2. 华南师范大学脑科学与康复医学研究院//华南师范大学心理应用研究中心//华南师范大学广东省心理健康与认知科学重点实验室脑研究所,广东 广州 510631
2. Institute for Brain Research and Rehabilitation/Guangdong Key Laboratory of Mental Health and Cognitive Science/Center for Studies of Psychological Application, South China Normal University, Guangzhou 510631, China
非小细胞肺癌主要有两种亚型:肺腺癌和肺鳞状细胞癌(LUSC)[1-3]。不同亚型的非小细胞肺癌与不同的分子生物学特征有关[4-6]。如发生在远端气道的肺腺癌常以KRAS基因突变有关,而LUSC则常以染色体3p的缺失出现在气道的近端[7-8]。关于两种非小细胞肺癌亚型各自的诊疗机制目前还没有很明确的研究报导,临床上也没有明确的诊疗方案,以至于两者的临床治疗反应存在明显差异。如经过Ipilimumab治疗,LUSC患者比肺腺癌患者有更长的生存时间[9],靶向EGFR激酶突变的药物gefitinib更适合于肺腺癌病人的治疗[10]。近期有研究发现microRNA可以区分肺癌中的鳞状细胞癌和腺癌[11-12]。因此,针对两种非小细胞肺癌亚型的转录组特征进行深度分析,寻找其各自更有效的分子诊断标志物及其潜在的可用于鉴别和治疗肺腺癌和LUSC的信号通路和生物学标记尤为重要。
本研究旨在从Gene Expression Omnibus数据库分析肺腺癌和LUSC的差异表达基因。通过使用生物信息学方法,确定microRNA和mRNA联合作为区分肺腺癌和LUSC潜在的分子诊断标记物。研究结果显示信号通路(Osteoarthritis和LXR/RXR)、转录因子(GATA4,RELA,YBX1,TP63和MBD2)、细胞因子(WNT5A和IL1A)和microRNAs(miR-34a,miR-15a和miR-181b)是区别肺腺癌和LUSC的潜在靶点。因此,结合转录组分析可为早期诊断和临床治疗做出可靠和有价值的选择。
1 资料和方法 1.1 微阵列数据集基因表达谱分析数据集(GSE27262,GSE19188,GSE33479)获自Gene Expression Omnibus。本研究使用肺腺癌组织和正常组织(GSE27262,GSE19188)、肺鳞状细胞癌组织和正常组织(GSE33479,GSE19188)的数据。GSE27262的微阵列数据包括25例肺腺癌组织和25例相邻的正常组织。GSE19188的微阵列数据包括45例肺腺癌组织和65例正常组织,27例肺鳞状细胞癌组织和65例正常肺组织。GSE33479的微阵列数据包括14例肺鳞状细胞癌组织和13例相邻的正常肺组织。
1.2 临床肺癌病人数据采集和检测课题组收集了临床23例肺鳞癌肿瘤组织和正常肺组织样。纳入标准:经病理学诊断确诊为肺癌的患者;经细胞学检测,确诊为肺鳞癌的患者;经KRAS基因突变检测为阴性患者。排除标准:发生多发肿瘤;肿瘤发病灶不能明确是来源于肺肿瘤;其他器官肿瘤转移至肺部者。样本由广州医科大学第一附属医院采集,样本采集由广州医科大学临床研究伦理委员会批准。所有病人均签署样本授权使用同意书。
实验采用荧光定量PCR分析miR-181b,WNT5A和MBD2的表达。RNA提取使用Trizol,然后使用iScript Reverse Transcription Supermix kit逆转录成cDNA,使用SYBR Green检测基因表达。引物使用如下:WNT5A(Forward:5'-ccggtactagctaactccaa-3';Reverse: 5'-caccattccacagagagaga-3)',MBD2(Forward:5'-gcggg aagaggatggatt-3';Reverse:5'-tgaacttcttaccacttggactga-3),' β- actin(Forward:5'- tcatcaccattggcaatgag-3';Reverse:5'- cactgtgttggcgtacaggt-3)。' miR-181b-5p的检测使用TaqMan方法(Thermo Fisher)。
1.3 差异表达基因鉴定使用R语言的在线Gene Expression Omnibus 2R处理原始数据。通过t检验来鉴定差异表达基因,倍数变化大于2.0(上调)或小于0.5(下调)。通过维恩图显示所鉴定的差异表达基因的可视化。
1.4 生物信息学分析和TCGA数据验证使用Database for Annotation,Visualization and Integrated Discovery(DAVID)数据库(https://david.ncifcrf.gov/)[13]进行生物学功能和gene ontology分析。根据gene ontology,使用DAVID基因注释工具分析差异表达基因的功能,P < 0.05和FDR < 0.05为差异具有统计学意义。使用Ingenuity Pathway Analysis(IPA)分析转录组特征,包括信号通路,上游调控分子和细胞因子。P < 0.05(-log(P)>1.3)为差异具有统计学意义。采用StarBase v2.0数据库分析TCGA数据中的肺腺癌和LUSC样本的差异表达基因[14]。
1.5 统计学分析使用SPSS16.0软件进行数据的统计分析,差异基因表达分析使用t检验。miR-181b-5p和WNT5A/MBD2的相关性分析采用Pearson相关分析,P < 0.05为差异具有统计学意义。
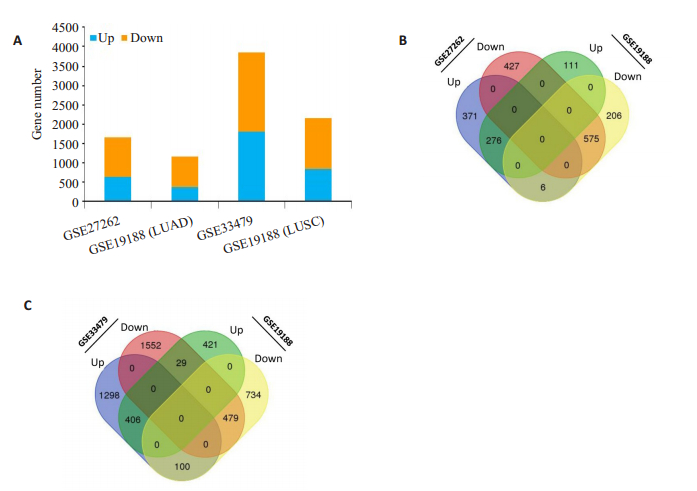
2 结果 2.1 LUAD和LUSC的差异表达基因本研究收集了来自Gene Expression Omnibus 2R的3个数据集:GSE27262、GSE19188和GSE33479,其包含肺腺癌或肺鳞癌及其相邻的正常组织基因表达谱。GSE19188分别包含45例肺腺癌患者,27例肺鳞癌患者和66例健康人。GSE27262仅含有50例肺腺癌患者,而GSE33479仅含有27例肺鳞癌患者。其中鉴定出的差异表达基因包括:肺腺癌:GSE27262(653个上调的DEGs和1002个下调的DEGs);GSE19188(387个上调DEGs和787个下调DEGs);肺鳞癌:GSE33479(1804个上调的DEGs和2060个下调的DEGs)和GSE19188(856个上调的DEGs和1313个下调的DEGs)(图 1A)。将肺腺癌和LUSC数据集合并在一起,以进一步分析两种类型的NSCLC之间的差异表达基因,数据显示:肺腺癌中有851个DEGs,其中276个(占32.4%)上调DEGs,575个(占67.6%)下调DEGs(图 1B)。肺鳞状细胞癌中共发现885个DEGs,其中406个(占45.9%)上调DEGs,479个(占54.1%)下调DEGs(图 1C),这些共同表达的DEGs将进一步用于生物信息学分析。
|
图 1 肺腺癌与肺鳞癌差异表达基因在不同数据库的鉴定 Fig.1 Identification of differentially expressed genes (DEGs) between lung adenocarcinoma and lung squamous cell carcinoma. A: Gene numbers of the 4 sets of gene expression microarray data; B: Venn diagram of 2 LUAD datasets of gene expression microarray data (GSE27262 and GSE19188); C: Venn diagram of 2 LUSC datasets of gene expression microarray data (GSE33479 and GSE19188). |
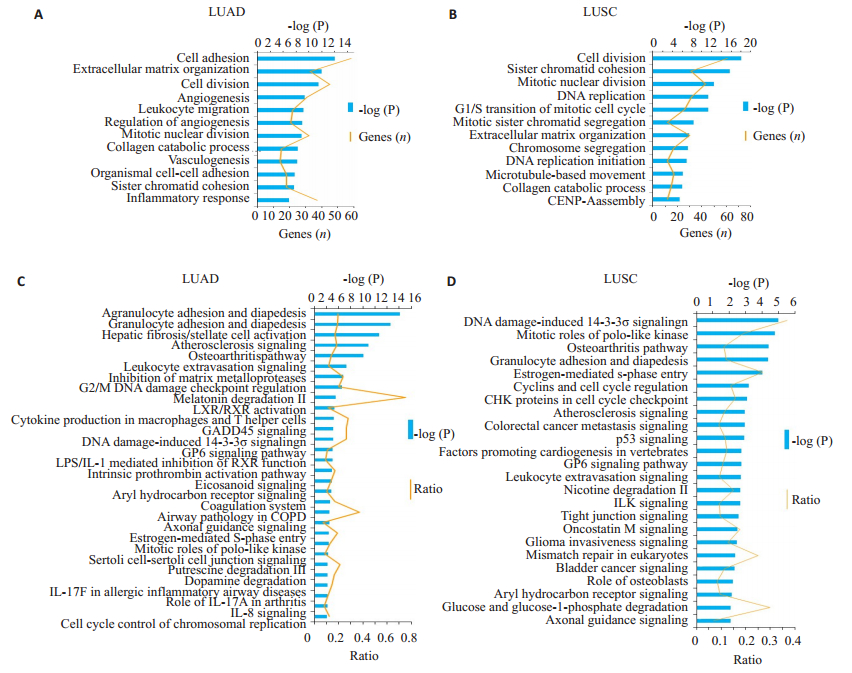
使用数据库DAVID分析了上调和下调的DEGs。通过DAVID中的gene ontology分析,发现生物过程中的细胞分裂,细胞粘附,基质组织,胶原分解代谢过程和细胞周期是肺腺癌和LUSC中最富集的信号通路(图 2A、B)。结果显示白细胞迁移和炎症反应在肺腺癌中比LUSC富集程度更高,表明肺腺癌相邻的与炎症反应有更强的相关性(图 2A)。
|
图 2 生物功能和IPA信号通路分析 Fig.2 Biological function and ingenuity pathway analysis. A: Biological functions in lung adenocarcinoma; B: Biological functions in lung squamous carcinoma; C: Canonical pathways in lung adenocarcinoma; D: Canonical pathways in lung squamous carcinoma. |
其次,使用IPA分析差异表达基因的信号通路。根据差异基因表达,鉴定出在肺腺癌中显著富集最高的3种信号通路分别与炎症信号通路相关:粒细胞粘附和渗透;粒细胞粘附和渗出;以及肝纤维化/肝星状细胞活化(图 2C)。而在LUSC中显著富集的3种信号通路分别与细胞周期相关:G2/M期DNA损伤检查点;细胞周期调控的染色体复制和肝纤维化/肝星状细胞活化相关(图 2D)。根据差异基因表达水平,基于Z值确定了肺腺癌和LUSC中显著性富集的相同和不同的信号通路,如:GADD45,Osteoarthritis pathway和Atherosclerosis signaling在两种肿瘤中都显著性富集。Osteoarthritis pathway在肺腺癌中是抑制状态,而在LUSC中显示是激活状态。Nanog在哺乳动物胚胎干细胞中的作用多能性在LUSC中被预测为抑制,而在肺腺癌中未发现其有显著性富集。此外,对肺腺癌和LUSC之间的信号途径进行了IPA比较分析,结果显示,4种信号通路TREM1 signaling、PPARα/RXRα Activation、Role of BRCA1 in DNA Damage Response和Inhibition of Angiogenesis by TSP1分别在肺腺癌和LUSC中的富集状态呈相反状态。
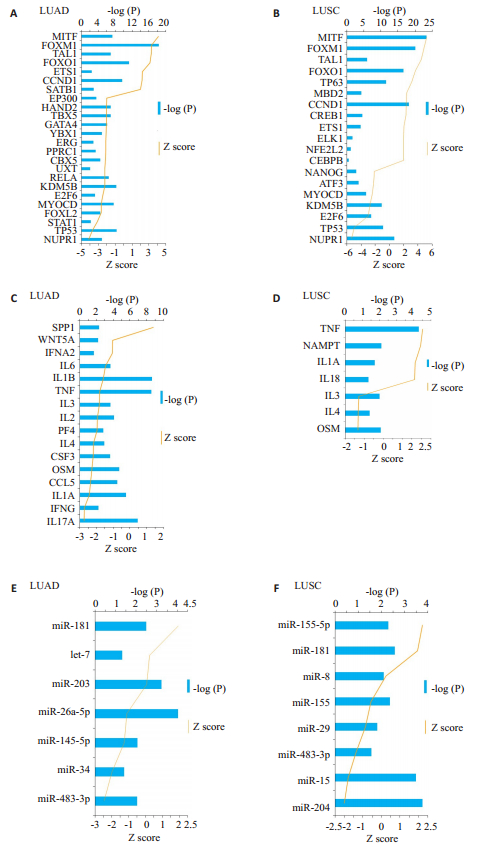
2.3 转录因子、细胞因子和microRNA分析肿瘤患者中不同细胞因子和转录因子的表达水平与化疗的敏感性反应有关。因此,使用IPA对上游信号通路Upstream analysis的重要细胞因子和转录因子进行分析。结果显示,肺腺癌中鉴定出7个激活和和17个抑制状态的转录因子(图 3A),而LUSC中鉴定出10个激活和7个抑制状态的的转录因子(图 3B)。在激活状态的转录因子中,MITF、FOXM1、TAL1和FOXO1是肺腺癌和LUSC中最重要和最常见的上游信号分子(图 3A、B)。TP63和MBD2在LUSC是激活状态,而在肺腺癌中没有发现有显著性变化(图 3B)。在抑制状态的转录因子中,NUPR1、TP53、MYOCD和E2F6在两种肿瘤中最常见(图 3A、B)。转录因子GATA4、YBX1和RELAZ在肺腺癌中呈现显著性抑制状态(图 3A)。这些转录因子可用于区分LUSC和肺腺癌。
|
图 3 IPA分析肺腺癌和肺鳞癌中的差异表达转录因子和细胞因子 Fig.3 Transcription factor and cytokines analysis in LUAD and LUSC. Transcription factor of the differential expression genes signature in LUAD (A) and LUSC (B); Cytokines of the differential expression genes signature in LUAD (C) and LUSC (D); microRNAs associated with differential expression genes signature in LUAD (E) and LUSC (F). |
大多数细胞因子富集在肺腺癌中而不在LUSC(图 3C)。16个细胞因子被鉴定出在肺腺癌中富集(P < 0.05),而7个细胞因子在LUSC中富集(图 3D)。IL-3、IL-4和OSM是肺腺癌和LUSC中呈现抑制性状态的最常见细胞因子。特别是,IL-1B和IL-1A在肺腺癌和LUSC之间显示出相反的富集表达,说明IL-1B和IL-1A是肺腺癌和LUSC潜在的差异表达分子标记物。此外,SPP1和WNT5A是肺腺癌中最重要的激活细胞因子,而在LUSC中未发现。结果显示细胞因子IL-3、IL-4、OSM、IL-1B、IL-1A、SPP1和WNT5A是肺腺癌和LUSC之间的重要的潜在区别因素。
除转录因子和细胞因子外,microRNA也可以区分肺癌中的鳞状细胞癌和腺癌。利用IPA软件根据差异表达基因和microRNA靶点预测进行了miRNA差异表达富集分析。尽管基于Z-score并未发现太多的microRNA具有统计学意义,但结果显示miR-181、miR-155、miR-34、miR-15、miR-204和miR-483在肺腺癌和LUSC中有显著性差异(图 3E、F)。细胞因子IL-1A是miR-34的潜在靶基因之一,WNT5A是miR-181b的潜在靶基因之一。
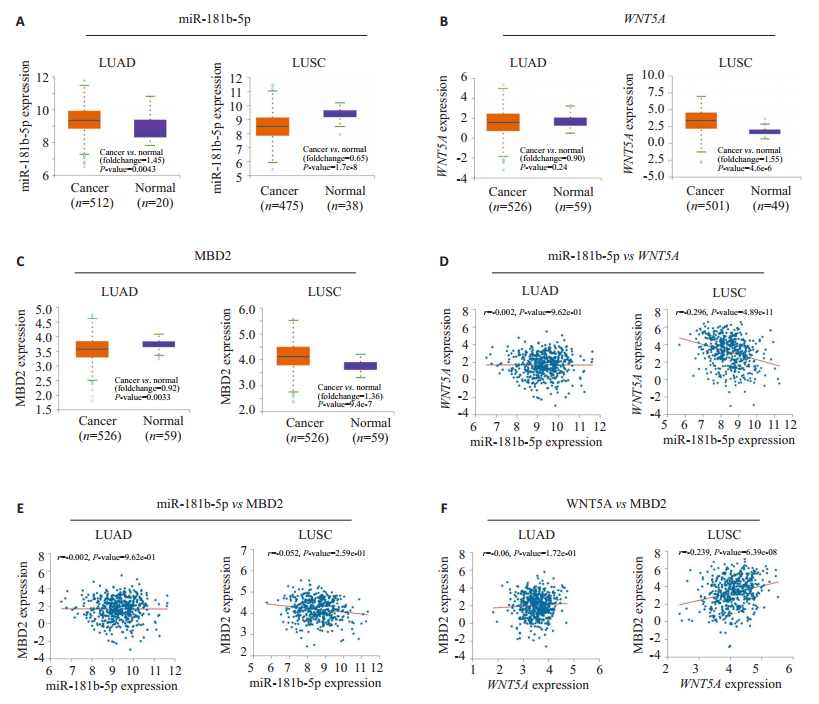
2.4 miR-181/WNT5A/MBD2在TCGA数据和临床肺癌样本中的表达为验证转录组数据分析中鉴定的miRNA-mRNA的有效性,本研究挑选了miR-181和它的2个靶基因WNT5A和MBD2进行了验证。首先,使用StarBase数据库分析了肺腺癌和肺鳞状细胞癌的TCGA临床样品,结果发现miR-181b-5p在肺腺癌中的表达高于正常肺组织,而在LUSC中表达降低(图 4A)。它的两个靶基因WNT5A和MBD2在LUSC中表达上调,但在肺腺癌的肿瘤样本和正常肺组织相比没有显著性差异(图 4B、C)。此外,miR-181b表达与肺鳞状细胞癌中的WNT5A表达呈负相关(r2=-0.296,P < 0.000,图 4D)。尽管MBD2和miR-181b的表达没有显著相关性(图 4E、F),但MBD2与WNT5A在LUSC中呈正相关(r=0.239,P < 0.000,图 4F)。
|
图 4 TCGA数据分析miR-181b-5p和靶基因WNT5A,MBD2在肺腺癌和肺鳞癌的表达 Fig.4 miR-181b-5p and expression of its target genes in clinical LUAD and LUSC patients from TCGA. The relative expression of miR-181b-5p (A), WNT5A (B) and MBD2 (C) in the tumors and its adjacent tissues of LUAD and LUSC patients from TCGA; D: Correlation analysis of miR-181b-5p vs WNT5A in LUAD and LUSC; E: Correlation analysis of miR-181b-5p vs MBD2 in LUAD and LUSC; F: Correlation analysis of WNT5A vs MBD2 in LUAD and LUSC; Spearman's rank correlation analysis was performed. |
为进一步验证miR-181b-5p/WNT5A/MBD2是否可用作LUSC的生物标志物,本研究从肺鳞癌患者中收集了23例肺鳞癌患者的肺肿瘤组织(实验组)及其邻近癌旁肺组织(23例癌旁肺组织,对照组),并检测了miR- 181b-5p和其靶基因WNT5A和MBD2的表达。与邻近正常肺组织相比,miR-181b-5p在肺肿瘤组织中的表达显著性降低,而WNT5A和MBD2表达升高(图 5A);miR-181b-5p与WNT5A在肿瘤组织中的表达呈负相关(r2=0.2146,P=0.026,图 5B);MBD2和miR-181b-5p的表达没有相关性(r2=0.04724,P=0.319,图 5C)。实验结果支持miR-181b-5p的表达上升和WNT5A的表达下调可视为诊断LUSC的分子标记物。
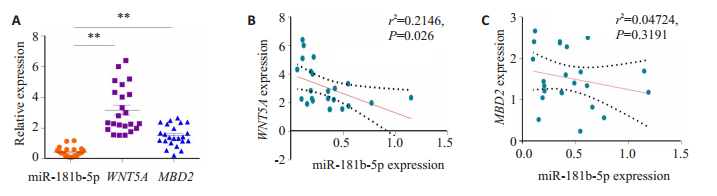
|
图 5 临床肺鳞状细胞癌样本中miR-181b-5p和其靶基因的表达 Fig.5 miR-181b-5p and expression of its target genes in clinical squamous cell carcinoma patients. A: The relative expression of miR-181b-5p, WNT5A, and MBD2 in the tumors and its adjacent tissues of lung squamous cell carcinoma patients, **P < 0.01; B, C: Spearman's rank correlation analysis of the correlation between miR-181b-5p and WNT5A and between miR-181b-5p and MBD2 in lung squamous cell carcinoma. |
除了miR-181b-5p/WNT5A/MBD2,miR-34a和其靶基因IL-1A在肺腺癌中表达呈相反趋势,在肺腺癌和LUSC中的表达亦呈现相反的表达趋势。miR-15a在肺腺癌的表达中显著性上调,LUSC中显著性下调,其靶基因YBX1则成相反趋势,根据分析结果在临床肺癌样本中做进一步实验验证(表 1)。结果进一步证实miRNA和其靶基因表达在LUSC和肺腺癌中呈现相反趋势,可作为区分鉴定LUSC和肺腺癌的分子诊断标记物。
| 表 1 配对miRNA-mRNA在TCGA肺腺癌和肺鳞癌样本中的表达 Tab.1 Expression level of paired miRNA-mRNA in LUAD and LUSC from TCGA |
本研究揭示了肺腺癌和LUSC的转录组学特征的差异,主要包括富集的信号通路、上游调控转录因子、细胞因子和miRNA。实验以正常肺组织样本为对照组,是缺乏可作为肺鳞癌对照的足够的鳞化上皮或基底细胞,基于正常肺组织与异常的癌组织之间基因差异表达而言,虽有扩大范围之嫌,但是分析出肺腺癌和LUSC的差异表达基因来源于Gene Expression Omnibus 2R的3个数据集:GSE27262,GSE19188和GSE33479,其包含肺腺癌或肺鳞癌及其相邻的正常组织基因表达谱,正如有的文献也是以正常肺组织作为肺鳞癌的对照[15-16]。组织样品是含有大量的间质细胞,正常组织样品与癌组织样品相比,间质细胞的异常表达亦可作为癌组织的生物标记,与相关研究文献一致[17-18]。与相关研究报道[16, 19-20]相比,本实验通过分析肺腺癌和LUSC富集的差异表达基因,发现了差异表达的配对miRNA和其靶mRNA可用于鉴别肺腺癌和LUSC。生物功能分析结果显示,炎症相关性信号通路如:Osteoarthritis pathway,COPD, IL-17和IL-8等主要富集于肺腺癌,而LUSC主要富集的信号通路是细胞周期和DNA损伤。本研究验证了两种信号通路Osteoarthritis pathway和LXR/RXR,分别在肺腺癌和LUSC差异表达基因中的富集呈相反状态。有文献报道LXR/RXR信号通路的激活在鳞状细胞癌中表达呈下调状态[9];LXRα/β的缺失导致小鼠自发性外周鳞状细胞肺癌的研究也证明了LXR/RXR在鳞状细胞癌中发挥的关键作用[21]。本实验验证结果与文献报道相符,表明在LUSC中LXR/RXR的表达呈抑制状态,而在肺腺癌中略微上调。因此,进一步分析Osteoarthritis pathway和LXR/RXR通路的相关信号分子将为鉴别诊断肺腺癌和LUSC提供更可靠的验证数据。
基于转录组的上游分析,本研究通过鉴定GATA4、RelA、YBX1、TP63和MBD2分析,发现各转录因子均在肺腺癌和LUSC中差异表达,其中TP53和TP63更集中于肺鳞状细胞癌。TP63是一种众所周知的鳞状细胞癌标记物[22-24],本实验结果表明,TP63在LUSC中表达增加,但在肺腺癌中的表达无明显差异。有研究证明TP53和TP63均可调节细胞增殖并在鳞状细胞癌中发挥关键作用[25-27],结果表明细胞周期相关分子在鳞状细胞癌的发展过程中起重要作用。不同的是炎症相关转录因子和细胞因子在肺腺癌中的表达存在差异。例如,RelA是NF-κB/Rel家族的主要成员之一,文献研究不仅揭示了RelA在肿瘤相关骨髓细胞中的关键作用[28-30],还证明RelA是多种免疫细胞中炎症反应的主要介质[31-32]。NF-κB/RelA在肿瘤细胞中可被激活,而且它在骨髓细胞激活的肿瘤进展中比在上皮衍生的肿瘤细胞中具有更重要的作用[32]。本实验数据显示RelA在肺腺癌中被显著激活,这表明NF-κB相关炎症反应和细胞因子在肺腺癌中的重要作用。综上,TP53和TP63主要在肺腺癌中表达,而RelA和NF-κB主要表达在LUSC中。
miRNA在NSCLC中的作用已得到广泛研究[33-35],研究表明miRNA可用作肺癌的诊断和判断预后的潜在生物标志物[36-37],但具体关于肺腺癌和LUSC的有效分子诊断标志物及潜在的鉴别诊断的信号通路或生物学标记则尚未有明确的文献报道。基于转录组数据分析,我们鉴定出3个miRNA,包括miR-34a、miR-15a和miR-181b,它们在肺腺癌中均显著上调,而在LUSC中下调,而且其潜在的靶基因大多在相同类型的肺癌中表达相反。因此,可采用成对的miRNA和其靶基因表达的相反趋势作为区分鉴定LUSC和肺腺癌的分子诊断标记物。后续采用更多更广泛的临床肺腺癌和肺鳞癌肿瘤样本做进一步的检测分析,将有助于联合使用miRNA和其靶基因的差异表达鉴别诊断不同的肺癌亚型。
| [1] |
Yang H, Linandy Y. Treatment of lung carcinosarcoma and other rare histologic subtypes of non-small cell lung cancer[J]. Curr Treat Options Oncol, 2017, 18(9): 54-65. DOI:10.1007/s11864-017-0494-9 |
| [2] |
Lim SL, Jia ZA, Lu YH, et al. Metabolic signatures of four major histological types of lung cancer cells[J]. Metabolomics, 2018, 14(9): 118-27. DOI:10.1007/s11306-018-1417-x |
| [3] |
Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013[J]. J Natl Compr Canc Netw, 2013, 11(6): 645-53. DOI:10.6004/jnccn.2013.0084 |
| [4] |
Bjorkqvist AM, Husgafvel-Pursiainen K, Anttila S, et al. DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma[J]. Genes Chromosomes Cancer, 1998, 22(1): 79-82. |
| [5] |
Cooper WA, Lam DC, O'toole SA, et al. Molecular biology of lung cancer[J]. J Thorac Dis, 2013, 5(5): S479-90. |
| [6] |
Ceppi P, Volante M, Saviozzi S, et al. Squamous cell carcinoma of the lung compared with other histotypes shows higher messenger RNA and protein levels for thymidylate synthase[J]. Cancer, 2006, 107(7): 1589-96. DOI:10.1002/(ISSN)1097-0142 |
| [7] |
Davidson MR, Gazdarandb AF. E. clarke. the pivotal role of pathology in the management of lung cancer[J]. J Thorac Dis, 2013, 37(5): 463-78. |
| [8] |
Langer CJ, Besse B, Gualberto A, et al. The evolving role of histology in the management of advanced non-small-cell lung cancer[J]. J Clin Oncol, 2010, 28(36): 5311-20. DOI:10.1200/JCO.2010.28.8126 |
| [9] |
Tomasini P, Khobta N, Greillier LA. Ipilimumab: its potential in non-small cell lung cancer[J]. Ther Adv Med Oncol, 2012, 4(2): 43-50. DOI:10.1177/1758834011431718 |
| [10] |
Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy[J]. Science, 2004, 304(5676): 1497-500. DOI:10.1126/science.1099314 |
| [11] |
Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival[J]. Clin Cancer Res, 2009, 15(19): 6192-200. DOI:10.1158/1078-0432.CCR-09-1467 |
| [12] |
Patnaik S, Mallick R, Kannisto E, et al. MiR-205 and MiR-375 MicroRNA assays to distinguish squamous cell carcinoma from adenocarcinoma in lung cancer biopsies[J]. J Thorac Oncol, 2015, 10(3): 446-53. |
| [13] |
da WH, Shermanandr BT, Lempicki A. Systematic and integrative analysis of large gene lists using David bioinformatics resources[J]. Nat Protoc, 2009, 4(1): 44-57. DOI:10.1038/nprot.2008.211 |
| [14] |
Li JH, Liu S, Zhou H, et al. Starbase v2.0: decoding miRNAceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data[J]. Nucleic Acids Res, 2014, 42(Database issue): D92-7. |
| [15] |
Li Y, Gu J, Xu FK, et al. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data[J]. Sci Rep, 2018, 8(1): 15834-45. |
| [16] |
Xie LX, Dang YF, Guo JS, et al. High KRT8 expression independently predicts poor prognosis for lung adenocarcinoma patients[J]. Genes (Basel), 2019, 10(1): 1279-92. |
| [17] |
Zhao ZX, Liu BJ, Sun J, et al. Baicalein inhibits orthotopic human Non-Small cell lung cancer xenografts via Src/Id1 pathway[J]. Evid Based Complement Alternat Med, 2019, 98(6): 162-73. |
| [18] |
庄铭锴, 陈丰霖. 上皮间质转化相关的生物标志在胃癌中的研究进展[J]. 世界华人消化杂志, 2013, 21(30): 3204-10. |
| [19] |
Liu SY, Wang XJ, Qin WY, et al. Transcription factors contribute to differential expression in cellular pathways in lung adenocarcinoma and lung squamous cell carcinoma[J]. Interdiscip Sci, 2018, 10(4): 836-47. DOI:10.1007/s12539-018-0300-9 |
| [20] |
Li Y, Sun N, Lu ZL, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer[J]. Cancer Lett, 2017, 393(1): 40-51. |
| [21] |
Dai YB, Miao YF, Wu WF, et al. Ablation of liver X receptors alpha and beta leads to spontaneous peripheral squamous cell lung cancer in mice[J]. Proc Natl Acad Sci USA, 2016, 113(27): 7614-9. DOI:10.1073/pnas.1607590113 |
| [22] |
Yang Z, Zhuan B, Yan Y, et al. Integrated analyses of copy number variations and gene differential expression in lung squamous-cell carcinoma[J]. Biol Res, 2015, 48(7): 108-16. |
| [23] |
Zhang S, Li MF, Ji HB, et al. Landscape of transcriptional deregulation in lung cncer[J]. BMC Genomics, 2018, 19(1): 435-49. DOI:10.1186/s12864-018-4828-1 |
| [24] |
Shi YX, Wang Y, Li X, et al. Genome-wide DNA methylation profiling reveals novel epigenetic signatures in squamous cell lung cancer[J]. BMC Genomics, 2017, 18(11): 901-15. |
| [25] |
Cheng H, Yang XP, Si H, et al. Genomic and transcriptomic characterization links cell lines with aggressive head and neck cancers[J]. Cell Rep, 2018, 25(5): 1332-43. DOI:10.1016/j.celrep.2018.10.007 |
| [26] |
Yoshida M, Yokota E, Sakuma T, et al. Development of an integrated CRISPRi targeting DeltaNp63 for treatment of squamous cell carcinoma[J]. Oncotarget, 2018, 9(49): 29220-32. |
| [27] |
Eun YG, Lee D, Lee YC, et al. Clinical significance of YAP1 activation in head and neck squamous cell carcinoma[J]. Oncotarget, 2017, 8(67): 111130-43. |
| [28] |
Mishra A, Bharti AC, Varghese P, et al. Differential expression and activation of NF-kappa B family proteins during oral carcinogenesis: role of high risk human papillomavirus infection[J]. Int J Cancer, 2006, 119(12): 2840-50. DOI:10.1002/(ISSN)1097-0215 |
| [29] |
Taggart CC, Cryan SA, Weldon S, et al. Secretory leucoprotease inhibitor binds to NF-kappa B binding sites in monocytes and inhibits p65 binding[J]. J Exp Med, 2005, 202(12): 1659-68. DOI:10.1084/jem.20050768 |
| [30] |
Huang X, Guo B, Liu S, et al. Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment[J]. Nat Commun, 2018, 19(21): 2741-56. |
| [31] |
Liu C, Zhou Y, Li M, et al. Absence of GdX/UBL4A protects against inflammatory diseases by regulating NF-small ka, CyrillicB signaling in macrophages and dendritic cells[J]. Theranostics, 2019, 9(5): 1369-84. DOI:10.7150/thno.32451 |
| [32] |
Jagannathan L, Jose CC, Arita AA, et al. Nuclear factor kappa B1/RelA mediates inflammation in human lung epithelial cells at atmospheric oxygen levels[J]. J Cell Physiol, 2016, 231(7): 1611-20. DOI:10.1002/jcp.25262 |
| [33] |
Chen YJ, Guo YN, Shi K, et al. Down-regulation of microRNA-144-3p and its clinical value in non-small cell lung cancer: a comprehensive analysis based on microarray, miRNA-sequencing, and quantitative real-time PCR data[J]. Respir Res, 2019, 20(7): 48-59. |
| [34] |
Li G, Wang K, Wang J, et al. miR-497-5p inhibits tumor cell growth and invasion by targeting SOX5 in non-small-cell lung cancer[J]. J Cell Biochem, 2019, 283(12): 1570-83. |
| [35] |
Zhuangandy B, Cheng M. MicroRNA629 inhibition suppresses the viability and invasion of nonsmall cell lung cancer cells by directly targeting RUNX3[J]. Mol Med Rep, 2019, 11(4): 315-26. |
| [36] |
Wang H. Predicting MicroRNA biomarkers for cancer using phylogenetic tree and microarray analysis[J]. Int J Mol Sci, 2016, 17(5): 257-68. |
| [37] |
Gyoba J, Shan S, Roa W, et al. Diagnosing lung cancers through examination of Micro-RNA biomarkers in blood, plasma, serum and sputum: a review and summary of current literature[J]. Int J Mol Sci, 2016, 17(4): 494-508. DOI:10.3390/ijms17040494 |
 2019, Vol. 39
2019, Vol. 39

