2. Department of Urology, Fifth Affiliated Hospital of Southern Medical University, Guangzhou 510900, China
2. 南方医科大学第五附属医院泌尿外科,广东 广州 510900
Prostate cancer (PCa) is one of the most common malignancies in men worldwide and causes over 300 000 cancer-related deaths each year[1]. Radical prostatectomy and androgen deprivation therapy are currently the primary options of treatment for localized PCa[2], but for metastatic PCa, which is estimated to occur eventually in of 90% of the patients[3], these treatments can be futile and the overall survival of the patients is only 12-15 months [4, 5].
The technology of next-generation sequencing has accelerated our understanding of the cancer-related genes underlying the development and progression of PCa, such as the mutations of PIK3CA, FOXM1, SPOP, MED12; the copy number loss in the PTEN (10q) and NKX3.1 (8p) or gain in the androgen receptor (AR) (Xq12); and gene fusion of ETS transcription factors with androgen-responsive promoters[6-8]. However, these massive sequencing data on PCa also bring new challenges. How to identify the actually functional "driver" genes from the background of "passenger" alterations? How can the functional relevance of these genes be established and integrated with the mechanistic insights gained in cell culture and animal model systems? The answers to these questions are crucial to the comprehension of the new findings from these next-generation sequencing data [9].
Citron Rho-interacting serine/threonine kinase (CIT) is a downstream substrate of Rho protein[10]. During cytokinesis, CIT plays an important role in the formation of midbody by regulating multiple molecular networks[11]. In the contractile process, CIT binds to RhoA and maintains RhoA localization at the cleavage site. The CIT-RhoA interaction increases RhoA activity and promotes contractile ring dynamics[12]. In addition, recent studies have suggested a pathogenic role of CIT. Loss of CIT leads to cytokinesis failure and apoptosis in mammalian neuronal progenitors and germ cells, causing microcephaly and testicular hypoplasia[13]. CIT is also associated with the development of cancer in human: the expression of CIT is found to be up-regulated in colon cancer, and knockdown of CIT reduces cancer cell proliferation via p53 signaling pathway[10]. Similarly, in live cancer, CIT regulates the G2/M transition and loss of it inhibits tumor growth[14]. Whitworth et al performed RNA interference (RNAi) phenotypic screening and found that CIT deficiency suppressed the growth of both androgen-dependent and castration-resistant PCa cells [15]; but the role of CIT in metastasis, the most fatal cancer stage, remains unknown.
In this study, we acquired the next generation sequencing data of PCa from the Cancer Genome Atlas (TCGA) and Memorial Sloan-Kettering Cancer Center (MSKCC) datasets to identify new oncogenes that correlate with tumor metastasis. We also assessed the effects of CIT silencing on the biological behaviors of PC-3 cells, and these changes provide insights into the role of CIT in the tumorigenesis and progression of PCa.
MATERIALS AND METHODS Bioinformatics analysis of CIP expression and clinical dataWe analyzed the expression of CIT in PCa tissues using the transcriptomic data from TCGA PRAD dataset, which includes 51 pairs of PCa tissues and matched adjacent prostate tissues (https://portal.gdc.cancer.gov). The fold change of CIT expression in PCa relative to the matched adjacent samples was calculated as described previously [16].
The correlations of CIT gene with the clinicopathological features of the patients were analyzed in a clinical transcriptome study (Memorial Sloan Kettering Cancer Centre, MSKCC), available through the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal)[17]. This collection contains 150 PCa samples. The correlations of CIT gene expression patterns with the clinicopathological features (tumor grade, Gleason scores, and invasion status) were analyzed with R/Bioconductor.
Cell cultureThe PCa cell line PC-3 was obtained from American Type Culture Collection (ATCC). The cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and incubated at 37 ℃ in 5% CO2.
RNA interferenceTwo pairs of small interfering RNAs (siRNAs) targeting human CIT mRNA were used:
siRNA1:sense, 5'-GGGAGAUGUUGAAGUUCAAAU-3', anti-sense, 5'-UUGAACUUCAACAUCUCCCCA-3';
siRNA 2: sense, 5'-GCGACAGAAUGUCAGCAUAAA-3', anti-sense, 5'-UAUGCUGACAUUCUGUCGCUU-3'.
These siRNAs (synthesized by Ribobio, Guangzhou, China) were transfected into PC-3 cells using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA), with a scrambled siRNA sequence serving as the negative control (NC). All the procedures of siRNA transfection were carried out according to the manufacturer's instructions. The efficiency of RNAi was evaluated by detecting CIT mRNA expression in the cells using quantitative real-time PCR (qRT-PCR) at 48 h after siRNA transfection. Briefly, the total RNA were extracted from the transfected PC-3 cells using RNAiso reagent (Takara, Tokyo, Japan) and reverse transcribed using PrimeScript RT Master Mix (Takara) according to the manufacturer's protocol. qRT-PCR analysis of CIT mRNA expression was conducted using the SYBR Green I qPCR Mix (Takara) on the Roche 480 System (Roche, Basel, Switzerland). The data were analyzed using 2-ΔΔCt method. GAPDH was used as the internal control. The following primers were used:
CIT (NM_0, 17, 409. 3),
5'-CAGGCAAGATTGAGAACG-3',
5'-GCACGATTG AGACAGGGA-3';
GAPDH (NM_00, 12, 56, 799. 1),
5'-TGACTTCAACAGCGACACCCA-3',
5'-CAC CCT GTT GCT GTA GCC AAA-3'.
Western blottingAt 48 h after transfection with siRNAs, PC-3 cells were lysed using RIPA lysis buffer. The proteins were then separated on SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking with 5% skim milk, the blots were incubated with the primary antibodies, followed by incubation with HRP-conjugated goat anti-rabbit secondary antibody (Proteintech, SA00001-2; dilution 1:5000). The bands were visualized using enhanced chemiluminescence (ECL; Millipore). The primary antibodies used were shown in Tab. 1.
| Tab.1 The primary antibodies for Western blotting. |
PC-3 cells were cultured in 96-well plates at 1000 per well. The adherent cells were transfected with siRNAs, and CCK-8 solution (Keygen, Nanjing, China) was added to each well at 0, 24, 48 and 72 h after siRNA transfection. After incubation for 1 h at 37 ℃, the absorbance at 450 nm (A450 nm)was read to determine the cell viability on a microplate reader (Monecular device, California, USA).
Wound healing assayPC-3 cells were seeded in 6-well plates at 3×105 per well and cultured in RPMI 1640 medium without FBS. After incubation at 37 ℃ overnight, the adherent cells were transfected with siRNAs for 48 h. The cell monolayer was then scratched with a 10 μL pipette and incubated with 1% serum-containing medium for another 48 h. The images of the cell monolayer were captured under a phase-contrast microscope (Nikon, Tokyo, Japan) and the relative migration distance of the cells was calculated.
Transwell migration and invasion assayFor cell migration assay, PC-3 cells (1×105) transfected with siRNAs for 48 h were seeded into the upper chamber, and the lower chamber contained RPMI 1640 supplemented with 10% FBS. After incubation at 37 ℃ for 48 h, the membranes were stained with crystal violet solution. The cells on the top surface of the insert were removed and counted under a microscope in 5 random fields. Cell invasion assay were performed following similar procedures using the Transwell membranes coated with Matrigel.
Statistical analysisThe data are presented as Mean±SD and analyzed using SPSS 20.0 software (SPSS, Chicago, IL, USA). The differences among the groups were tested using unpaired Student's t-test and one-way ANOVA as appropriate. The correlation between CIT expression and the clinicopathological features of PCa patients was evaluated by Fisher's exact test. P < 0.05 was considered to indicate a statistically significant difference.
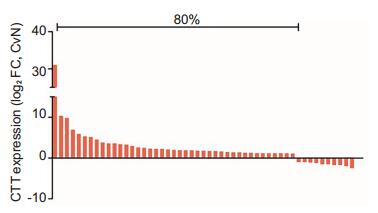
RESULTS CIT was upregulated in PCa in correlation with disease progressionTo investigate the role of CIT in PCa, we assessed the RNA-seq data of CIT gene from 51 paired PCa tissue samples based on TCGA database. The results showed that the expression of CIT gene was significantly upregulated in 41 (80%) of the PCa tissues compared to their non-cancerous counterparts (Fig. 1).
|
Fig.1 CIT expression was upregulated in (41/51) 80% of PCa tissues (TCGA cohort).Data are presented as log2 (T/NT), the log2 ratio of CIT expression levels between PCa tissues and the adjacent non-cancerous normal tissues. Upregulation was defined as log2 (T/NT) > 0. |
To determine the involvement of CIT in the progression of PCa, we investigated the correlation between CIT expression and the clinicopathological features of the patients based on MSKCC database. MSKCC database analysis showed that upregulation of CIT was associated with advanced N stage (P=0.001), M stage (P < 0.001), Gleason score (P=0.010) and PSA level (P=0.004; Tab. 2).
| Tab.2 Clinical features and driver alterations in 150 prostate cancer patients from MSKCC database (No. of patients) |
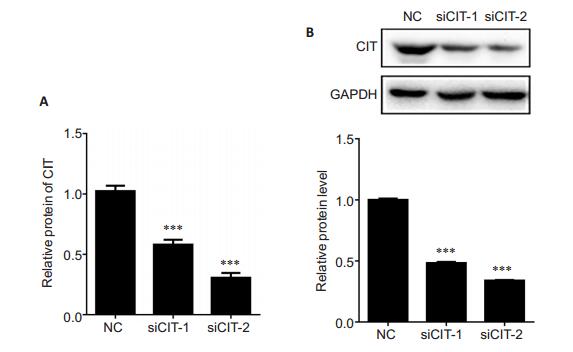
To explore the role of CIT in the tumorigenesis of PCa, we knocked down CIT expression in PC-3 cells by transfecting the cells with siRNAs, and verified the efficiency of CIT knockdown using qRT-PCR and Western blotting. As shown in Fig. 2, at 48 h after transfection with siRNAs, both the mRNA and protein levels of CIT were significantly decreased in PC-3 cells in comparison with the cells in NC group.
|
Fig.2 CIT expression was significantly lowered in PC- 3 cells at 48 after siRNA transfection.A: The mRNA level of CIT detected by qRT-PCR. Data are presented as Mean±SD from 3 independent experiments. ***P < 0.001 vs NC group; B: The protein level of CIT detected by Western blotting with GAPDH as the control. ***P < 0.001 vs NC group. |
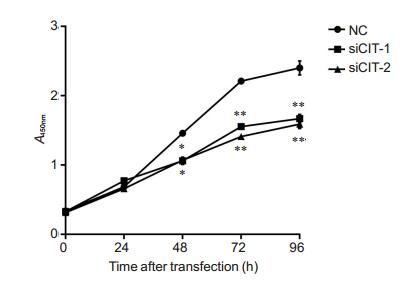
CCK-8 assay showed that transfection with the CIT-specific siRNAs significantly inhibited the growth of PC-3 cells at 48, 72 and 96 h (Fig. 3).
|
Fig.3 CIT knockdown significantly suppressed cell proliferation in PCa. Cell vitality of PC-3 cells was examined by CCK-8 assay after siRNAs transfection for 0, 24, 48, 72, 96 h, respectively. *P < 0.05, **P < 0.01 vs NC group. |
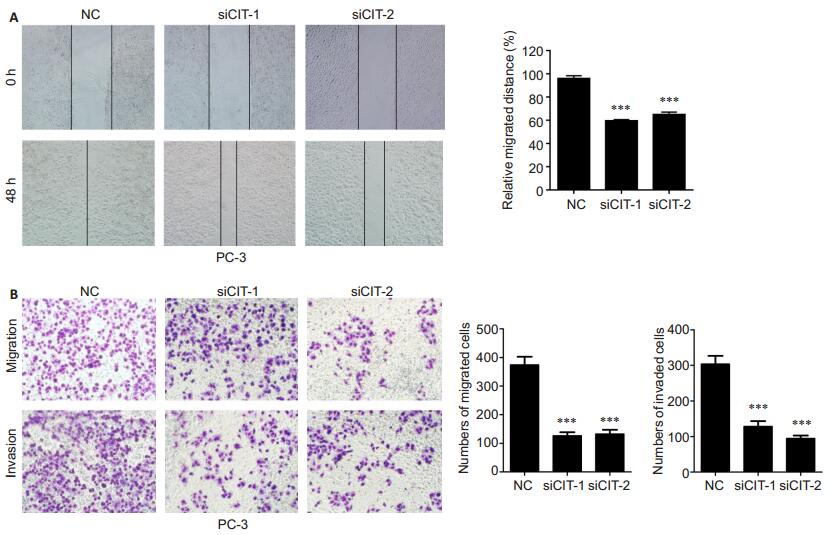
We tested the effect of CIT knockdown silencing on the migration and invasion of PC-3 cells using wound healing and Transwell assays. The results showed that transfection of the cells with the CIT-specific siRNAs significantly reduced both the migration distance and the number of migrating cells at 48 h (P < 0.01; Fig. 4). Invasion assay demonstrated that CIT silencing also significantly suppressed the invasion of PC-3 cells as compared with the control cells (P < 0.01).
|
Fig.4 CIT knockdown significantly suppressed migration and invasion of PCa cells. A: Cell migration capacity of PC-3 cells assessed by wound healing assay; B: Cell migration and invasion capacity of PC-3 cells assessed by Transwell migration and invasion assays. Data are presented as Mean±SD from 3 independent experiments. ***P < 0.001 vs NC group. |
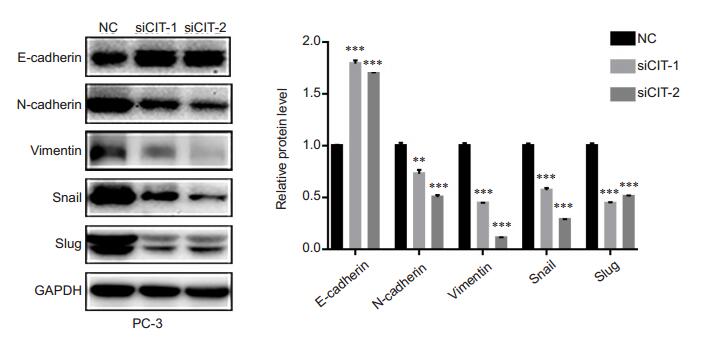
We performed Western blotting to detect the effect of CIT knockdown in PC-3 cells on the protein expressions of N-cadherin, vimentin, Snail, Slug and E-cadherin. Transfection with the CIT-specific siRNAs resulted in significantly increased expression of E-cadherin and decreased expressions of N-cadherin, vimentin, Snail, and Slug in PC-3 cells compared with the control cells (Fig. 5), indicating the reversal of the malignant EMT phenotype of PC-3 cells after CIT silencing.
|
Fig.5 CIT silencing significantly inhibited EMT in PC-3 cells. The protein level of E-cadherin, N-cadherin, Vimentin, Snail, and Slug were detected by Western blotting in the cells after transfection with CIT-specific siRNAs, using GAPDH as the control. **P < 0.01, ***P < 0.001 vs NC group. |
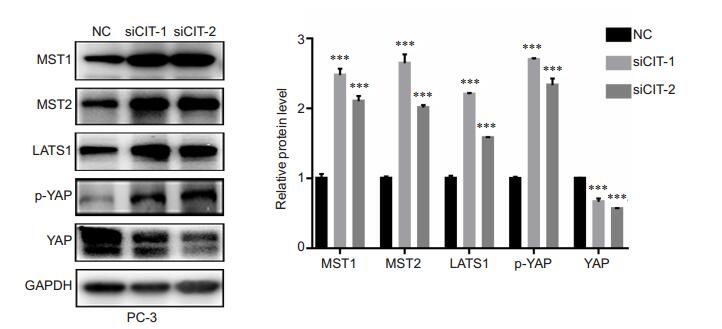
As shown in Fig. 6, CIT silencing in PC-3 cells caused significantly increased expressions of MST1/2 (which activates LATS), LATS1 (which phosphorylates and inhibits YAP), and phosphorylation levels of YAP (p-YAPser397), while reduced the expression levels of total YAP in the cells. These results suggested that CIT knockdown blocked Hippo-YAP pathway by partially modulating the upstream regulators MST1/2 and LATS1, and subsequently enhanced the phosphorylation of YAP.
|
Fig.6 CIT silencing significantly blocked Hippo- YAP pathway in PC-3 cells. The protein level of MST1, MST2, LATS1, phospholated-YAP-Ser397, and YAP were detected by Western blotting in PC-3 cells after transfection with CIT siRNAs, using GAPDH as the control. Data are presented as Mean ± SD from 3 independent experiments. ***P < 0.001 vs NC group. |
From TCGA and TSKCC PCa data, we found that CIT was upregulated in PCa tissues and positively correlated with the metastatic features of the malignancy. In the loss-of-function study, we demonstrated that CIT silencing inhibited the cell growth, migration and invasion, and reversed EMT in PC-3 cells. More importantly, our results indicate that CIT might function through the Hippo-YAP pathway to regulate the metastasis of PCa. Taken together, these findings support the hypothesis that CIT contributes to the development of PCa partially via the Hippo-YAP signaling pathway and may provide a promising therapeutic target for PCa treatment.
EMT is thought to be an important biological mechanism that plays critical roles in cancer cell invasion and metastasis, and has been widely studied in PCa [18, 19]. However, the key mediators and molecular circuits underlying EMT in PCa remain poorly understood. In this study, we examined the correlation between EMT and CIT in PC-3 cells and demonstrated that CIT knockdown suppressed the migration and invasion potentials and reversed EMT phenotypes to MET characteristics in the cells. These results suggest that CIT performs its regulatory roles on PCa invasion and metastasis by altering the process of EMT.
Several pathways have been identified to regulate EMT, including NF-κB, Wnt, and PI3k/Akt [20]. Accumulating evidence has shown that the Hippo pathway also cross-talks with EMT. The Hippo pathway is an emerging signaling pathway involved in organ size control, stem cell homeostasis and tumorigenesis[21, 22]. In mammals, the Hippo pathway comprises a kinase cascade including MST1/2, LATS1, and the downstream effectors: transcriptional co-activator with PDZ-binding motif (TAZ, also known as WWTR1) and yes-associated protein (YAP). Upon Hippo activation, MST1/2 phosphorylates LATS1 and in turn phosphorylates and inactivates TAZ/YAP by their cytoplasmic retention and proteasome-mediated degradation. Notably, deregulated YAP activity has been reported frequently in a broad range of cancers[21]. Aberrant YAP overexpression was implicated in fundamental cellular processes, such as cell proliferation, migration, invasion and EMT[23-25]. In PCa, YAP is found to be overexpressed in tumor tissues and regulates tumor cell motility, invasion, and castration-resistant growth[26, 27]. Here we show that CIT is upregulated in PCa by bioinformatics analysis of the TCGA dataset. Given that CIT is involved in the generation of specific neuronal precursors, which are also regulated by the activation of YAP gene, we assume that CIT executes its oncogenic functions in PCa by modulating the Hippo pathway[13, 28]; this assumption is supported by our finding that CIT knockdown activates the MST-LATS-YAP phosphorylation cascade and suppresses the expression of YAP. Taken together, our results provide evidence that CIT, as an oncogene, regulates PCa cell growth, metastasis, and EMT through the Hippo-YAP pathway.
| [1] | Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012[J]. CA Cancer J Clin, 2015, 65(2): 87-108. DOI: 10.3322/caac.21262. |
| [2] | Kohli M, Tindall DJ. New developments in the medical management of prostate cancer[J]. Mayo Clin Proc, 2010, 85(1): 77-86. DOI: 10.4065/mcp.2009.0442. |
| [3] | Carlin B I, Andriole G L. The natural history, skeletal complications, and management of bone metastases in patients with prostate carcinoma[J]. Cancer, 2000, 88(12 Suppl): 2989-94. |
| [4] | de Bono J S, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial[J]. Lancet, 2010, 376(9747): 1147-54. DOI: 10.1016/S0140-6736(10)61389-X. |
| [5] | Li H, Zhang Y, Zhang Y, et al. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker[J]. Tumour Biol, 2014, 35(6): 5771-6. DOI: 10.1007/s13277-014-1766-7. |
| [6] | Baca SC, Garraway LA. The genomic landscape of prostate cancer[J]. Front Endocrinol (Lausanne), 2012, 3: 69. |
| [7] | Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer[J]. Nat Genet, 2012, 44(6): 685-9. DOI: 10.1038/ng.2279. |
| [8] | Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer[J]. Nature, 2012, 487(7406): 239-43. DOI: 10.1038/nature11125. |
| [9] | Barbieri CE, Bangma CH, Bjartell A, et al. The mutational landscape of prostate cancer[J]. Eur Urol, 2013, 64(4): 567-76. DOI: 10.1016/j.eururo.2013.05.029. |
| [10] | Wu Z, Zhu X, Xu W, et al. Up-regulation of CIT promotes the growth of colon cancer cells[J]. Oncotarget, 2017, 8(42): 71954-64. |
| [11] | Bassi ZI, Audusseau M, Riparbelli MG, et al. Citron kinase controls a molecular network required for midbody formation in cytokinesis[J]. Proc Natl Acad Sci USA, 2013, 110(24): 9782-7. DOI: 10.1073/pnas.1301328110. |
| [12] | Bassi ZI, Verbrugghe KJ, Capalbo L, et al. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis[J]. J Cell Biol, 2011, 195(4): 595-603. DOI: 10.1083/jcb.201105136. |
| [13] | Di Cunto F, Imarisio S, Hirsch E, et al. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis[J]. Neuron, 2000, 28(1): 115-27. DOI: 10.1016/S0896-6273(00)00090-8. |
| [14] | Fu Y, Huang J, Wang K S, et al. RNA interference targeting CITRON can significantly inhibit the proliferation of hepatocellular carcinoma cells[J]. Mol Biol Rep, 2011, 38(2): 693-702. |
| [15] | Whitworth H, Bhadel S, Ivey M, et al. Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen[J]. PLoS One, 2012, 7(6): e38950. DOI: 10.1371/journal.pone.0038950. |
| [16] | Ye G, Huang K, Yu J, et al. MicroRNA-647 targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer[J]. Theranostics, 2017, 7(13): 3338-53. DOI: 10.7150/thno.20512. |
| [17] | Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal[J]. Sci Signal, 2013, 6(269): pl1. |
| [18] | Montanari M, Rossetti S, Cavaliere C, et al. Epithelial-mesenchymal transition in prostate cancer: an overview[J]. Oncotarget, 2017, 8(21): 35376-89. |
| [19] | Lo UG, Lee CF, Lee MS, et al. The role and mechanism of epithelial-tomesenchymal transition in prostate cancer progression[J]. Int J Mol Sci, 2017, 18(10). |
| [20] | Maier J, Traenkle B, Rothbauer U. Visualizing epithelial-mesenchymal transition using the chromobody technology[J]. Cancer Res, 2016, 76(19): 5592-96. DOI: 10.1158/0008-5472.CAN-15-3419. |
| [21] | Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer[J]. Nat Rev Cancer, 2013, 13(4): 246-57. DOI: 10.1038/nrc3458. |
| [22] | Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal[J]. Nat Cell Biol, 2011, 13(8): 877-83. DOI: 10.1038/ncb2303. |
| [23] | Chan SW, Lim CJ, Loo LS, et al. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation[J]. J Biol Chem, 2009, 284(21): 14347-58. DOI: 10.1074/jbc.M901568200. |
| [24] | Overholtzer M, Zhang J, Smolen GA, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon[J]. Proc Natl Acad Sci USA, 2006, 103(33): 12405-10. DOI: 10.1073/pnas.0605579103. |
| [25] | Zhang H, Liu CY, Zha ZY, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition[J]. J Biol Chem, 2009, 284(20): 13355-62. DOI: 10.1074/jbc.M900843200. |
| [26] | Zhang L, Yang S, Chen X, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells[J]. Mol Cell Biol, 2015, 35(8): 1350-62. DOI: 10.1128/MCB.00102-15. |
| [27] | Collak FK, Demir U, Ozkanli S, et al. Increased expression of YAP1 in prostate cancer correlates with extraprostatic extension[J]. Cancer Biol Med, 2017, 14(4): 405-13. DOI: 10.20892/j.issn.2095-3941.2017.0083. |
| [28] | Fernandez LA, Northcott PA, Dalton J, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation[J]. Genes Dev, 2009, 23(23): 2729-41. DOI: 10.1101/gad.1824509. |
 2019, Vol. 39
2019, Vol. 39

