2. Second Department of Breast Cancer, Cancer Center, Guangdong Provincial People's Hospital/Guangdong Academy of Medical Sciences, Guangzhou 510080, China
2. 广东省人民医院乳腺二科,广东 广州 510080
It is estimated that 15%-20% of breast cancers express low levels of estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2) and highly express several basal cell markers. These breast cancers are collectively known as basal-like breast cancer (BLBC), which represents a heterogeneous subtype of breast cancer that is insensitive to endocrine or HER2-targeted therapies, prone to metastasize at early stage and has low 5-year survival rates[1, 2]. The molecular mechanism of invasion and metastasis of BLBC cells still remains unclear, and currently no effective targeted therapies have been available for treating this aggressive subtype of breast cancer.
Histone deacetylase 11 (HDAC11) was first identified by Gao et al in 2002[3]. Among the total of 18 HDACs in 4 classes, HDAC11 is the only member of Class Ⅳ, which shares sequence similarities with both Class Ⅰ and Ⅱ, and dominates negative regulation of interleukin 10 expression in antigen-presenting cells[4]. HDAC11 is associated with the survival of motor neuron complex and participates in mRNA splicing process[5]. Buglio et al[6] found that HDAC11 knockdown induced OX40 ligand expression in Hodgkin lymphoma. Some non-histone proteins have been implicated as the binding partners or substrates of HDAC11 [7-9]. Recently, HDAC11 was also identified to have fatty-acid de-acylase activity [10, 11]. Although recent studies have indicated the pathological roles of HDAC11 in neuroblastoma[12] and pituitary tumor [13], the functional role and molecular mechanism of HDAC11 in regulating the invasion and metastasis of BLBC remain unclear. In this study, we aimed to explore the role of HDAC11 in modulating the invasion and metastasis of BLBC cells and the underlying mechanism by antagonizing the function of Twist protein.
MATERIALS AND METHODS Cell cultureBreast cancer cell line SUM1315 was cultured in Ham's F-12 medium supplemented with 5% fetal bovine serum (FBS, GIBCO, USA), 10 ng/mL epidermal growth factor (EGF, Peprotech, USA), and 10 μg/mL insulin. BT549 and T47D cells were maintained in RPMI-1640 supplemented with 10% FBS. HMLE cells were cultured in DMEM/F-12mediumcontaining 10ng/mLEGF, 10μg/mL insulin and 0.5 μg/mL hydrocortisone. Other breast cancer cell lines (MDA-MB-231, MCF7, BT474, ZR75, MDA-MB-157, MDA-MB-435, SUM149 and SUM159) were grown in DMEM/F12 medium with 10% FBS. Lentivirus for HDAC11 expression or the short hairpin RNA (shRNA) plasmids (Sigma, USA) were transfected into HEK293T cells for virus package. The breast cancer cell lines were selected with puromycin (2 μg/mL) or blasticidin (5 μg/mL), and the stable cell polyclones were constructed and the transfection efficiency was examined using Western blotting.
ReagentsAntibodies for Twist, HDAC11 and HA tag were respectively purchased from Abcam (Cambridge, MA), GeneTex (Irvine, CA) and Roche (Basel, Switzerland). The shRNA against HDAC11 and the antibodies against Flag tag and β-actin were purchased from Sigma-Aldrich (St. Louis, MO). Smart pool siRNA against human HDAC11, HAS2 and Twist were obtained from Dharmacon (Chicago, IL). The lentiviral expression plasmid of HDAC11 was from GeneCopoeia (Guangzhou, China).
Immunoprecipitation, immunoblotting and real-time PCRFor immunoblotting, the cells were harvested and lysed, and the protein concentrations were determined using BCA protein assay kit (Keygentec, China). Equal amounts of the purified proteins were separated by 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were incubated with the primary antibodies for HDAC11 (1:1000), Twist (1:1000), GAPDH (Proteintech; 1:1000) and β-actin (Sigma; 1:5000). The anti- rabbit or anti- mouse secondary antibody (Earthox) was used at the dilution of 1:5000. The Western signals were detected using enhanced chemiluminescence kit (FDbio Science). For immunoprecipitation, the cell lysates were extracted using IP buffer (50 mmol/L TrisHCl [pH7.4], 150 mmol/L NaCl, 0.2 mmol/L EDTA, 0.2% NP40, 10% glycerol and protease inhibitors) and immunoprecipitated with the indicated antibodies and protein G-sepharose (Thermo, USA). Pulldown protein complexes were analyzed by Western blotting using antiTwist, HDAC11, HA and Flag antibodies. For real-time PCR (RT- PCR), the total RNA was extracted from the cells by RNeasy Mini Kit (#74104, Qiagen, Valencia, CA), and 1 μg of RNA was reverse transcribed by SuperScriptR Ⅲ reverse transcriptase (#18080044, Thermo Fisher Scientific, Waltham, MA). Real-time PCR was analyzed using Power SYBR Green Master Mix (Applied Biosystems, USA). The relative expressions were calculated using the ΔΔCT method. GAPDH was used as the housekeeping gene for normalization. The results were represented as relative fold change. The primers of HAS2 gene for RT-PCR were: 5'-gggggagatgtc cagatttt-3' and 5'-atgcactgaacacacccaaa-3'.
Chromatin immunoprecipitationApproximately 1 × 106 control and HDAC11-overexpressing SUM1315 and BT549 cells were fixed with cross-link solution and collected. ChIP assays were performed using Imprint Chromatin Immunoprecipitation Kit (Sigma, #CHP1) according to the manufacturer's instructions. Twist antibody-immunoprecipitated DNA was analyzed by real-time PCR. The specific primers for the HAS2 promoter were: 5'-gaccagcgcgatctttttag-3' and 5'-tgtggcgctgaagtaatttg-3'. The unspecific primers for CHIP experiment were: 5'-ccgttggctgtgctgtattt-3' and 5'-agcttcacattttgggcagg-3'.
Luciferase reporter assayWe cloned the human HAS2 gene promoter region (-1438 bp upstream of the translation initiation site) into pGL3 reporter plasmid and generated its promoter-luciferase construct. HEK293T cells were seeded in 60 mm dishes and transfected with the plasmids using FuGene 6 transfection reagent (Roche) for 24 h. The cell lysates were extracted with passive lysis buffer (Promega, Madison, WI), and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). All the experiments were performed for 3 times in triplicate. The relative luciferase activities were calculated as folds of induction compared with the vector control.
Invasion assayEach insert (8-μm pore size, Falcon) was coated with Matrigel (BD biosciences, San Jose, CA) overnight. 1×105 control or clone cells were re-suspended in 150μL serumfree medium and seeded in the upper Boyden chamber coated with Matrigel while the bottom chambers were filled with 600 μL serum-free medium containing 100 nmol/L LPA. After 24-48 h, the non-invasive cells on the apical side of the membrane were removed using wet cotton swabs, and the invasive cells were stained with crystal violent and counted under a microscope. The experiments were performed for 3 times in triplicate.
Hyaluronan detection AssayAn equal number of control and treated cells were seeded in culture dishes, and 2 days later the culture media were collected to determine hyaluronan level using Quantikin ELISA kit following the manufacturer's protocol (R&D system, #DHYAL0). The amount of hyaluronan was determined using a microplate reader for measuring the absorbance at 450 nm with a correction wave length at 570 nm. Fresh culture medium was used as the negative control to determine the baseline concentration of hyaluronan.
Gene expression correlation analysisMicroarray gene expression data for breast carcinoma patients from 4 studies, including Bos et al (GSE12276, n=204), Loi et al (GSE9195, n=77), Pawitan et al (GSE1456, n=159) and Schmidt et al (GSE11121, n= 200), were downloaded from the Oncomine database. Pearson's correlation coefficient was calculated to analyze the correlation between Twist and HAS2 expression. The P value was calculated based on the hypothesis that Pearson's correlation coefficient equals zero, i.e. the expressions of the genes are independent.
Metastasis modelThe animal experiments were performed with the approval of and following the guidelines of the Southern Medical University Animal Care and Use Committee. Female BALB/c nude mice (4-6 weeks) were purchased from Guangdong Medical Laboratory Animal Center. The mice were housed in autoclaved, ventilated cages and provided with autoclaved water. The mice were subjected to injections of 2 × 105 vector control or HDAC11- overexpressing MDA-MB-231 cells via the tail vein (n=7 in each group). After 35 days, the mice were euthanized, and the metastatic nodules in the lung were counted and photographed.
Differential gene expression analysisHDAC11 expression levels in breast cancers of different molecular subtypes were analyzed. The gene expression data and the corresponding clinical information were obtained from NCBI Gene Expression Omnibus (GEO) database, Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) and TCGABRCA datasets. Multiple comparisons were made by Nemenyi-tests of the mean rank sums of independent samples. P values were determined using Kruskal-Wallis rank sum test performed in R Environment Version 3.4.1.
Statistical AnalysisThe data of the continuous variables are presented as Mean±SD. Student's t test (two-tailed) was used for comparison of data in normal distribution with homogeneous variance between two groups. Multiple comparisons were performed using one-way ANOVA and Welch's test for data with unequal variance. A P value < 0.05 was considered to indicate a statistically significant difference.
RESULTS HDAC11 is lowly expressed in BLBCSince previous study suggested that promoter methylation of HDAC11 gene is highly associated with poor outcome of ovarian cancer[14], we were interested in the expression status and functional role of HDAC11 in basal like breast cancer. Differential gene expression analysis of breast cancer specimens indicated that the expression level of HDAC11 in basal-like subtype is significantly lower than that in luminal-A or luminal- B breast cancer (Fig. 1A). We also examined protein expression of several HDAC family members in normal and breast cancer cell lines. HDAC11 expression was found to decrease exclusively in BLBC cells compared to other cells, while the protein levels of other HDAC family members including HDAC2, HDAC3 and HDAC6 were relatively constant in all the cell lines (Fig. 1B). These observations suggest a specific pathological role of HDAC11 in BLBC.
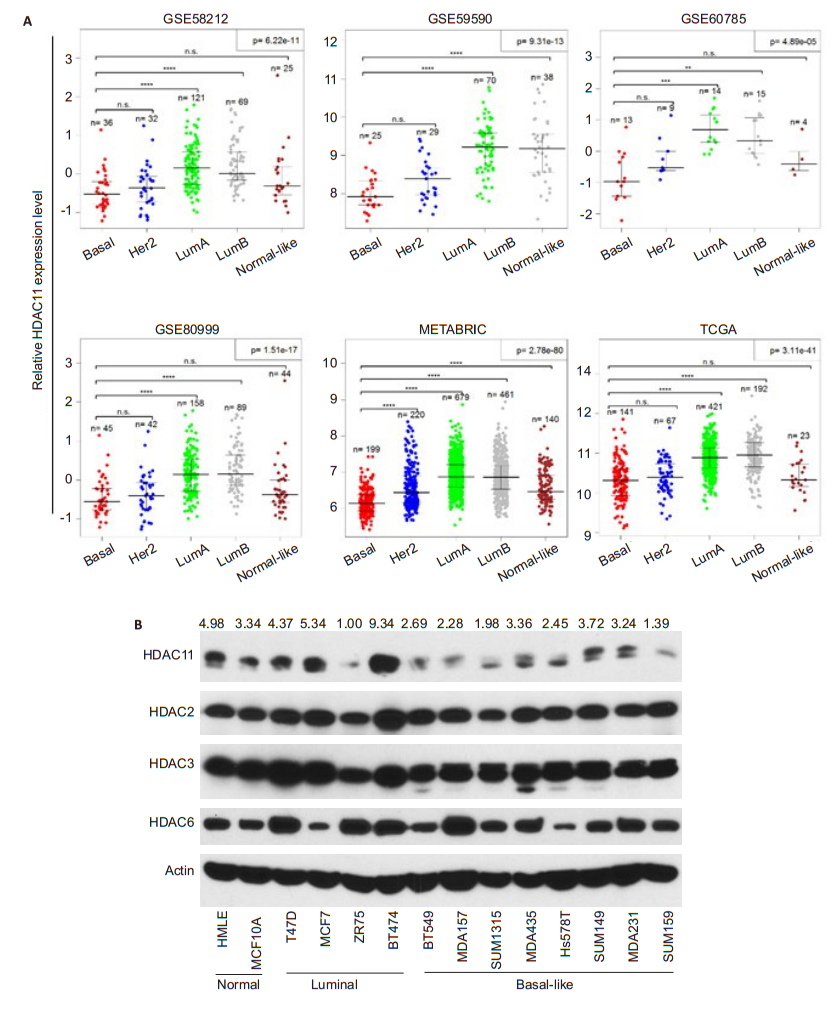
|
Fig.1 HDAC11 is lowly expressed in BLBC. A: Differential gene expression analysis derived from 6 databases containing clinical information of patients with breast cancer of Luminal A, Luminal B, HER2+, basal-like and Normal-like subtypes; B: Protein levels of HDAC11 and other HDAC family members analyzed by Western blotting in 2 normal breast cell lines, 4 luminal breast cancer and 8 BLBC cell lines. |
To explore the role of HDAC11 in BLBC progression, we over- expressed HDAC11 in BLBC cell lines BT549 and SUM1315 (Fig. 2A). HDAC11 overexpression did not obviously affect the growth potential of the tumor cells (data not shown), but Transwell invasion analysis showed that overexpression of HDAC11 significantly inhibited the cell invasion (Fig. 2B). To study whether HDAC11 is involved in the regulation of cell metastasis, we constructed stable HDAC11-overexpressing MDA-MB- 231 clones (Fig. 2C). Transwell experiments again confirmed that ectopic expression of HDAC11 inhibited the cell invasive ability (Fig. 2D). The in vivo studies in mice showed that intravenous injection of HDAC11- overexpressing MDA-MB-231 cells resulted in significantly fewer lung metastatic nodules compared to vector control group (Fig. 2E). These results demonstrate that HDAC11 overexpression suppresses the invasion and metastasis of BLBC cells.
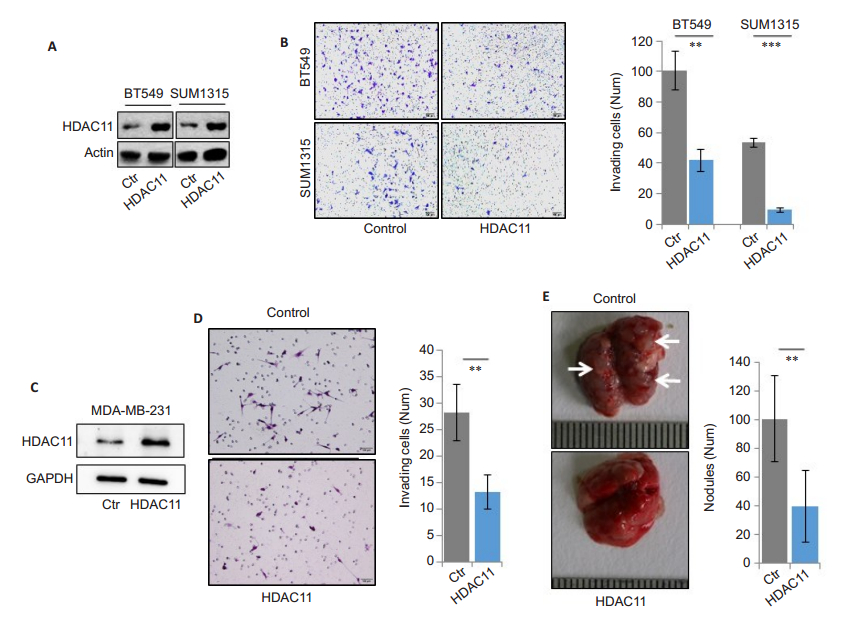
|
Fig.2 HDAC11 inhibits BLBC invasion and metastasis. A: Measurement of HDAC11 over-expression in SUM1315 and BT549 cells. Polyclonal HDAC11-overexpressing cells are selected and representative Western blotting results from 3 independent transfection experiments are shown; B: Transwell assay for assessment of invasive ability of BT549 cell (**P < 0.01) and SUM1315 cells (***P < 0.001). Typical images of invading cells (left panel) and statistical data (right panel) are shown; C: Detection of stable HDAC11 overexpression in MDA-MB-231 cells; D: Transwell assay for assessing cell invasive ability. Typical images of invading cells (left panel) and statistical data (right panel) are shown (**P < 0.01); E: MDA-MB-231 vector control or HDAC11-overexpressing cells are injected into the tail vein of BALB/c nude mice, and the photographs show the metastatic nodules in the lungs (left panel). The statistical data are shown in the right panel (**P < 0.01). |
To identify the molecular mechanism underlying the inhibitory effect of HDAC11 on tumor cell metastasis, we pulled down endogenous Twist and Snail proteins that are essential to tumor cell invasion and metastasis[15]. The results revealed the presence of HDAC11, but not HDAC6, in the immunoprecipitated Twist complex (Fig. 3A); Snail did not form a complex with HDAC11 (data not shown), indicating that HDAC11 forms an endogenous protein complex with Twist in BLBC cells. We further constructed wild-type and deletion mutant plasmids of Twist and also of HDAC11 to determine the specific binding regions between the two proteins (Fig. 3B). After co-expression of the wild-type and mutant Twist plasmids with wild-type HDAC11, only TDL4 mutant completely lost while the others retained strong interaction with HDAC11 (Fig. 3C), suggesting that the latter 102 amino acids in Twist protein contains the potential HDAC11-binding motif. Interestingly, this region also contains the bHLH motif, which is responsible for DNA binding [16]. To investigate the specific Twist-binding domain in HDAC11 protein, we co-transfected wild-type Twist along with full-length HDAC11 and its mutants (Fig. 3D). The pull-down experiments showed that HDL2 and HDL3 completely lost interaction with Twist, suggesting that the first 17 amino acids in the N-terminus of HDAC11 is responsible for Twist binding. Because ectopic expression of Twist was shown to enhance the invasive ability in epithelial cells and Twist is able to bind to HDAC11, we overexpressed Twist and simultaneously knocked down endogenous HDAC11 to test whether HDAC11 modulates the biological function of Twist (Fig. 3E). As expected, Twist expression itself obviously promoted cell invasion, and this effect was significantly enhanced by HDAC11 knockdown (Fig. 3F). On the other hand, HDAC11 knockdown alone promoted the cell invasion, and this effect was partially attenuated by co-knockdown of Twist (Fig. 3G-H), validating that HDAC11 inhibits BLBC cell invasion by blocking Twist activity.
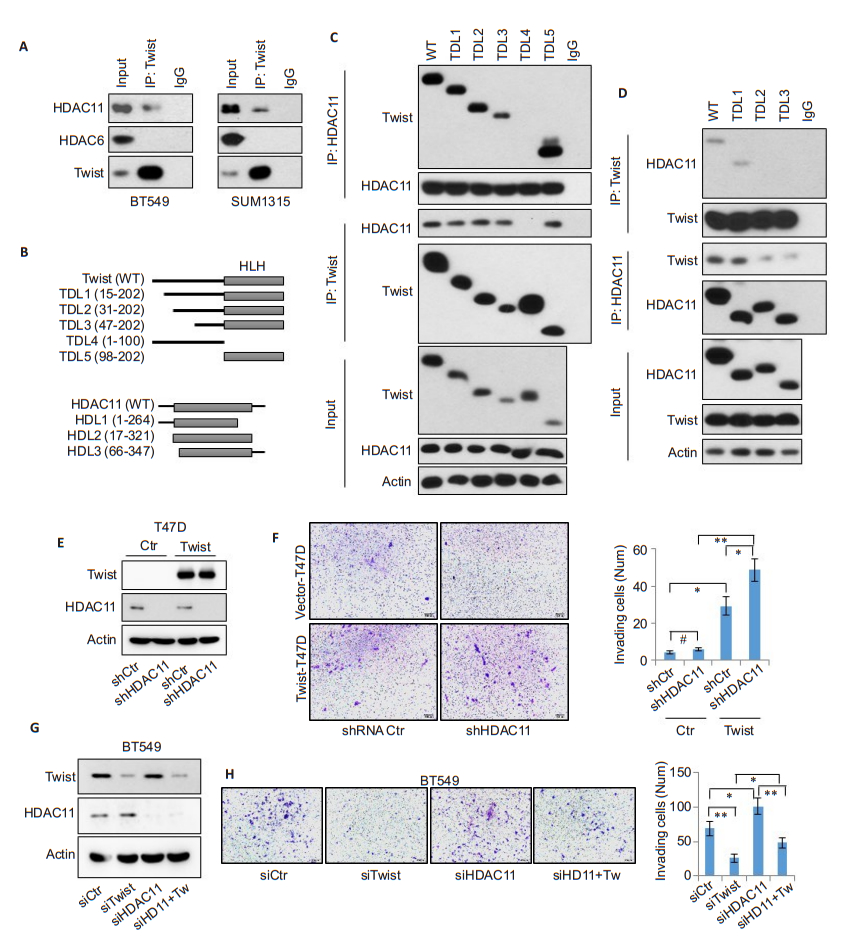
|
Fig.3 HDAC11 interacts with Twist and antagonizes its function. A: Endogenous Twist protein is immunoprecipitated from SUM1315 and BT549 cells, and HDAC11 pulldown is examined by Western blotting; B: Schematic depiction of full-length Twist and its 5 deletion mutants used (top panel) [including TDL1 (15-202 aa), TDL2 (31-202 aa), TDL3 (47-202 aa), TDL4 (1-100 aa) and TDL5 (98-202 aa)], and full-length HDAC11 and its 3 deletion mutants used (bottom panel) [including HDL1 (1-264 aa), HDL2 (17-321 aa) and HDL3 (66-347 aa)]; C: HA-tagged wild-type and 5 deletion mutants of Twist are co-expressed with Flag-tagged wild-type HDAC11 in HEK293T cells. Immunoprecipitation is performed using anti-HA or anti-Flag antibody, and Western blotting is used to detect the bound HDAC11 or Twist; D: Flag-tagged wild-type and 3 deletion mutants of HDAC11 are co-expressed with HA-tagged wild-type Twist in HEK293T cells. After immunoprecipitation with anti-HA or anti-Flag antibody, the bound HDAC11 or Twist is examined by Western blotting; E: Knockdown of endogenous HDAC11 by shRNA plasmid in Twist-overexpressing T47D cells. The expression of Twist and HDAC11 is detected by Western blotting; F: Transwell assay for assessing cell invasive ability. Typical images of invading cells (left panel) and statistical data (right panel) are shown. (*P < 0.05, **P < 0.01); G: Endogenous Twist and/or HDAC11 are knocked down by siRNA in BT549 cells; H: Transwell assay of cell invasive ability (*P < 0.05, **P < 0.01). |
We further studied whether HDAC11 modulates the expression of the target gene of Twist. Previously we performed cDNA microarray analysis of T47D/Twist cells (GSE53222) and found that Twist overexpression significantly enhanced the mRNA expression of hyaluronan synthase 2 (HAS2), suggesting that the latter is a likely target of Twist[15]. In this study, we found that Twist overexpression strongly induced HAS2 expression, which was suppressed by HDAC11 overexpression (Fig. 4A). Because HAS2 is responsible for hyaluronan production, we further studied whether Twist can modulate endogenous hyaluronan level. We found that Twist overexpression significantly enhanced hyaluronan production, and such effect was significantly blocked by HDAC11 overexpression (Fig. 4B). Intriguingly, HDAC11 overexpression also reduced HAS2 expression, which was partially reversed by co-expression of Twist (Fig. 4C). These data indicate that Twist and HDAC11 interplay to regulate the expression of HAS2. To verify that HAS2 is a direct target gene of Twist, we cloned human HAS2 gene promoter region and generated its promoter-luciferase construct. The results showed that Twist strongly induced luciferase activity; HDAC11 coexpression significantly suppressed the effect of Twist (Fig. 4D). Moreover, chromatin immunoprecipitation results showed that Twist was associated with HAS2 gene promoter, and overexpression of HDAC11 reduced the binding of Twist to HAS2 gene promoter (Fig. 4E). Analysis of the mRNA expressions of HAS2 and Twist in the gene expression datasets revealed a positive correlation between them (Fig. 4F). Consistent with previous observations, knockdown of HAS2 resulted in a significant reduction of hyaluronan production and suppressed the cell invasion (Fig. 4G-H). These data confirm that HDAC11 inhibits Twist/HAS2 signaling.
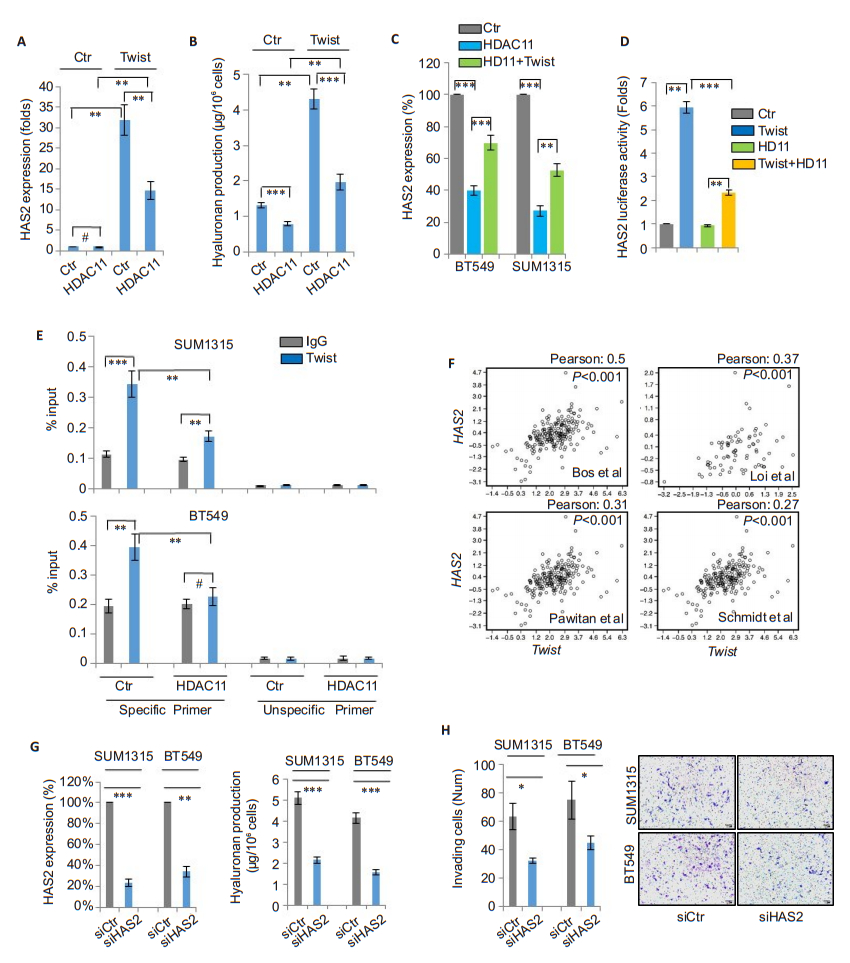
|
Fig.4 HDAC11 represses the expression of Twist-targeted gene HAS2. A: HAS2 mRNA levels are detected by real-time PCR in T47D, T47D-Twist cells and their HDAC11 overexpressing clones (**P < 0.01); B: Hyaluronan levels are detected in T47D, T47DTwist cells and their HDAC11 overexpressing clones (**P < 0.01, ***P < 0.001); C: HAS2 mRNA levels are measured in SUM1315 and BT549 cells with or without HDAC11 or Twist over-expression (**P < 0.01, ***P < 0.001); D: Detection of HAS2 promoter luciferase reporter activity in HEK293T cells co-transfected with plasmids of HDAC11 and Twist. Data are shown as Mean ± SD (**P < 0.01, ***P < 0.001); E: Chromatin immunoprecipitation for vector control and HDAC11-overexpressing SUM1315 and BT549 cells using anti-Twist antibody, and results are analyzed by real-time PCR (**P < 0.01, ***P < 0.001); F: Analysis of mRNA expression correlation between TWIST and HAS2 in 4 gene expression datasets. Corresponding Pearson correlation coefficients were shown; G: siRNAmediated HAS2 knockdown in SUM1315 and BT549 cells detected by real-time PCR (left panel). Hyaluronan levels are measured by hyaluronan assay (right panel) (**P < 0.01, ***P < 0.001); H: Assessment of cell invasion of control and HAS2-knockdown SUM1315 and BT549 cells. Statistical data (left panel) (*P < 0.05) and representative images of invading cells (right panel) are shown. |
Our data support the suppressive role of HDAC11 in modulating the metastasis of BLBC. Due to its high rate of early metastasis and the lack of effective targeted therapies, BLBC is considered the most deadly subtype of breast cancer, and understanding of the underlying signaling molecules that modifies the metastatic capacity of BLBC cells in the early stage can be of crucial importance. We found that the expression level of HDAC11 decreased specifically in BLBC cells in comparison with other breast cancer cells and normal breast cells. Over-expression of HDAC11 strongly inhibited the invasion as well as metastasis of BLBC cells both in vitro and in vivo, while HDAC11 knockdown promoted their invasion and metastasis. These data suggest that a drug that enhances HDAC11 expression may confer therapeutic benefit in the treatment of BLBC.
We show that HDAC11 recognizes and interacts with the DNA binding region of Twist by its N-terminal fragment, leading to the inhibition of Twist transcriptional activity and also the expression of its target gene HAS2. Knockdown of HDAC11 in T47D cells potentiated Twist-induced cell invasion, and silencing of Twist partially reversed cell invasion induced by HDAC11 knockdown in the cells. Twist is essential for tumor progression for its roles in initiating the epithelialmesenchymal transition (EMT) process and inducing cell invasion and metastasis[16-20]. However, targeted inhibition of Twist is difficult as it lacks an obvious ligand binding domain. Here we identify HDAC11 as an endogenous modulator of Twist by physically occupying the DNAbinding motif of Twist, indicating that HDAC11 suppresses BLBC metastasis by directly targeting to Twist protein, which shed light on a strategy to antagonize the pro-oncogenic activity of Twist. But as a de-acetylase, HDAC11 has been implicated to have several downstream protein substrates besides Twist and participate in several physiological processes[7-9], thus direct pharmacological targeting of HDAC11 may potentially cause cytotoxic and unspecific effects. The discovery of small molecules that specifically stabilize HDAC11-Twist interaction might be a safer strategy to block the oncogenic activity of Twist.
Our findings also identify HAS2 as a novel target gene of Twist. Knowledge from studies of Drosophila mesoderm development indicates that Twist is a master transcriptional factor that governs multiple physiological processes [21]. Our microarray analysis implicated several hundred of transcriptionally active genes in Twistoverexpressing cells, which are the likely target genes of Twist[15]. HAS2 is a member of vertebrate hyaluronan synthase family. The catalytic product of HAS2 is hyaluronan or hyaluronic acid, a high-molecular-mass polysaccharide in the extracellular matrix and plays key roles in cell migration[22, 23]. Recently, HAS2 was reported to participate in EMT and tumor cell invasion [24-27], but how tumor cells manipulate HAS2 expression and hyaluronan production is still unknown. The identification of HAS2 as a new target gene of Twist helps to clarify how tumor cells modulate their endogenous hyaluronan synthesis at the transcriptional level and enriches our understanding of Twist signaling.
AcknowledgementWe thank Dr. Edward Seto (H. Lee Moffitt Cancer Center and Research Institute) for providing the plasmids of fulllength and deletion mutants of HDAC11.
| [1] |
Rakha EA, El-Sayed ME, Reis-Filho J, et al. Patho-biological aspects of basal-like breast cancer[J]. Breast Cancer Res Treat, 2009, 113(3): 411-22. DOI:10.1007/s10549-008-9952-1 |
| [2] |
Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triplenegative breast cancer-the road to new treatment strategies[J]. Lancet, 2017, 389(10087): 2430-42. DOI:10.1016/S0140-6736(16)32454-0 |
| [3] |
Gao L, Cueto MA, Asselbergs F, et al. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family[J]. J Biol Chem, 2002, 277(28): 25748-55. DOI:10.1074/jbc.M111871200 |
| [4] |
Villagra A, Cheng F, Wang HW, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance[J]. Nat Immunol, 2009, 10(1): 92-100. DOI:10.1038/ni.1673 |
| [5] |
Joshi P, Greco TM, Guise AJ, et al. The functional interactome landscape of the human histone deacetylase family[J]. Mol Syst Biol, 2003, 9: 672. |
| [6] |
Buglio D, Khaskhely NM, Voo KS, et al. HDAC11 plays an essential role in regulating OX40 ligand expression in Hodgkin lymphoma[J]. Blood, 2011, 117(10): 2910-7. DOI:10.1182/blood-2010-08-303701 |
| [7] |
Glozak MA, Seto E. Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1[J]. J Biol Chem, 2009, 284(17): 11446-53. DOI:10.1074/jbc.M809394200 |
| [8] |
Feng W, Lu Z, Luo RZ, et al. Multiple histone deacetylases repress tumor suppressor gene ARHI in breast cancer[J]. Int J Cancer, 2007, 120(8): 1664-8. DOI:10.1002/ijc.v120:8 |
| [9] |
Lozada EM, Andrysik Z, Yin M, et al. Acetylation and deacetylation of Cdc25A constitutes a novel mechanism for modulating Cdc25A functions with implications for cancer[J]. Oncotarget, 2016, 7(15): 20425-39. |
| [10] |
Moreno-Yruela C, Galleano I, Madsen AS, et al. Histone deacetylase 11 is an epsilon-N-myristoyllysine hydrolase[J]. Cell Chem Biol, 2018, 25(7): 849-56. DOI:10.1016/j.chembiol.2018.04.007 |
| [11] |
Kutil Z, Novakova Z, Meleshin M, et al. Histone deacetylase 11 is a fatty-acid deacylase[J]. ACS Chem Biol, 2018, 13(3): 685-93. DOI:10.1021/acschembio.7b00942 |
| [12] |
Thole TM, Lodrini M, Fabian J, et al. Neuroblastoma cells depend on HDAC11 for mitotic cell cycle progression and survival[J]. Cell Death Dis, 2017, 8(3): e2635. DOI:10.1038/cddis.2017.49 |
| [13] |
Wang W, Fu L, Li S, et al. Histone deacetylase 11 suppresses p53 expression in pituitary tumor cells[J]. Cell Biol Int, 2017, 41(12): 1290-5. DOI:10.1002/cbin.v41.12 |
| [14] |
Dai W, Zeller C, Masrour N, et al. Promoter CpG island methylation of genes in key cancer pathways associates with clinical outcome in highgrade serous ovarian cancer[J]. Clin Cancer Res, 2013, 19(20): 5788-97. DOI:10.1158/1078-0432.CCR-13-1217 |
| [15] |
Shi J, Wang Y, Zeng L, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer[J]. Cancer Cell, 2014, 25(2): 210-25. DOI:10.1016/j.ccr.2014.01.028 |
| [16] |
Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis[J]. Cell, 2004, 117(7): 927-39. DOI:10.1016/j.cell.2004.06.006 |
| [17] |
Qin Q, Xu Y, He T, et al. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms[J]. Cell Res, 2012, 22(1): 90-106. |
| [18] |
Yang MH, Hsu DS, Wang HW, et al. Bmi1 is essential in Twist1- induced epithelial-mesenchymal transition[J]. Nat Cell Biol, 2010, 12(10): 982-92. DOI:10.1038/ncb2099 |
| [19] |
Eckert MA, Lwin TM, Chang AT, et al. Twist1-induced invadopodia formation promotes tumor metastasis[J]. Cancer Cell, 2011, 19(3): 372-86. DOI:10.1016/j.ccr.2011.01.036 |
| [20] |
Yang MH, Wu MZ, Chiou SH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis[J]. Nat Cell Biol, 2008, 10(3): 295-305. DOI:10.1038/ncb1691 |
| [21] |
Sandmann T, Girardot C, Brehme M, et al. A core transcriptional network for early mesoderm development in drosophila melanogaster[J]. Genes Dev, 2007, 21(4): 436-49. DOI:10.1101/gad.1509007 |
| [22] |
Itano N, Kimata K. Mammalian hyaluronan synthases[J]. IUBMB Life, 2002, 54(4): 195-9. DOI:10.1080/15216540214929 |
| [23] |
Moustakas A, Heldin P. TGFbeta and matrix-regulated epithelial to mesenchymal transition[J]. Biochim Biophys Acta, 2014, 1840(8): 2621-34. DOI:10.1016/j.bbagen.2014.02.004 |
| [24] |
Udabage L, Brownlee GR, Waltham M, et al. Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer[J]. Cancer Res, 2005, 65(14): 6139-50. DOI:10.1158/0008-5472.CAN-04-1622 |
| [25] |
Li Y, Li L, Brown TJ, et al. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells[J]. Int J Cancer, 2007, 120(12): 2557-67. DOI:10.1002/(ISSN)1097-0215 |
| [26] |
Bernert B, Porsch H, Heldin P. Hyaluronan synthase 2 (HAS2) promotes breast cancer cell invasion by suppression of tissue metalloproteinase inhibitor 1 (TIMP-1)[J]. J Biol Chem, 2011, 286(49): 42349-59. DOI:10.1074/jbc.M111.278598 |
| [27] |
Porsch H, Bernert B, Mehic M, et al. Efficient TGFbeta-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2[J]. Oncogene, 2013, 32(37): 4355-65. DOI:10.1038/onc.2012.475 |
 2019, Vol. 39
2019, Vol. 39

