 33(27): 2999-3007. doi:10.1200/jco.2015.62.1888 |
| [25] | Saltzman AF, Cost NG, Romao RLP. Wilms tumor[J]. Urol Clin North Am, 2023, 50(3): 455-64. doi:10.1016/j.ucl.2023.04.008 |
| [26] | 向 彬. CircRACGAP1/miR-486-3p/CTNNBIP1调控轴促进肾母细胞瘤增殖侵袭和迁移的机制研究 [D], 重庆医科大学, 2023. |
| [27] | Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRNAs in cancer and oncology[J]. Nat Rev Clin Oncol, 2022, 19(3): 188-206. doi:10.1038/s41571-021-00585-y |
| [28] | Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs[J]. Nat Rev Genet, 2019, 20(11): 675-91. doi:10.1038/s41576-019-0158-7 |
| [29] | Beilerli A, Gareev I, Beylerli O, et al. Circular RNAs as biomarkers and therapeutic targets in cancer[J]. Semin Cancer Biol, 2022, 83: 242-52. doi:10.1016/j.semcancer.2020.12.026 |
| [30] | Lasda E, Parker R. Circular RNAs: diversity of form and function[J]. RNA, 2014, 20(12): 1829-42. doi:10.1261/rna.047126.114 |
| [31] | Zhang F, Zou HM, Li XY, et al. CircRNA_0017076 acts as a sponge for miR-185-5p in the control of epithelial-to-mesenchymal transition of tubular epithelial cells during renal interstitial fibrosis[J]. Hum Cell, 2023, 36(3): 1024-40. doi:10.1007/s13577-023-00877-8 |
| [32] | Wang L, Zhou Y, Jiang L, et al. CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway[J]. Mol Cancer, 2021, 20(1): 43. doi:10.1186/s12943-021-01332-8 |
| [33] | Xu F, Xiao Q, Du WW, et al. CircRNA: functions, applications and prospects[J]. Biomolecules, 2024, 14(12): 1503. doi:10.3390/biom14121503 |
| [34] | Dong J, Zeng Z, Huang Y, et al. Challenges and opportunities for circRNA identification and delivery[J]. Crit Rev Biochem Mol Biol, 2023, 58(1): 19-35. doi:10.1080/10409238.2023.2185764 |
| [35] | Xu K, Park D, Magis AT, et al. Small molecule KRAS agonist for mutant KRAS cancer therapy[J]. Mol Cancer, 2019, 18(1): 85. doi:10.1186/s12943-019-1012-4 |
| [36] | Casagrande GMS, Silva MO, Reis RM, et al. Liquid biopsy for lung cancer: up-to-date and perspectives for screening programs[J]. Int J Mol Sci, 2023, 24(3): 2505. doi:10.3390/ijms24032505 |
| [37] | Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6[J]. Cancer Cell Int, 2018, 18: 116. doi:10.1186/s12935-018-0602-3 |
| [38] | Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer[J]. Clin Chim Acta, 2017, 466: 167-71. doi:10.1016/j.cca.2017.01.025 |
| [39] | Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer[J]. World J Gastroenterol, 2017, 23(34): 6330-8. doi:10.3748/wjg.v23.i34.6330 |
| [40] | Chen Z, Xu W, Zhang D, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression[J]. Mol Ther Nucleic Acids, 2021, 23: 1120-35. doi:10.1016/j.omtn.2020.12.013 |
| [41] | Wang Y, Li X, Wang H, et al. CircCAMSAP1 promotes non-small cell lung cancer proliferation and inhibits cell apoptosis by sponging miR-1182 and regulating BIRC5[J]. Bioengineered, 2022, 13(2): 2428-39. doi:10.1080/21655979.2021.2011639 |
| [42] | Luo Z, Lu L, Tang Q, et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway[J]. J Cell Mol Med, 2021, 25(8): 3793-802. doi:10.1111/jcmm.16254 |
| [43] | Zhou C, Liu HS, Wang FW, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis[J]. Mol Ther, 2020, 28(3): 914-28. doi:10.1016/j.ymthe.2019.12.008 |
| [44] | Wang Y, Yan Q, Mo Y, et al. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumori-genesis via a SERPINH1/c-Myc positive feedback loop[J]. Mol Cancer, 2022, 21(1): 62. doi:10.1186/s12943-022-01502-2 |
| [1] | 马思源, 张博超, 浦春. Circ_0000437通过靶向let-7b-5p/CTPS1轴促进乳腺癌细胞的增殖、侵袭、迁移及上皮间质转化[J]. 南方医科大学学报, 2025, 45(8): 1682-1696. |
| [2] | 蔡蕊, 黄卓, 贺文霞, 艾添红, 宋晓伟, 胡淑婷. 剪接因子HNRNPH1通过调控Circ-MYOCD的反向剪接影响心肌肥厚的发生[J]. 南方医科大学学报, 2025, 45(3): 587-594. |
| [3] | 高志强, 林 洁, 洪 鹏, 胡再宏, 董军君, 石秦林, 田小毛, 刘 丰, 魏光辉. 基于高通量 RNA 测序分析 Wilms 瘤中关键基因对预后及免疫应答的影响[J]. 南方医科大学学报, 2024, 44(4): 727-738. |
| [4] | 张 铃, 赵春雨, 许瑶瑶, 陈炎森, 蔡志雄, 林浩伟, 蔡巧燕. 环状RNA hsa_circ_0006834可作为肝细胞癌患者预后的潜在生物标志物[J]. 南方医科大学学报, 2023, 43(11): 1850-1856. |
| [5] | 周佩涛, 程炳霖, 孙一宁, 吴德华, 陈宇翰. 环状RNA circRSF1结合HuR促进辐射诱导的肝星状细胞炎性表型[J]. 南方医科大学学报, 2023, 43(1): 46-51. |
| [6] | 詹炜杰, 严 涛, 高嘉文, 宋旻恺, 王 婷, 林 菲, 周海榆, 李 栎, 张 超. 环状RNA在免疫相关疾病中的作用[J]. 南方医科大学学报, 2022, 42(2): 163-170. |
| [7] | 左学良, 蔡 娟, 陈志强, 李艳娜, 张斗峰. CircPCSK5在胃癌中高表达并促进胃癌细胞的增殖、侵袭和上皮间质转化[J]. 南方医科大学学报, 2022, 42(10): 1440-1451. |
| [8] | 刘 磊, 杜成友, 魏续福, 廖 锐. 癌旁circWDR25与肝细胞癌根治性切除术后的预后相关[J]. 南方医科大学学报, 2021, 41(9): 1388-1393. |
| [9] | 张朝霞, 王 璋, 金黎明, 谭小军, 王钊颖, 沈炼桔, 魏光辉, 何大维. piRNA NU13对肾母细胞瘤细胞增殖、凋亡、迁移和侵袭的影响[J]. 南方医科大学学报, 2021, 41(2): 184-192. |
| [10] | 曹 娟, 孙丽萍, 安建红, 张 欢, 何潇潇, 申 洪. miR-200c-3p靶向CCNE2抑制肾母细胞瘤细胞增殖[J]. 南方医科大学学报, 2020, 40(09): 1246-1252. |
| [11] | 罗 鑫,董军君,何兴跃,沈炼桔,龙春兰,刘 丰,刘 星,林 涛,何大维,魏光辉. MiR-155-5p:在肾母细胞瘤中表达下调并抑制肾母细胞瘤细胞的增殖、迁移及促进细胞凋亡[J]. 南方医科大学学报, 2019, 39(12): 1476-1481. |
| [12] | 肖斌,温嘉欣,赵超然,陈丽丹,孙朝晖,李林海. Luminal亚型乳腺癌细胞与正常乳腺细胞的circRNA表达谱差异分析[J]. 南方医科大学学报, 2018, 38(08): 1014-. |
| [13] | 赵永斌, 郑少斌, 毛向明, 周海宽, 张鹏. 肾母细胞瘤术前化疗疗效分析[J]. 南方医科大学学报, 2004, 24(06): 722-724. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||