南方医科大学学报 ›› 2024, Vol. 44 ›› Issue (11): 2184-2191.doi: 10.12122/j.issn.1673-4254.2024.11.15
• • 上一篇
收稿日期:2024-06-12
出版日期:2024-11-20
发布日期:2024-11-29
通讯作者:
陈莉
E-mail:17720502549@163.com;chenli1978@nju.edu.cn
作者简介:齐淇,在读硕士研究生,E-mail: 17720502549@163.com
基金资助:
Qi QI1( ), Zheng ZHOU2, Jinzhao MA1, Bing YAO1, Li CHEN1(
), Zheng ZHOU2, Jinzhao MA1, Bing YAO1, Li CHEN1( )
)
Received:2024-06-12
Online:2024-11-20
Published:2024-11-29
Contact:
Li CHEN
E-mail:17720502549@163.com;chenli1978@nju.edu.cn
摘要:
目的 结合三代测序技术(TGS)与胚胎植入前遗传学检测(PGT)成功阻断一个由SPAST基因突变引起的遗传性痉挛性截瘫(HSP),探讨PGT-M与TGS在遗传性痉挛性截瘫中的应用价值。 方法 选取一个遗传性痉挛性截瘫家系进行全外显子检测(WES),发现SPAST基因c.1699G>T突变,对该家系中先证者的SPAST基因突变位点c.1699G>T进行Sanger测序验证,同时应用三代测序技术在该家系SPAST基因突变位点两侧选择单核苷酸多态性(SNP)位点作为遗传连锁标记,进而构建携带基因突变的家系SNP单体型。行拮抗剂方案刺激卵巢以获取卵母细胞,并进行卵细胞质内单精子注射 (ICSI)及胚胎培养,对囊胚滋养层细胞进行活检。应用PGT-M技术后,选择其中不携带致病基因的胚胎进行移植。 结果 本周期共获取卵母细胞20枚,其中正常受精18枚,最后形成可供活检的囊胚12枚。基因检测结果显示,12枚囊胚均成功扩增,且均为整倍体,12枚囊胚中8枚不携带父源突变,4枚胚胎携带父源突变。挑选其中1枚不携带父源突变的优质整倍体胚胎进行冻融胚胎移植(FET),成功妊娠后羊水检测显示胎儿不携带父源突变后产下一健康女婴。 结论 对于家系不全的遗传病案例,应用三代测序及PGT-M技术可有效阻断SPAST基因突变向子代垂直传递,也可避免非整倍体胚胎妊娠导致的流产,帮助常染色体显性遗传性痉挛性截瘫家庭获得健康子代。
齐淇, 周征, 马金召, 姚兵, 陈莉. 胚胎植入前遗传学检测结合三代测序在阻断遗传性痉挛性截瘫中的成功应用[J]. 南方医科大学学报, 2024, 44(11): 2184-2191.
Qi QI, Zheng ZHOU, Jinzhao MA, Bing YAO, Li CHEN. Successful application of preimplantation genetic testing combined with third-generation sequencing for blocking hereditary spastic paraplegia[J]. Journal of Southern Medical University, 2024, 44(11): 2184-2191.
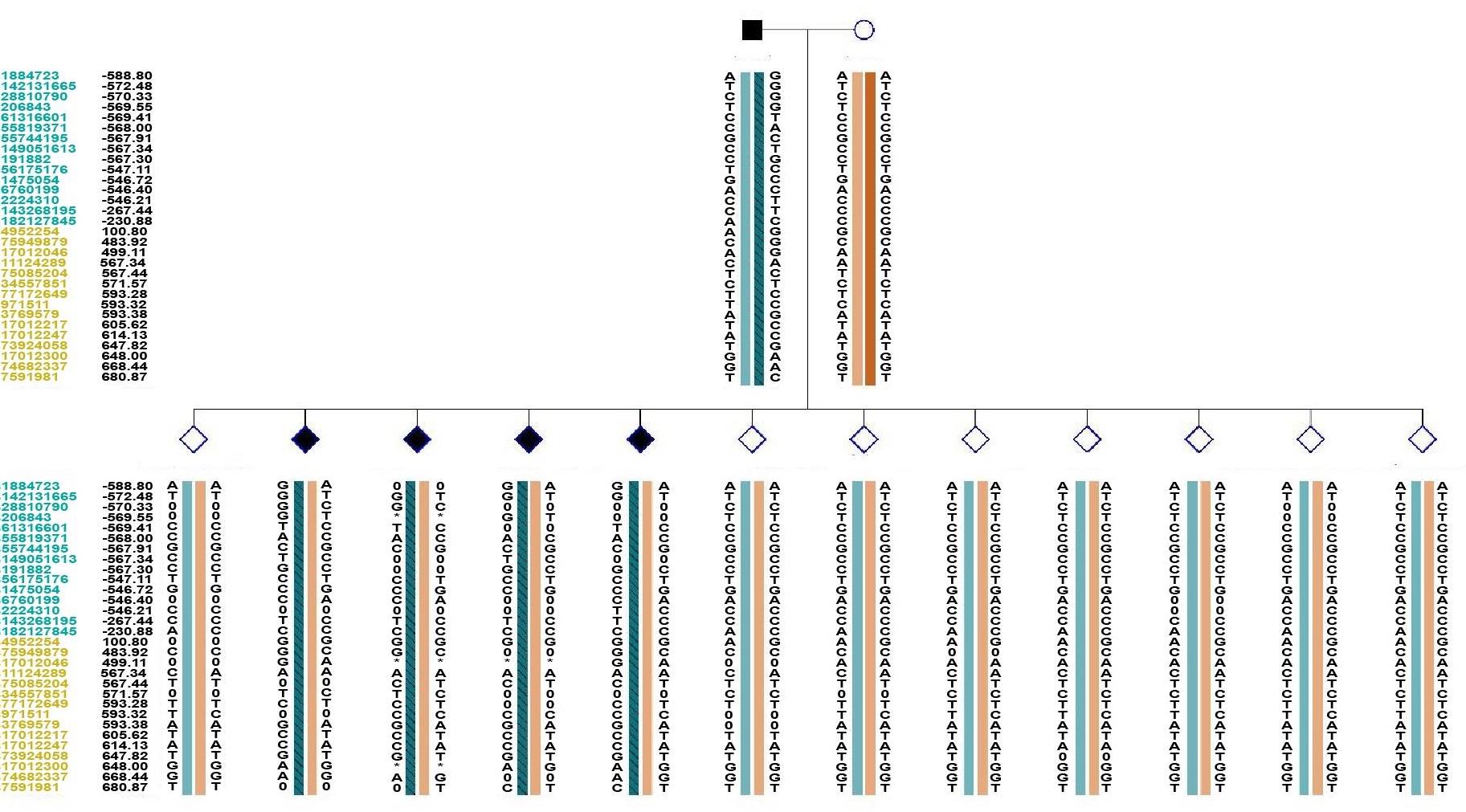
图3 该家系的SNP单体型连锁分析结果
Fig.3 Single nucleotide polymorphisms (SNP)‑based genetic linkage map of the pedigree. ■: Affected male carrying pathogenic mutations; ○: Normal female not carrying pathogenic mutations; ◇: Unaffected, of unknown gender, not carrying pathogenic mutations; ◆: Affected, of unknown gender, carrying pathogenic mutations; *: ADO, allele drop out; 0: Missing data; Number: Distance (Kb) to the upstream or downstream of the gene; Blue: Location of SNP in the upstream of the chromosomal position of the target gene; Orange: Location of SNP in the downstream of the chromosomal position of the target gene; Left slash inside the haplotype: haplotype linked to maternal mutation; Right slash inside the haplotype: haplotype linked to paternal mutation.
| Probe ID | Chr | Pos | Male | Female | E-1 | E-2 | E-3 | E-4 | E-5 | E-6 | E-7 | E-8 | E-9 | E-10 | E-11 | E-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1884723 | 2 | 31477017 | A G | A A | A A | G A | 0 0 | G A | G A | A A | A A | A A | A A | A A | A A | A A |
| rs142131665 | 2 | 31493330 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | T T | T T | T T | T T |
| rs28810790 | 2 | 31495488 | C G | C C | 0 0 | G C | G C | 0 0 | 0 0 | C C | C C | C C | C C | C C | 0 0 | C C |
| rs206843 | 2 | 31496262 | T G | T T | 0 0 | G T | ** | G T | 0 0 | T T | T T | T T | T T | T T | 0 0 | T T |
| rs61316601 | 2 | 31496406 | C T | C C | C C | T C | T C | 0 0 | T C | C C | C C | C C | C C | C C | C C | C C |
| rs55819371 | 2 | 31497812 | C A | C C | C C | A C | A C | A C | A C | C C | C C | C C | C C | C C | C C | C C |
| rs55744195 | 2 | 31497904 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs149051613 | 2 | 31498476 | C T | C C | C C | T C | 0 0 | T C | 0 0 | C C | C C | C C | C C | C C | C C | C C |
| rs191882 | 2 | 31498514 | C G | C C | C C | G C | 0 0 | G C | G C | C C | C C | C C | C C | C C | C C | C C |
| rs56175176 | 2 | 31516496 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs1475054 | 2 | 31516895 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs6760199 | 2 | 31517208 | A C | A A | 0 0 | C A | C A | 0 0 | C A | A A | A A | A A | A A | 0 0 | A A | A A |
| rs2224310 | 2 | 31517397 | C T | C C | C C | 0 0 | 0 0 | 0 0 | T C | C C | C C | C C | C C | 0 0 | C C | C C |
| rs143268195 | 2 | 31796171 | C T | C C | C C | T C | T C | T C | T C | C C | C C | C C | C C | C C | C C | C C |
| rs182127845 | 2 | 31832726 | A C | C C | A C | C C | C C | C C | C C | A C | A C | A C | A C | A C | A C | A C |
| rs4952254 | 2 | 32258435 | A G | G G | 0 0 | G G | G G | G G | G G | A G | A G | A G | A G | A G | A G | A G |
| rs75949879 | 2 | 32641555 | C G | C C | C C | G C | G C | 0 0 | G C | C C | C C | 0 0 | C C | C C | C C | C C |
| rs17012046 | 2 | 32656752 | A G | A A | 0 0 | G A | ** | ** | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs11124289 | 2 | 32724978 | C A | A A | C A | A A | A A | A A | A A | C A | C A | C A | C A | C A | C A | C A |
| rs75085204 | 2 | 32725079 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs34557851 | 2 | 32729211 | C T | C C | 0 0 | T C | T C | 0 0 | 0 0 | C C | 0 0 | C C | C C | C C | C C | C C |
| rs77172649 | 2 | 32750915 | T C | T T | T T | C T | C T | 0 0 | C T | T T | T T | T T | T T | T T | T T | T T |
| rs971511 | 2 | 32750955 | T C | C C | T C | 0 0 | C C | C C | C C | 0 0 | T C | T C | T C | T C | T C | T C |
| rs3769579 | 2 | 32751020 | A G | A A | A A | G A | G A | G A | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs17012217 | 2 | 32763264 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs17012247 | 2 | 32771766 | A C | A A | A A | C A | C A | C A | C A | A A | A A | A A | A A | A A | A A | A A |
| rs73924058 | 2 | 32805458 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | 0 0 | T T | T T | T T |
| rs17012300 | 2 | 32805642 | G A | G G | G G | A G | ** | A G | A G | G G | G G | G G | G G | G G | G G | G G |
| rs74682337 | 2 | 32826076 | G A | G G | G G | A G | A G | 0 0 | A G | G G | G G | G G | G G | G G | G G | G G |
| rs7591981 | 2 | 32838505 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
表1 SNP位点检测结果
Tab.1 Single nucleotide polymorphism haplotype analysis
| Probe ID | Chr | Pos | Male | Female | E-1 | E-2 | E-3 | E-4 | E-5 | E-6 | E-7 | E-8 | E-9 | E-10 | E-11 | E-12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1884723 | 2 | 31477017 | A G | A A | A A | G A | 0 0 | G A | G A | A A | A A | A A | A A | A A | A A | A A |
| rs142131665 | 2 | 31493330 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | T T | T T | T T | T T |
| rs28810790 | 2 | 31495488 | C G | C C | 0 0 | G C | G C | 0 0 | 0 0 | C C | C C | C C | C C | C C | 0 0 | C C |
| rs206843 | 2 | 31496262 | T G | T T | 0 0 | G T | ** | G T | 0 0 | T T | T T | T T | T T | T T | 0 0 | T T |
| rs61316601 | 2 | 31496406 | C T | C C | C C | T C | T C | 0 0 | T C | C C | C C | C C | C C | C C | C C | C C |
| rs55819371 | 2 | 31497812 | C A | C C | C C | A C | A C | A C | A C | C C | C C | C C | C C | C C | C C | C C |
| rs55744195 | 2 | 31497904 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs149051613 | 2 | 31498476 | C T | C C | C C | T C | 0 0 | T C | 0 0 | C C | C C | C C | C C | C C | C C | C C |
| rs191882 | 2 | 31498514 | C G | C C | C C | G C | 0 0 | G C | G C | C C | C C | C C | C C | C C | C C | C C |
| rs56175176 | 2 | 31516496 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs1475054 | 2 | 31516895 | G C | G G | G G | C G | C G | C G | C G | G G | G G | G G | G G | G G | G G | G G |
| rs6760199 | 2 | 31517208 | A C | A A | 0 0 | C A | C A | 0 0 | C A | A A | A A | A A | A A | 0 0 | A A | A A |
| rs2224310 | 2 | 31517397 | C T | C C | C C | 0 0 | 0 0 | 0 0 | T C | C C | C C | C C | C C | 0 0 | C C | C C |
| rs143268195 | 2 | 31796171 | C T | C C | C C | T C | T C | T C | T C | C C | C C | C C | C C | C C | C C | C C |
| rs182127845 | 2 | 31832726 | A C | C C | A C | C C | C C | C C | C C | A C | A C | A C | A C | A C | A C | A C |
| rs4952254 | 2 | 32258435 | A G | G G | 0 0 | G G | G G | G G | G G | A G | A G | A G | A G | A G | A G | A G |
| rs75949879 | 2 | 32641555 | C G | C C | C C | G C | G C | 0 0 | G C | C C | C C | 0 0 | C C | C C | C C | C C |
| rs17012046 | 2 | 32656752 | A G | A A | 0 0 | G A | ** | ** | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs11124289 | 2 | 32724978 | C A | A A | C A | A A | A A | A A | A A | C A | C A | C A | C A | C A | C A | C A |
| rs75085204 | 2 | 32725079 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs34557851 | 2 | 32729211 | C T | C C | 0 0 | T C | T C | 0 0 | 0 0 | C C | 0 0 | C C | C C | C C | C C | C C |
| rs77172649 | 2 | 32750915 | T C | T T | T T | C T | C T | 0 0 | C T | T T | T T | T T | T T | T T | T T | T T |
| rs971511 | 2 | 32750955 | T C | C C | T C | 0 0 | C C | C C | C C | 0 0 | T C | T C | T C | T C | T C | T C |
| rs3769579 | 2 | 32751020 | A G | A A | A A | G A | G A | G A | G A | 0 0 | A A | A A | A A | A A | A A | A A |
| rs17012217 | 2 | 32763264 | T C | T T | T T | C T | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| rs17012247 | 2 | 32771766 | A C | A A | A A | C A | C A | C A | C A | A A | A A | A A | A A | A A | A A | A A |
| rs73924058 | 2 | 32805458 | T G | T T | T T | G T | G T | G T | G T | T T | T T | T T | 0 0 | T T | T T | T T |
| rs17012300 | 2 | 32805642 | G A | G G | G G | A G | ** | A G | A G | G G | G G | G G | G G | G G | G G | G G |
| rs74682337 | 2 | 32826076 | G A | G G | G G | A G | A G | 0 0 | A G | G G | G G | G G | G G | G G | G G | G G |
| rs7591981 | 2 | 32838505 | T C | T T | T T | 0 0 | C T | C T | C T | T T | T T | T T | T T | T T | T T | T T |
| Embryo No. | Embryo grade | Detection result | Gene mutation site detection |
|---|---|---|---|
| E-1 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-2 | 4AB (D5) | Euploid | Carrying paternal mutations |
| E-3 | 4BB (D5) | Euploid | Carrying paternal mutations |
| E-4 | 3BB (D5) | Euploid | Carrying paternal mutations |
| E-5 | 4BA (D5) | Euploid | Carrying paternal mutations |
| E-6 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-7 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-8 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-9 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-10 | 4BA (D6) | Euploid | Not carrying paternal mutations |
| E-11 | 4BC (D6) | Euploid | Not carrying paternal mutations |
| E-12 | 4BB (D6) | Euploid | Not carrying paternal mutations |
表2 囊胚检测结果
Tab.2 Detection results of the blastocysts
| Embryo No. | Embryo grade | Detection result | Gene mutation site detection |
|---|---|---|---|
| E-1 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-2 | 4AB (D5) | Euploid | Carrying paternal mutations |
| E-3 | 4BB (D5) | Euploid | Carrying paternal mutations |
| E-4 | 3BB (D5) | Euploid | Carrying paternal mutations |
| E-5 | 4BA (D5) | Euploid | Carrying paternal mutations |
| E-6 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-7 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-8 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-9 | 4BB (D5) | Euploid | Not carrying paternal mutations |
| E-10 | 4BA (D6) | Euploid | Not carrying paternal mutations |
| E-11 | 4BC (D6) | Euploid | Not carrying paternal mutations |
| E-12 | 4BB (D6) | Euploid | Not carrying paternal mutations |
| 1 | de Souza PVS, de Rezende Pinto WBV, de Rezende Batistella GN, et al. Hereditary spastic paraplegia: clinical and genetic hallmarks[J]. Cerebellum, 2017, 16(2): 525-51. |
| 2 | Meyyazhagan A, Kuchi Bhotla H, Pappuswamy M, et al. The puzzle of hereditary spastic paraplegia: from epidemiology to treatment[J]. Int J Mol Sci, 2022, 23(14): 7665. |
| 3 | 郑备红, 孙 艳, 邱淑敏, 等. 全外显子测序技术鉴定一个常染色体显性遗传性痉挛性截瘫3A型家系ATL1基因突变[J]. 实用医学杂志, 2021, 37(13): 1761-4. |
| 4 | Ho NJ, Chen X, Lei Y, et al. Decoding hereditary spastic paraplegia pathogenicity through transcriptomic profiling[J]. Zool Res, 2023, 44(3): 650-62. |
| 5 | Zhu Z, Zhang C, Zhao G, et al. Novel mutations in the SPAST gene cause hereditary spastic paraplegia[J]. Parkinsonism Relat Disord. 2019, 69: 125-33. |
| 6 | Murala S, Nagarajan E, Bollu PC. Hereditary spastic paraplegia[J]. Neurol Sci, 2021, 42(3): 883-94. |
| 7 | Shribman S, Reid E, Crosby AH, et al. Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches[J]. Lancet Neurol, 2019, 18(12): 1136-46. |
| 8 | Elert-Dobkowska E, Stepniak I, Radziwonik-Fraczyk W, et al. SPAST intragenic CNVs lead to hereditary spastic paraplegia via a haploinsufficiency mechanism[J]. Int J Mol Sci, 2024, 25(9): 5008. |
| 9 | Chen R, Du SY, Yao YY, et al. A novel SPAST mutation results in spastin accumulation and defects in microtubule dynamics[J]. Mov Disord, 2022, 37(3): 598-607. |
| 10 | Parodi L, Fenu S, Barbier M, et al. Spastic paraplegia due to SPAST mutations is modified by the underlying mutation and sex[J]. Brain. 2018,141(12): 3331-42. |
| 11 | Mo A, Saffari A, Kellner M, et al. Early-onset and severe complex hereditary spastic paraplegia caused by de novo variants in SPAST[J]. Mov Disord, 2022, 37(12): 2440-6. |
| 12 | Cui F, Sun LQ, Qiao J, et al. Genetic mutation analysis of hereditary spastic paraplegia: a retrospective study[J]. Medicine, 2020, 99(23): e20193. |
| 13 | Panza E, Meyyazhagan A, Orlacchio A. Hereditary spastic paraplegia: genetic heterogeneity and common pathways[J]. Exp Neurol, 2022, 357: 114203. |
| 14 | Méreaux JL, Banneau G, Papin M, et al. Clinical and genetic spectra of 1550 index patients with hereditary spastic paraplegia[J]. Brain, 2022, 145(3): 1029-37. |
| 15 | Lallemant-Dudek P, Durr A. Clinical and genetic update of hereditary spastic paraparesis[J]. Rev Neurol, 2021, 177(5): 550-6. |
| 16 | Meyyazhagan A, Orlacchio A. Hereditary spastic paraplegia: an update[J]. Int J Mol Sci, 2022, 23(3): 1697. |
| 17 | 姜红芳, 余永林, 阮雯聪, 等. 儿童遗传性痉挛性截瘫临床与康复进展[J]. 中国实用儿科杂志, 2023, 38(1): 19-23. |
| 18 | 何秉涛, 黄美欢, 贠国俊, 等. 遗传性痉挛性截瘫基因分型、分子诊断和治疗的研究进展[J]. 中国优生与遗传杂志, 2022, 30(3): 516-9. |
| 19 | Bellofatto M, de Michele G, Iovino A, et al. Management of hereditary spastic paraplegia: a systematic review of the literature[J]. Front Neurol, 2019, 10: 3. |
| 20 | Di Ludovico A, Ciarelli F, la Bella S, et al. The therapeutic effects of physical treatment for patients with hereditary spastic paraplegia: a narrative review[J]. Front Neurol, 2023, 14: 1292527. |
| 21 | Parikh F, Athalye A, Madon P, et al. Genetic counseling for pre-implantation genetic testing of monogenic disorders (PGT-M)[J]. Front Reprod Health, 2023, 5: 1213546. |
| 22 | Madero JI, Manotas MC, García-Acero M, et al. Preimplantation genetic testing in assisted reproduction[J].Minerva Obstet Gynecol. 2023,75(3): 260-72. |
| 23 | Cornelisse S, Zagers M, Kostova E, et al. Preimplantation genetic testing for aneuploidies (abnormal number of chromosomes) in in vitro fertilisation[J]. Cochrane Database Syst Rev, 2020, 9(9): CD005291. |
| 24 | Yan L, Cao Y, Chen ZJ, et al. Chinese experts' consensus guideline on preimplantation genetic testing of monogenic disorders[J]. Hum Reprod, 2023, 38(): ii3-13. |
| 25 | Fan XY, Tang D, Liao YH, et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing[J]. PLoS Biol, 2020, 18(12): e3001017. |
| 26 | Cheng C, Fei ZJ, Xiao PF. Methods to improve the accuracy of next-generation sequencing[J]. Front Bioeng Biotechnol, 2023, 11: 982111. |
| 27 | Niu WB, Wang LL, Xu JW, et al. Improved clinical outcomes of preimplantation genetic testing for aneuploidy using MALBAC-NGS compared with MDA-SNP array[J]. BMC Pregnancy Childbirth, 2020, 20(1): 388. |
| 28 | Scarano C, Veneruso I, de Simone RR, et al. The third-generation sequencing challenge: novel insights for the omic sciences[J]. Biomolecules, 2024, 14(5): 568. |
| 29 | Searle B, Müller M, Carell T, et al. Third-generation sequencing of epigenetic DNA[J]. Angew Chem Int Ed, 2023, 62(14): e202215704. |
| 30 | Tan VJ, Liu TM, Arifin Z, et al. Third-generation single-molecule sequencing for preimplantation genetic testing of aneuploidy and segmental imbalances[J]. Clin Chem, 2023, 69(8): 881-9. |
| 31 | van Dijk EL, Naquin D, Gorrichon K, et al. Genomics in the long-read sequencing era[J]. Trends Genet, 2023, 39(9): 649-71. |
| 32 | de Rycke M, Berckmoes V. Preimplantation genetic testing for monogenic disorders[J]. Genes, 2020, 11(8): 871. |
| [1] | 杨 群, 刘晓鑫, 蒋玉娜, 马进安. 乙醛脱氢酶2基因rs671位点多态性与化疗所致的恶心呕吐相关[J]. 南方医科大学学报, 2023, 43(6): 1017-1022. |
| [2] | 穆 杰, 徐永申, 朱 辉. AF4/FMR2、IL-10基因多态性与强直性脊柱炎的遗传易感性和免疫浸润相关[J]. 南方医科大学学报, 2023, 43(5): 741-748. |
| [3] | 周小红, 王子贤, 秦 敏, 钟诗龙. 线粒体基因位点G12630A与冠心病患者他汀类药物诱导的肌痛相关[J]. 南方医科大学学报, 2020, 40(12): 1747-1752. |
| [4] | 李珍珍, 周兰庭, 翟立红, 肖 娟, 程正江. 胃癌中Piwil1基因启动子区rs28416520位点单核苷酸多态性及其临床意义[J]. 南方医科大学学报, 2020, 40(10): 1373-1379. |
| [5] | 徐惠玲,刘彦慧,晏平,何怡,秦佳纯,娄季武,周万军. 囊胚细胞WGA结合短片段Gap-PCR的α-地贫SEA突变快速植入前遗传学诊断[J]. 南方医科大学学报, 2018, 38(10): 1250-. |
| [6] | 何瑛,糟秀梅,魏学花. MDR1和CYP3A5基因多态性对伊马替尼治疗慢性骨髓性白血病预后的影响[J]. 南方医科大学学报, 2018, 38(01): 34-. |
| [7] | 向阳,郭静,彭友帆,蓝艳,黄华佗,韦叶生. 精氨酸加压素基因rs66818855A/G和rs1078152C/T在广西健康人群中的分布[J]. 南方医科大学学报, 2016, 36(07): 927-. |
| [8] | 黄香,邱勋定,孙明磊,何升腾. 海南汉族PLAG1 基因rs6474051 多态性与腮腺良性肿瘤的关联性[J]. 南方医科大学学报, 2015, 35(09): 1360-. |
| [9] | 黄文森,施亚雄,杨鑫娜,林莞蓉. 2型糖尿病患者PEDF基因启动子区多态性与非酒精性脂肪肝病的相关性[J]. 南方医科大学学报, 2015, 35(07): 1019-. |
| [10] | 周林,宗璐,张璐璐,邓聪,赵存友. GABRB2基因启动子区单核苷酸多态性与精神分裂症的关联[J]. 南方医科大学学报, 2015, 35(02): 256-. |
| [11] | 夏征,胡亚卓,张红红,韩志涛,白洁,傅淑宏,邓新立,何耀. 维生素D受体基因FokⅠ及BsmⅠ多态性与老年男性2型糖尿病脂代谢异常的相关性[J]. 南方医科大学学报, 2014, 34(11): 1562-. |
| [12] | 章俊,马亚琼,邱敏姿,杨蕾,卜阳,汤珣. 中国汉族TMEM39A基因单核苷酸多态性与系统性红斑狼疮的相关性[J]. 南方医科大学学报, 2014, 34(04): 556-. |
| [13] | 张鑫,孙学华,周振华,李曼,高月求. SRC同源区2结构域蛋白基因多态性与慢性乙型肝炎易感性的关系[J]. 南方医科大学学报, 2014, 34(04): 468-. |
| [14] | 熊兴东,成捷,刘新光,唐韶景,罗喜平. miR-124 rs531564基因多态性与宫颈癌遗传易感有密切关系[J]. 南方医科大学学报, 2014, 34(02): 210-. |
| [15] | 李虹,胡兆霆,谭珍妮,侯庆臻,彭健. GNAS1基因T393C单核苷酸多态性与非瓣膜性房颤的相关性[J]. 南方医科大学学报, 2013, 33(10): 1508-. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||