Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (2): 387-394.doi: 10.12122/j.issn.1673-4254.2025.02.20
Yan LIU( ), Jianyu YANG, Yujiao ZHOU, Wenbo DING, Xianyu ZHANG, Linran GAO, Beizhen PAN, Jifei YANG, Yundong ZHAO(
), Jianyu YANG, Yujiao ZHOU, Wenbo DING, Xianyu ZHANG, Linran GAO, Beizhen PAN, Jifei YANG, Yundong ZHAO( )
)
Received:2024-10-18
Online:2025-02-20
Published:2025-03-03
Contact:
Yundong ZHAO
E-mail:1269343452@qq.com;18604498530@163.com
Yan LIU, Jianyu YANG, Yujiao ZHOU, Wenbo DING, Xianyu ZHANG, Linran GAO, Beizhen PAN, Jifei YANG, Yundong ZHAO. A rapid method for detecting prfA and hly toxin genes of Listeria monocytogenes using double nucleic acid colloidal gold strips[J]. Journal of Southern Medical University, 2025, 45(2): 387-394.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.02.20
| Specific primer | Specific primer sequence (5'-3') |
|---|---|
| prfA | F:(6-FAM)-GCTGATTACCGAGAAGGGAA |
| R:(Biotin)-CCGCTAACGAGTGGATAAGA | |
| hly | F:(Digoxin)-TATACCACGGAGATGCAGTG |
| R:(Biotin)-AGCACCTGGATATGTTAGGC |
Tab.1 Specific primers for PCR amplification of Listeria monocytogenesprfA and hly genes
| Specific primer | Specific primer sequence (5'-3') |
|---|---|
| prfA | F:(6-FAM)-GCTGATTACCGAGAAGGGAA |
| R:(Biotin)-CCGCTAACGAGTGGATAAGA | |
| hly | F:(Digoxin)-TATACCACGGAGATGCAGTG |
| R:(Biotin)-AGCACCTGGATATGTTAGGC |
| Constituent | Quantity/Unit | Effect | Storage mode |
|---|---|---|---|
| ddH2O | 4 mL | DNA extraction and blank control | -20 ℃ |
| PCR tube | 20 | PCR amplification | -20 ℃ |
| Positive standard solution | 20 μL | Positive control | -20 ℃ |
| Negative standard solution | 20 μL | Negative control | -20 ℃ |
| Colloidal gold test strip | 20 | Visualization | -20 ℃ |
Tab.2 Composition of the double toxin detection kit
| Constituent | Quantity/Unit | Effect | Storage mode |
|---|---|---|---|
| ddH2O | 4 mL | DNA extraction and blank control | -20 ℃ |
| PCR tube | 20 | PCR amplification | -20 ℃ |
| Positive standard solution | 20 μL | Positive control | -20 ℃ |
| Negative standard solution | 20 μL | Negative control | -20 ℃ |
| Colloidal gold test strip | 20 | Visualization | -20 ℃ |
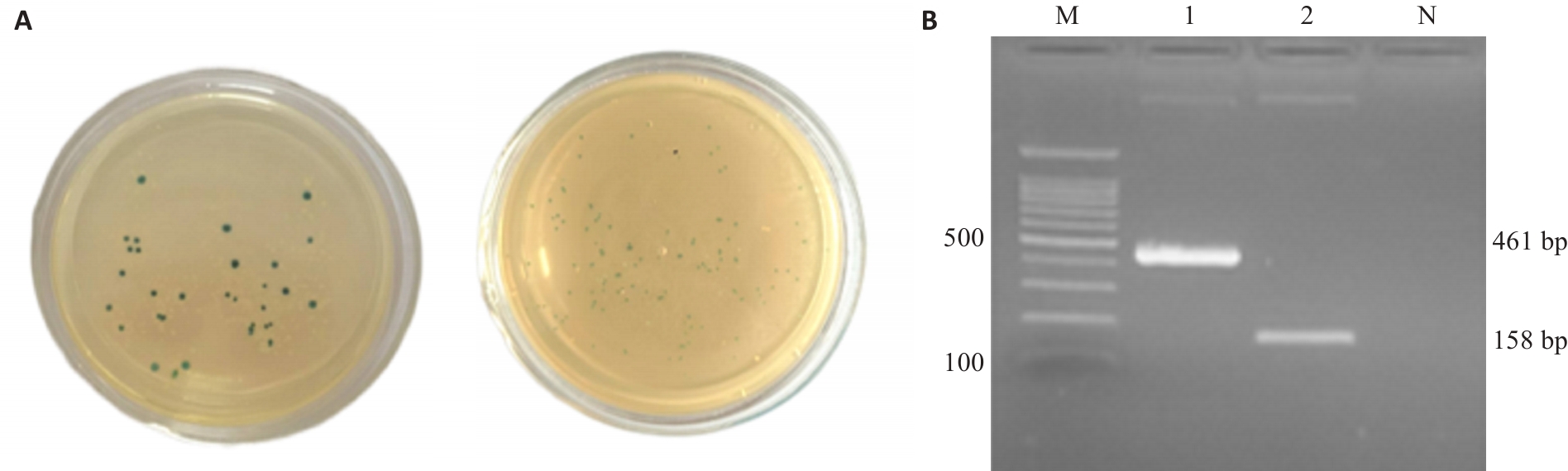
Fig.1 Electrophoresis of the cloned target gene fragments. A: Blue and white spot screening. B: Electrophoresis of prfA and hly plasmids. M: 100bp Marker; 1: Plasmid prfA; 2: Plasmid hly; N: Blank control.

Fig.4 Specificity assessment of the double virulence gene detection kit. 1: Listeria monocytogenes; 2: Staphylococcus aureus; 3: Escherichia coli; 4: Bacillus cereus; N: Blank control.
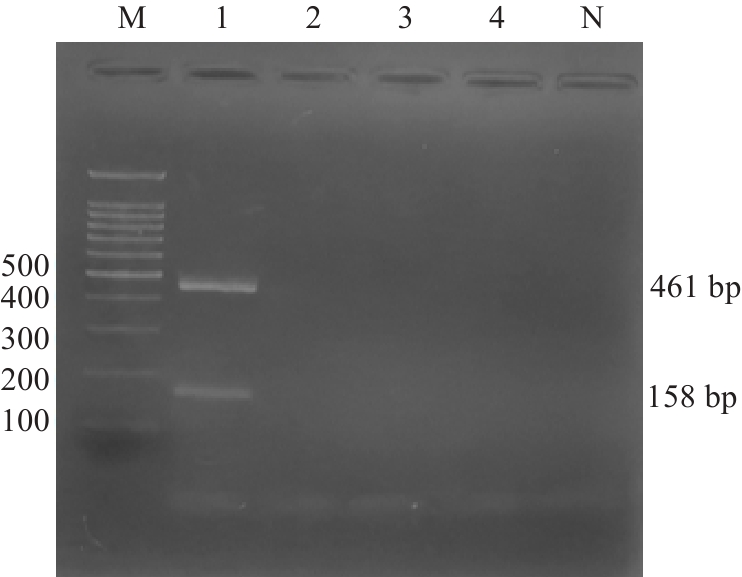
Fig.5 Specificity assessment of the kit with 1.5% agarose gel electrophoresis. M:100 bp Marker; 1: Listeria monocytogenes; 2: Staphylococcus aureus; 3: Escherichia coli; 4: Bacillus cereus; N: Blank control.

Fig.6 Electrophoretic sensitivity assessment of the dual virulence gene detection kit. M: 100 bp Marker; 1: 101 ng/μL; 2: 100 ng/μL; 3: 10-1 ng/μL; 4: 10-2 ng/μL; 5: 10-3 ng/μL; 6: 10-4 ng/μL; N: Blank control.
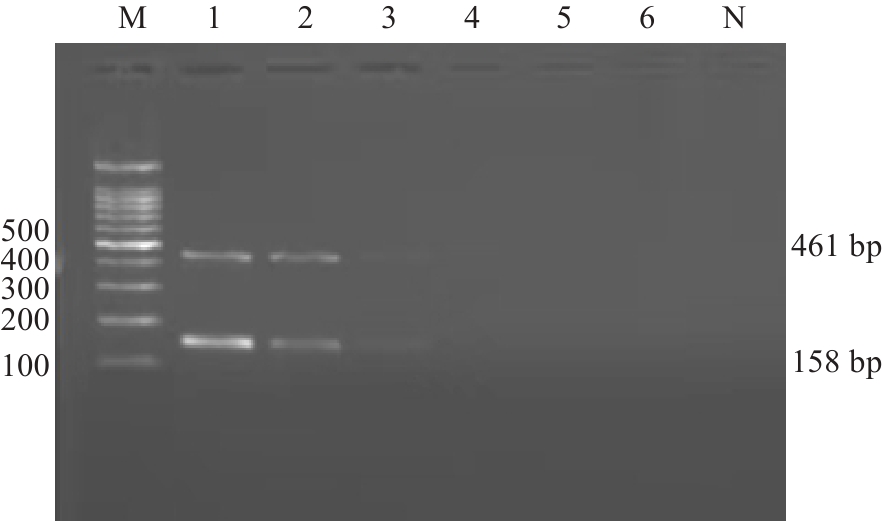
Fig.7 Sensitivity assessment of the kit with 1.5% agarose gel electrophoresis. M:100 bp Marker; 1: 101 ng/μL; 2: 100 ng/μL; 3: 10-1 ng/μL; 4: 10-2 ng/μL; 5: 10-3 ng/μL; 6: 10-4 ng/μL; N: Blank control.
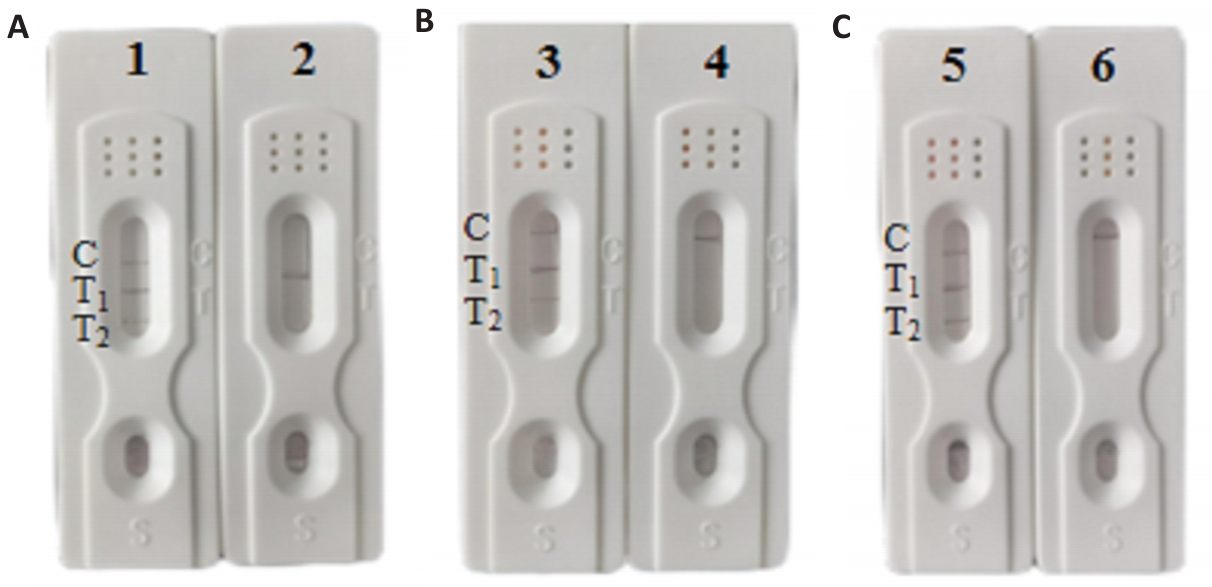
Fig.10 Stability test results of the double virulence gene test kit. A: 6th month. B: 9th month. C: 12th month. 1, 3, 5: Listeria monocytogenes; 2, 4, 6: Blank control.
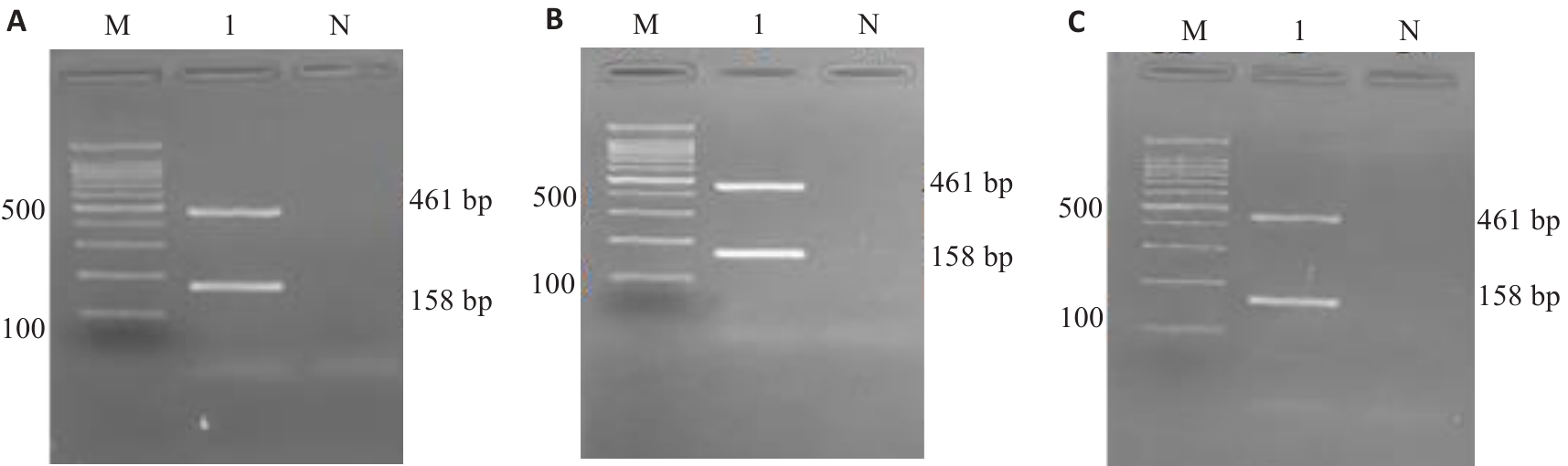
Fig.11 Stability test of the kit using 1.5% agarose gel electrophoresis. A: 6th month. B: 9th month. C: 12 th month. M: 100 bp Marker;1:Listeria monocytogenes; N: Blank control.

Fig.12 Results of dual virulence gene detection kit for contaminated samples. 1: 1.5×108 CFU/mL; 2: 1.5×107 CFU/mL; 3: 1.5×106 CFU/mL; 4: 1.5×105 CFU/mL; 5: 1.5×104 CFU/mL; 6: 1.5×103 CFU/mL; 7: 1.5×102 CFU/mL; 8: 1.5×101 CFU/mL; N: Blank control.
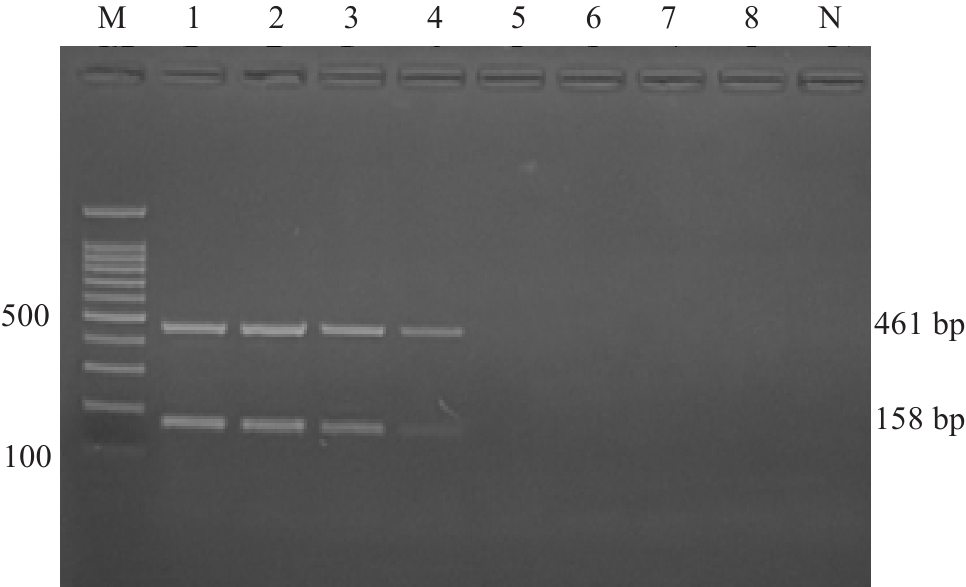
Fig.13 1.5% agarose gel electrophoresis for verification of the detection results using artificially contaminated samples. M:100 bp Marker; 1: 1.5×108 CFU/mL; 2: 1.5×107 CFU/mL; 3: 1.5×106 CFU/mL; 4: 1.5×105 CFU/mL; 5: 1.5×104 CFU/mL; 6: 1.5×103 CFU/mL; 7: 1.5×102 CFU/mL; 8: 1.5×101 CFU/mL; N: Blank control.

Fig.14 Detection results of actual samples using double virulence gene test kit. 1: Positive control; 2: Raw milk samples1; 3: Raw milk 2; 4: Raw milk 3; 5: Raw milk 4; 6: Raw beef; 7: Cooked meat product; 8: Quick-frozen aquatic products; 9: Vegetable salad; 10: Frozen drink; 11: Watermelon; 12: Blank control.
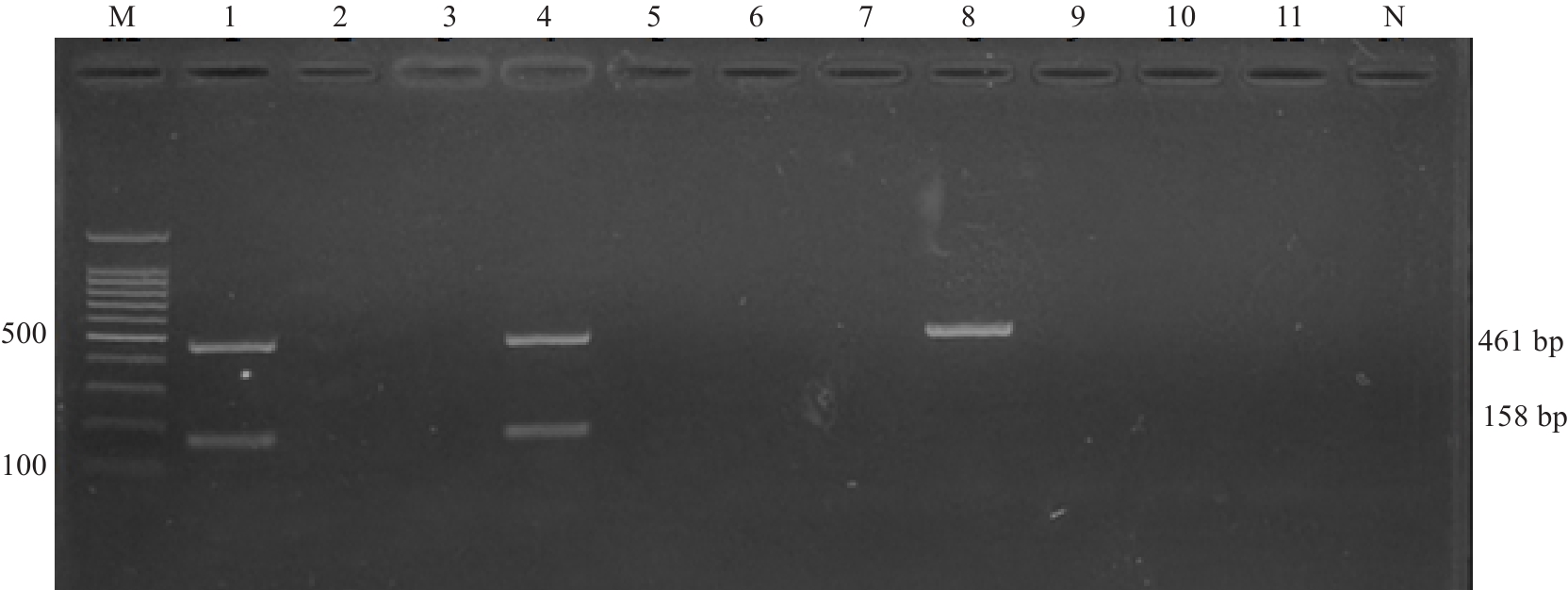
Fig.15 1.5% agarose gel electrophoresis for verification of the test results of using actual samples. M: 100 bp Marker; 1: Positive control; 2: Raw milk samples1; 3: Raw milk 2; 4: Raw milk 3; 5: Raw milk 4; 6: Raw beef; 7: Cooked meat product; 8: Quick-frozen aquatic products; 9: Vegetable salad; 10: Frozen drink; 11: Watermelon; N: Blank control.
| 1 | 龚云伟, 李月婷. 2010—2023年长春市市售食品中单核细胞增生李斯特菌检测分析[J]. 中国卫生工程学, 2024, 23(4): 473-4, 477. |
| 2 | Sun QF, Cheng JH, Lin RQ, et al. A novel multiplex PCR method for simultaneous identification of hypervirulent Listeria monocytogenes clonal complex 87 and CC88 strains in China[J]. Int J Food Microbiol, 2022, 366: 109558. |
| 3 | 孙婷婷, 王伟杰, 李 雪. 2020—2021年辽宁省食源性单核细胞增生李斯特菌毒力基因分布[J]. 中国食品卫生杂志, 2024, 36(4): 445-51. |
| 4 | 曾献莹, 吕素玲, 瞿 聪, 等. 广西单核细胞增生李斯特菌的流行率、毒力特征和分子分型研究[J]. 中国食品卫生杂志, 2023, 35(8): 1133-9. |
| 5 | 江玲丽, 高有领, 胡兴娟, 等. 检测产单核细胞李氏杆菌溶血素的胶体金试纸条制备及初步应用[J]. 畜牧兽医学报, 2022, 53(6): 1841-8. |
| 6 | 王凤军, 周叶熹, 陈丽敏, 等. TaqMan实时荧光PCR快速检测与鉴定单核细胞增生李斯特氏菌能力验证样品[J]. 保鲜与加工, 2024, 24(7): 84-90. |
| 7 | Gao LJ, Du BY, Ma QH, et al. Multiplex-PCR method application to identify duck blood and its adulterated varieties[J]. Food Chem, 2024, 444: 138673. |
| 8 | Lopes-Luz L, Silva-Filho E, Mendonça M, et al. Combined antibodies against internalins A and B proteins have potential application in immunoassay for detection of Listeria monocytogenes [J]. J Food Sci Technol, 2023, 60(1): 123-31. |
| 9 | Li YH, Gan ZY, Zhou X, et al. Accurate classification of Listeria species by MALDI-TOF mass spectrometry incorporating denoising autoencoder and machine learning[J]. J Microbiol Meth, 2022, 192: 106378. |
| 10 | 向婧姝, 周 倩, 周 藜, 等. 三种多重PCR技术检测食源性致病菌的对比研究[J]. 微量元素与健康研究, 2024, 41(5): 59-62. |
| 11 | 姚丽锋, 冯家望, 张 娟, 等. 重组酶介导等温扩增法检测食品中铜绿假单胞菌[J]. 食品安全质量检测学报, 2022, 13(17): 5695-701. |
| 12 | 国家卫生和计划生育委员会, 国家食品药品监督管理总局. 食品安全国家标准 食品微生物学检验 单核细胞增生李斯特氏菌检验: [S]. 北京: 中国标准出版社, 2016. |
| 13 | 饶晓虹, 陈 劲, 胡凤清, 等. 南平市2021—2023年市售食品中单核细胞增生李斯特菌污染状况及耐药性分析[J]. 海峡预防医学杂志, 2024, 30(3): 67-69. |
| 14 | 蔡 炯, 陈 潇, 朱虹霖, 等. 成都市冷藏即食预包装熟肉制品中单核细胞增生李斯特菌定量检测及污染风险研究[J]. 中国食品卫生杂志, 2024, 36(4): 426-32. |
| 15 | Xia L, Qiu S, Wang R, et al. Foodborne disease outbreaks in China from 2011 to 2020[J]. Wei Sheng Yan Jiu, 2023, 52(2): 226-31. |
| 16 | Sampedro F, Pérez-Rodríguez F, Servadio JL, et al. Quantitative risk assessment model to investigate the public health impact of varying Listeria monocytogenes allowable levels in different food commodities: a retrospective analysis[J]. Int J Food Microbiol, 2022, 383: 109932. |
| 17 | 张敬霞, 李 刚, 陈素明, 等. 单核细胞增生李斯特菌感染患者的临床特征分析[J]. 中国抗生素杂志, 2024, 49(10): 1114-20. |
| 18 | 马 力, 沈 锐. 多方法分离鉴定食品中单核细胞增生李斯特菌[J]. 食品工程, 2023(4): 44-6, 64. |
| 19 | Suebwongsa N, Jiemsup S, Santiyanont P, et al. MassARRAY: a high-throughput solution for rapid detection of foodborne pathogens in real-world settings[J]. Front Microbiol, 2024, 15: 1403579. |
| 20 | Dincer E. Detection of Listeria species by conventional culture-dependent and alternative rapid detection methods in retail ready-to-eat foods in Turkey[J]. J Microbiol Biotechnol, 2024, 34(2): 349. |
| 21 | Bai XK, Wang ZZ, Li WQ, et al. Portable sensor based on magnetic separation and enzyme-mediated immune nanomaterials for point-of-care testing of Listeria monocytogenes in food[J]. Anal Chim Acta, 2022, 1236: 340576. |
| 22 | 李志蕊.基于SRCA-NEMA-G-四链体的可视化及电化学生物传感器检测食品中单核细胞增生李斯特氏菌[D].河北农业大学, 2023. |
| 23 | 于 涵, 刘美琳, 王艺潼, 等.金钱白花蛇核酸试纸条快速鉴定及其试剂盒的研发[J].中药材, 2024, (6): 1390-6. |
| 24 | Wang N, Zhang J, Xiao B, et al. Amplification-free quantitative detection of genomic DNA using lateral flow strips for milk authentication[J]. Biosens Bioelectron, 2024, 252: 116140. |
| 25 | Lin F, Shen JY, Liu YL, et al. Rapid and effective detection of Macrobrachium rosenbergii nodavirus using a combination of nucleic acid sequence-based amplification test and immuno-chromatographic strip[J]. J Invertebr Pathol, 2023, 198: 107921. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||