2. 北京大学深圳医院神经外科,广东 深圳 518036;
3. 南方医科大学公共卫生学院 BSL-3实验室,广东 广州 510515;
4. 深圳大学第一附属医院,深圳市第二人民医院消化内科,广东 深圳 518036
2. Department of Neurosurgery, Peking University Shenzhen Hospital, Shenzhen 518036, China;
3. Biosafety Level-3 Laboratory, School of Public Health, Southern Medical University, Guangzhou 510515, China;
4. Department of Gastroenterology, First Affiliated Hospital of
无机多聚磷酸盐(polyphosphate, polyP)是由数十至数百个磷酸盐残基通过"高能"磷酸键结合而成的线性多聚体[1], 它广泛存在于生物体中, 不仅影响原核生物稳定生长期的生存、致病性及严谨反应的调控[2-13], 还对真核生物的血栓形成、癌症发生、钙盐沉积、免疫应答等方面具有重要作用[14-18]。大肠杆菌多聚磷酸盐激酶(Polyphosphate kinase, PPK)能催化polyP的合成[19]。为了深入研究polyP在肠出血型大肠埃希菌(Enterohemorrhagic Escherichia coli, EHEC) O157:H7中的生物学功能, 建立一种简便、可靠且特异性高的polyP定性和定量的检测方法十分迫切。
目前, 用于测定polyP方法有:(1)酶和离子交换法, 需要细胞标记32Pi, 分离polyP, 测定时需用酵母提取外切聚磷酸酶(scPPX1), 并将其分解为磷酸盐残基[20];(2)荧光素测定法, 利用polyP合成酶(ppk1), 催化polyP脱磷酸后, 与ADP结合生成ATP, 再用荧光素测定[21]; (3)异染性染色法, 应用甲苯胺蓝染色细胞内polyP [22-24], 蓝色染料将polyP染成紫色, 适用于polyP定性。上述方法步骤繁琐, 仪器设备要求高, 缺乏特异性, 无法测定小于60个磷酸盐残基的polyP, scPPX1不易获得, 难以推广使用。本文拟采用4', 6-二脒基-2-苯基吲哚(DAPI)染色方法直接定量测定大肠杆菌O157:H7内polyP含量。
DAPI广泛应用于双链DNA染色, 在360 nm波长激发下, 最大发射波长460 nm, 在荧光显微镜下呈蓝光[25]。DAPI与DNA结合主要依靠A-T间氢键和脒基间氢键, 还有双螺旋DNA磷酸基团与两个带正电荷的DAPI之间的静电作用[25]。
Allan等[26]首先在酿酒酵母菌中报道了用DAPI可以染色浓度比较高的polyP (50 μg/mL), DAPI-polyP在360 nm激发下呈黄绿色荧光。Aschar[27]优化了DAPI定量polyP的激发波长为415 nm、发射波长为550 nm, 应用酶标仪定量polyP。Diaz [28]将这种方法应用于强化的生物除磷过程(EBPR), 但仍采取先提取polyP再用DAPI定量的方式。Gomes等[29]将这种方法应用于寄生虫polyP半定量, 而应用DAPI定量致病性大肠杆菌polyP尚未见报道。
本研究旨在通过应用DAPI结合polyP形成DAPIpolyP复合物并发出黄绿色荧光的特征[26], 通过优化测定条件, 建立DAPI荧光染色法直接定量致病性大肠杆菌polyP的简便、可靠方法, 为深入研究polyP在肠出血型大肠埃希菌O157:H7致病过程中的作用提供技术支撑。
1 材料和方法 1.1 菌株和试剂EHEC O157:H7 EDL933由中国疾病预防控制中心惠赠; EHEC O157:H7 EDL933 ppk1缺失株(Δppk1)、EHEC O157:H7 EDL933 ppk1回补株(Cppk1)由本实验室构建、保存。4-羟乙基哌嗪乙磺酸(HEPES)、DAPI(nos. D9542)、多聚磷酸钠标准品(PolypType45) (nos.S4379)(Sigma-Aldrich)。其余试剂均为分析纯。
1.2 方法 1.2.1 DAPI-DNA与DAPI-polyP复合物发射光谱的扫描按照Magen细菌DNA提取试剂盒(Hi Pure Bacterial DNAKit)提取并纯化EHEC O157:H7 EDL933 WT株DNA。取其225 μL与HEPES溶液(20 mmol/L)675 μL混合, 加入DAPI(100 μmol/L) 100 μL, 充分混匀后, 室温避光放置10 min。取polyP标准品polyP45工作液(3 μmol/L) 225 μL与HEPES溶液(20 mmol/L) 675 μL混合, 加入DAPI (100 μmol/L) 100 μL, 充分混匀后, 室温避光孵育7.5 min, 再重复1次, 混匀、孵育。分别应用荧光分光光度计确定360 nm和415 nm激发光的发射光谱。
1.2.2 细菌前处理EHEC O157:H7 WT株常规培养后, 测定WT株A600值, 用不含抗生素的LB液体培养基稀释为A600=1.0。取1.5 mL细菌悬液, 4 ℃ 10 000 r/min离心15 min, 弃上清。加入20 mmol/L HEPES(pH= 7.0)1 mL涡旋重悬细菌, 4 ℃ 10 000 r/min离心15 min, 弃上清。加入20 mmol/L HEPES (pH=7.0)1 mL涡旋重悬细菌。各取上述重悬菌液1 mL, 分别进行6种方法处理, 即液氮速冻后室温下自然溶解、-80 ℃速冻溶解、-20 ℃速冻溶解、60 ℃加热10 min、Triton x-100作用及室温下不作处理, 以确定最佳的细菌前处理方法(表 1)。将6种方法处理后的细菌取100 μL进行倍比稀释到106, 涂LB平板, 确定不同处理方法对细菌生存的影响。6种方法处理后的菌液在原管中充分涡旋混匀, 取300 μL加入1.5 mL EP管; 再加入600 μL 20 mmol/LHEPES溶液; 实验组再加入100 μL DAPI溶液, 对照组均加100 μL ddH2O代替DAPI。溶液充分涡旋混匀, 室温避光孵育10 min。96孔板上样, 每孔200 μL。避光, 酶标仪荧光模块测量荧光值。激发波长为415 nm, 发射波长为550 nm。根据荧光值确定最佳的细菌前处理方法。
| 表 1 细菌前处理方法改善细胞膜通透性 Tab.1 Six pretreatment methods for improving the permeability of cells |
EHEC O157:H7 WT株常规培养后, 根据上一步确定的最佳细菌前处理方法处理细菌, 步骤同上。上述溶液充分涡旋混匀, 室温避光孵育10 min。离心, 4 ℃ 10 000 r/min离心3 min, 弃上清。HEPES洗1次后, 加入1 mL HEPES重悬。制作玻片, 取3~5 μL重悬液制作玻片, 置于激光共聚焦显微镜下, 60×油镜和10×目镜, 以405 nm和488 nm激发光激发, 用Nikon NIS Elements v4.5.观察荧光。余液应用荧光分光光度计确定360 nm和415 nm激发光的发射光谱。
1.2.4 polyP含量的直接测定按前述步骤配制3 μmol/LpolyP45工作液。按照表 2比例配制polyP标准系列。其中ru指900 μL (HEPES液+polyP标准品)混合液中标准品的体积比。混匀后, 室温避光孵育7.5 min, 再重复1次。96孔板每孔上样200 μL, 每个浓度至少做2个平行孔。测定polyP荧光值, 激发波长415 nm, 发射波长550 nm。EHEC O157:H7 WT、△ppk1、Cppk1均采用常规培养。
| 表 2 polyP标准系列配制 Tab.2 Dilution series of standard polyP45 for constructing the standard calibration curve |
前处理后的菌液充分混匀, 取300 μL, 置1.5 mL EP管中, 再加入600 μL 20 mmol/L HEPES溶液; 实验组再加入100 μL DAPI溶液, 对照组均加100 μL ddH2O代替DAPI。溶液充分涡旋混匀, 室温避光孵育5~7 min。96孔板上样, 每孔上样200 μL。避光, 测量荧光值, 激发波长415 nm, 发射波长550 nm。根据标准曲线推算各株细胞内polyP含量。
1.2.5 统计学处理计量数据用均数或均数±标准差描述, 6种处理方法及3株细菌polyP含量差异的比较应用单因素方差分析, 多重比较采用SNK and Bonferroni tests, 检验水准α=0.05。
2 结果 2.1 测定出DAPI-DNA与DAPI-polyP不同的最大发射波长EHEC O157:H7 WT株、纯化WT株DNA分别与DAPI作用后, 应用360 nm激发波长, 获得DAPI-DNA的发射光谱(图 1A、C)。再通过标准品polyP45、细菌WT株分别与DAPI作用后, 应用415 nm激发波长, 测定DAPI-polyP的发射光谱(图 1B、D), DAPI-polyP的最大发射波长是550 nm, 而DAPI-DNA的最大发射波长是460 nm。
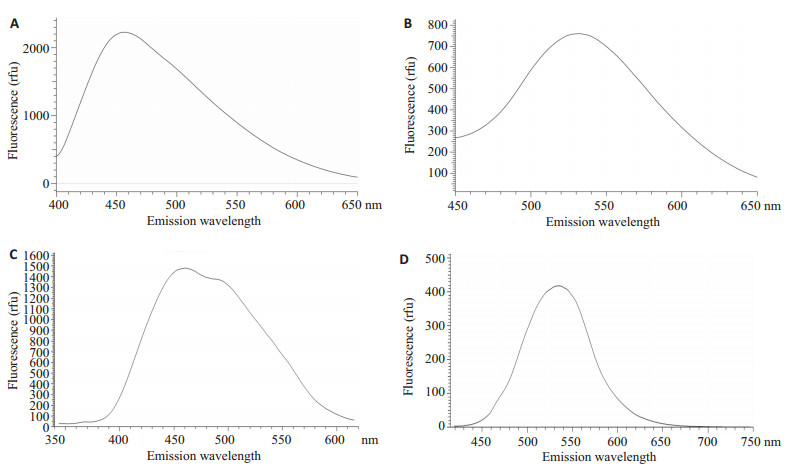
|
图 1 DAPI-DNA和DAPI-polyP的发射光谱 Fig.1 Emission spectra of DAPI-DNA and DAPI-polyP complexes. A: Emission spectra of DNA from the living EHEC WT with 360 nm excitation; B: Emission spectra of 0.25 relative unit (ru) of polyP 45 (equals 0.75 μmol/L P) excited at 415 nm; C: Emission spectra of living EHEC WT with 360 nm excitation; D: Emission spectra of living EHEC WT with 415 nm excitation. |
6种处理方法作用WT株后, 倍比稀释涂平板, 观察对细菌存活的影响。结果显示(表 3), 6种处理方法对细菌存活具有显著性差异(F=37.91, P=0.000);多重比较结果显示, 常温、-20 ℃、-80 ℃、0.5%Triton x-100等4种方法的菌落数无显著性差异。而与常温组相比, 60℃作用10 min及液氮速冻后自然溶解这2种方法直接导致细菌数量减少(细菌存活平均菌落数分别是0和15.25×106 cfu), 尤其60℃细菌作用10 min后细菌全部死亡(表 3)。
| 表 3 6种细菌前处理方法对细菌WT株存活影响的比较 Tab.3 Comparison of wild-type strain survival after 6 pretreatments |
对细菌存活无显著影响的4种处理方法与DAPI作用后, 测定并比较荧光值。结果显示, 4种处理方法的荧光值有显着性差异(F=78.67, P=0.000);多重比较, 结果发现最佳处理方法为-80 ℃速冻后室温下自然融化; 最差方法为室温下放置不作任何处理(表 4)。
| 表 4 4种细菌前处理方法与DAPI作用后荧光值比较(WT株菌) Tab.4 Comparison of fluorescence after 4 pretreatments and DAPI staining |
共聚焦显微镜观察, 结果显示, 当应用405 nm激发光, 在425~475 nm范围内收集到蓝色荧光, 为DAPIDNA。而当应用488 nm激发光时, 在500~560 nm范围收集到绿色荧光, 为DAPI-polyP (图 2)。
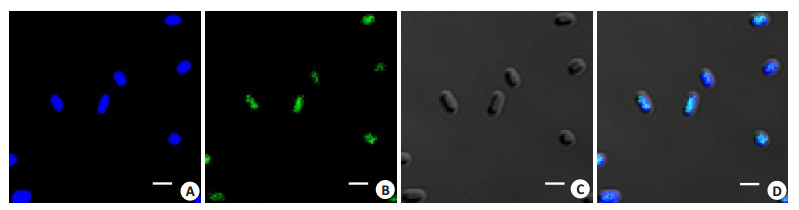
|
图 2 共聚焦显微镜观察DAPI-DNA和DAPI-polyP Fig.2 Confocal images of EHEC obtained with a Nikon A1R confocal microscope under the 60 × oil immersion objective with 10 × ocuLars. A: 405 nm laser line for detecting DNA (blue); B: 488 nm laser line for detecting polyP (green); C: No laser line for the Figure of cells; D: Merged images ofA, B and C. Scale bars: 2 µm |
利用标准品polyP45建立荧光值与其浓度的标准曲线。结果显示, 反应体系中, 10 μmol/L DAPI与polyP45结合的最大量是0.25 ru。因此polyP45在0~ 0.25 ru范围内, 荧光值与其成线性关系, 而随着polyP45量的增加, 荧光值不再增加(图 3A)。由此, 得出荧光值与polyP45相对单位(ru)的标准曲线Y=5546X+127.5, R2=0.991 (图 3B)。进一步根据表 2的换算获得荧光值与polyP45浓度(μmol/L)的标准曲线为Y=1849X + 127.5, R2=0.991 (图 3C)。
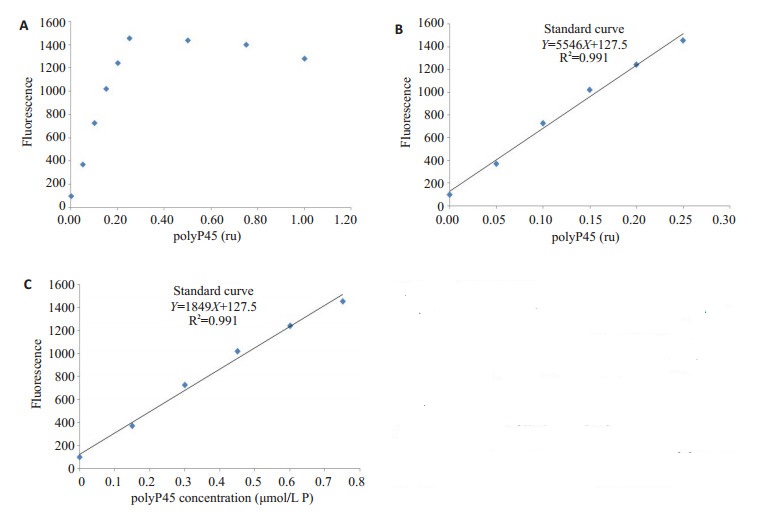
|
图 3 polyP45标准曲线 Fig.3 The relationship and standard calibration curve between polyP45 and fluorescence (measured emission at 550 nm when excited at 415 nm) with a final DAPI concentration of 10 μmol/L in each sample. A: The fluorescence increased gradually at polyP45 0-0.25 relative unit (ru), while it remained relatively stable at 0.25 to 1.0 ru; B: Standard curve of polyP45 ru and fluorescence; C: Standard curve of polyP45 concentration (based on B) and fluorescence. Both B and C had the same intercept β0 and R2 value of the regression line. |
各株细菌-80 ℃速冻, 室温下自然融化, 与DAPI作用, 测荧光值, 比较结果显示, 各株细菌polyP荧光值有显着性差异; 多重比较结果显示, Δppk1株的荧光值显著低于WT株和Cppk1株(表 5)。
| 表 5 各株细菌polyP荧光值比较 Tab.5 Comparison of fluorescence among different bacteria strains (EHEC WT, Δppk1 and Cppk1) |
根据标准曲线, 推算1 mLA600=1.0的各株菌所含polyP量(表 6)。
| 表 6 各株菌polyP浓度 Tab.6 polyP concentration (μmol/L Pi) deduced through standard calibration curve |
DAPI通常被用来检测细胞内DNA, 是否可应用于定性和定量检测大肠杆菌O157:H7内polyP尚不清楚。本研究通过荧光分光光度计测定固定激发波长时, DAPI-DNA及DAPI-polyP复合物的最大发射波长, 发现DAPI分别与polyP标准品polyP45及WT结合, 用415 nm波长激发时, 发射光谱最大波长是550 nm (图 1B、D); 而DAPI-DNA复合物用360 nm激发, 最大发射波长是460 nm (图 1A、C), 二者并无交叉。激光共聚焦显微镜观察发现, DAPI-DNA呈蓝色荧光, 而DAPI-polyP呈绿色荧光(图 2)。因此, 上述结果从定性和定量两方面说明DAPI可有效定量检测大肠杆菌polyP, 并可避免DNA的干扰。
应用DAPI直接定量polyP, DAPI可直接渗透入细菌内结合polyP, 而不需要将细菌杀死后提取polyP。polyP的DAPI荧光定量法和polyP提取法进行对比, 结果显示, polyP提取法比DAPI定量法低估了28~55% polyP含量[28, 30]。
DAPI与polyP的作用机制可能为[25]: DAPI与带负电荷的离子相互作用, 尤其DAPI通过N-H基团产生静电吸引力。polyP是带负电荷的磷酰基团, 它作为阴离子吸引大量DAPI分子结合, 并引起染料间的相互作用。
Zink等[31]认为DAPI穿透活细胞的效率比较低。为了获得DAPI渗透入细胞最佳的细菌前处理方法, 改善细胞膜通透性, 使DAPI易于进入活细胞, 我们进行了6种细菌前处理方法的比较(表 1)。细菌经前处理后, 取等量菌液倍比稀释后涂LB平板, 观察前处理方法对细菌生存的影响。结果显示, 6种处理方法对细菌的存活具有显著性差异, 细菌存活菌落数显著不同。常温、-20 ℃、-80 ℃、0.5% Triton x-100四种前处理方法未明显影响细菌的存活; 而液氮速冻后再自然溶解的方法导致部分细菌死亡, 60 ℃细菌作用10 min后出现细菌全部死亡, 分解的细胞将影响DAPI结合polyP。此外, 加热导致长链polyP裂解(长度 < 5个Pi), DAPI也无法结合[28]。
再将对细菌生存无显著影响的4种处理方法与DAPI作用后, 应用荧光分光光度计测定并比较荧光值。结果显示, 细菌前处理最好的方法是-80 ℃速冻后在室温下自然融化。另外, -20 ℃速冻后室温自然融化与-80 ℃的处理无显著性差异, 说明这些物理方法只是改变了细胞膜的通透性而未改变其形态; Triton x-100方法对于细菌渗透效果远没有真核细胞明显; 效果最差的处理方法是室温下不作任何处理。
DAPI荧光法定量检测方法的建立为polyP相关研究打下坚实基础。在大肠杆菌内, 多聚磷酸盐激酶(ppk1)催化polyP的合成, 为了验证本方法的适用性, 在EHEC O157:H7野生株(WT)基础上, 我们构建了ppk1基因缺失株(Δppk1)和回补株(Cppk1)。将3株细菌应用-80 ℃速冻自然融化, 进行DAPI染色, 测定荧光值进行比较。结果显示, 3株细菌荧光值有显著性差异; 多重比较显示, Δppk1株的荧光值显著低于WT株与Cppk1株, 说明ppk1基因缺失突变株细胞内显polyP的合成显著减少, 与理论预设相符, 也与致脑膜炎大肠杆菌K1 (RS218) [32]结果一致。
应用标准品polyP45建立的荧光值与标准品相对浓度的标准曲线, 即Y=1849X+127.5, R2=0.991说明自变量Y (荧光值)与因变量X (polyP标准品浓度)呈极线性相关。标准曲线的斜率和截距会随着不同实验室的实验条件而变化[28]。因此, 建议在不同实验室或应用不同的polyP标准品时构建新的标准曲线。
对于体外培养细菌, 我们建议用A600=1.0时1 mL细菌内polyP的浓度μmol/L Pi/mL为单位更为简单合理。将三株菌荧光值带入标准曲线, 换算成A600=1.0时1 mL菌液中polyP的浓度, 结果显示WT为4.25 μmol/L Pi/mL, Δppk1和Cppk1分别为1.06和4.64 μmol/L Pi/mL。细菌浓度无差异的情况下, 3株的polyP浓度具有显著性差异。WT和Cppk1显著高于Δppk1株, 与预期结果一致。
鉴于此, 本研究应用DAPI与polyP结合发出不同于DAPI-DNA荧光的特点, 建立了肠出血型大肠埃希菌polyP的DAPI直接定量荧光检测方法, 为进一步研究polyP及其催化酶在该菌生存、致病、进化过程中的作用提供技术支持。
| [1] |
Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival[J].
Annu Rev Biochem, 2009, 78: 605-47.
DOI: 10.1146/annurev.biochem.77.083007.093039. |
| [2] |
Kim KS, Rao NN, Fraley CD, et al. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp[J].
Proc Natl Acad Sci USA, 2002, 99(11): 7675-80.
DOI: 10.1073/pnas.112210499. |
| [3] |
Li L, Rao NN, Kornberg A. Inorganic polyphosphate essential for lytic growth of phages P1 and fd[J].
Proc Natl Acad Sci USA, 2007, 104(6): 1794-9.
DOI: 10.1073/pnas.0610763104. |
| [4] |
Rashid MH, Rao NN, Kornberg A. Inorganic polyphosphate is required for motility of bacterial pathogens[J].
J Bacteriol, 2000, 182(1): 225-7.
DOI: 10.1128/JB.182.1.225-227.2000. |
| [5] |
Zhang HY, Gomez-Garcia MR, Shi XB, et al. Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum with a role in cytokinesis[J].
Proc Natl Acad Sci USA, 2007, 104(42): 16486-91.
DOI: 10.1073/pnas.0706847104. |
| [6] |
Rohlfing AE, Ramsey KM, Dove SL. Polyphosphate kinase antagonizes virulence gene expression in francisella tularensis[J].
J Bacteriol, 2018, 200(3): e00460-17.
|
| [7] |
Grillo-Puertas M, Ariane Schurig-Briccio LA, Regina Rintoul M, et al. Copper tolerance mediated by polyphosphate degradation and low-affinity inorganic phosphate transport system in Escherichia coli[J].
BMC Microbiol, 2014, 14(3): 72.
|
| [8] |
Livermore TM, Chubb JR, Saiardi A. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum[J].
Proc Natl Acad Sci U S A, 2016, 113(4): 996-1001.
DOI: 10.1073/pnas.1519440113. |
| [9] |
Gray MJ, Jakob U. Oxidative stress protection by polyphosphate-- new roles for an old player[J].
Curr Opin Microbiol, 2015, 24: 1-6.
DOI: 10.1016/j.mib.2014.12.004. |
| [10] |
Alcantara C, Blasco A, Zuniga M, et al. Accumulation of polyphosphate in lactobacillus spp. and its involvement in stress resistance[J].
Appl Environ Microbiol, 2014, 80(5): 1650-9.
DOI: 10.1128/AEM.03997-13. |
| [11] |
Chen J, Su LJ, Wang XR, et al. Polyphosphate kinase mediatesantibiotic tolerance in extraintestinal pathogenic Escherichia coli PCNO33[J].
Front Microbiol, 2016, 7(5): 724.
|
| [12] |
Anand, Kumar, Dharanesh, et al. Polyphosphate and associated enzymes as global regulators of stress response and virulence in Campylobacter jejuni[J].
World J Gastroenterol, 2016, 22(33): 7402-14.
DOI: 10.3748/wjg.v22.i33.7402. |
| [13] |
Varas MA, Riquelme-Barrios S, Valenzuela C, et al. Inorganic polyphosphate is essential for salmonella typhimurium virulence and survival in dictyostelium discoideum[J].
Front Cell Infect Microbiol, 2018, 8(1): 8.
|
| [14] |
Angelova PR, Baev AY, Berezhnov AV. Role of inorganic polyphosphate in mammalian cells: from signal transduction and mitochondrial metabolism to cell death[J].
Biochem Soc Trans, 2016, 44(1): 40-5.
DOI: 10.1042/BST20150223. |
| [15] |
Kawazoe Y, Katoh S, Onodera Y, et al. Activation of the FGF signaling pathway and subsequent induction of mesenchymal stem cell differentiation by inorganic polyphosphate[J].
Int J Biol Sci, 2008, 4(1): 37-47.
|
| [16] |
Morrissey JH, Smith SA. Polyphosphate as modulator of hemostasis, thrombosis, and inflammation[J].
J Thromb Haemost, 2015, 13(1, SI): S92-7.
DOI: 10.1111/jth.12766. |
| [17] |
Labberton L, Kenne E, Long AT, et al. Neutralizing blood-borne polyphosphate in vivo provides safe thromboprotection[J].
Nat Commun, 2016, 7(9): 12616.
|
| [18] |
Smith SA, Morrissey JH. 2013 scientific sessions Sol Sherry distinguished lecture in thrombosis: polyphosphate: a novel modulator of hemostasis and thrombosis[J].
Arterioscler Thromb Vasc Biol, 2015, 35(6): 1298-305.
DOI: 10.1161/ATVBAHA.115.301927. |
| [19] |
Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein[J].
J Biol Chem, 1992, 267(31): 22556-61.
|
| [20] |
Rao NN, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response[J].
J Bacteriol, 1998, 180(8): 2186-93.
|
| [21] |
Ault-Riche D, Fraley CD, Tzeng CM, et al. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli[J].
J Bacteriol, 1998, 180(7): 1841-7.
|
| [22] |
Mullan A, Quinn JP, Mcgrath JW. A nonradioactive method for the assay of polyphosphate kinase activity and its application in the study of polyphosphate metabolism in Burkholderia cepacia[J].
Anal Biochem, 2002, 308(2): 294-9.
DOI: 10.1016/S0003-2697(02)00249-X. |
| [23] |
Werner TP, Amrhein N, Freimoser FM. Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase[J].
Arch Microbiol, 2005, 184(2): 129-36.
|
| [24] |
Freimoser FM, Huerlimann HC, Jakob CA, et al. Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism[J].
Genome Biol, 2006, 7(11): R109.
DOI: 10.1186/gb-2006-7-11-r109. |
| [25] |
Omelon S, Georgiou J, Habraken W. A cautionary (spectral) tail: redshifted fluorescence by DAPI-DAPI interactions[J].
Biochem Soc Trans, 2016, 44(1): 46-9.
DOI: 10.1042/BST20150231. |
| [26] |
Allan RA, Miller JJ. Influence of S-adenosylmethionine on DAPIinduced fluorescence of polyphosphate in the yeast vacuole[J].
Can J Microbiol, 1980, 26(8): 912-20.
DOI: 10.1139/m80-158. |
| [27] |
Aschar-Sobbi R, Abramov AY, Diao CA, et al. High sensitivity, quantitative measurements of polyphosphate using a new DAPIBased approach[J].
J Fluoresc, 2008, 18(5): 859-66.
DOI: 10.1007/s10895-008-0315-4. |
| [28] |
Diaz JM, Ingall ED. Fluorometric quantification of natural inorganic polyphosphate[J].
Environ Sci Technol, 2010, 44(12): 4665-71.
DOI: 10.1021/es100191h. |
| [29] |
Gomes FM, Ramos IB, Wendt C, et al. New insights into the in situ microscopic visualization and quantification of inorganic polyphosphate stores by 4', 6-diamidino-2-phenylindole (DAPI)-staining[J].
Eur J Histochem, 2013, 57(4): 227-35.
|
| [30] |
Kulakova AN, Hobbs D, Smithen M, et al. Direct quantification of inorganic polyphosphate in microbial cells using 4'-6-Diamidino-2- Phenylindole (DAPI)[J].
Environ Sci Technol, 2011, 45(18): 7799-803.
DOI: 10.1021/es201123r. |
| [31] |
Zink D, Sadoni N, Stelzer E. Visualizing chromatin and chromosomes in living cells[J].
Methods, 2003, 29(1): 42-50.
DOI: 10.1016/S1046-2023(02)00289-X. |
| [32] |
Peng L, Luo WY, Zhao T, et al. Polyphosphate kinase 1 is required for the pathogenesis process of meningitic Escherichia coli K1(RS218)[J].
Future Microbiol, 2012, 7(3): 411-23.
DOI: 10.2217/fmb.12.3. |
 2019, Vol. 39
2019, Vol. 39

