2. 中国科学院大学,北京 100049;
3. 宁波大学 理学院,浙江 宁波 315211
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. School of Science, Ningbo University, Ningbo 315211, China
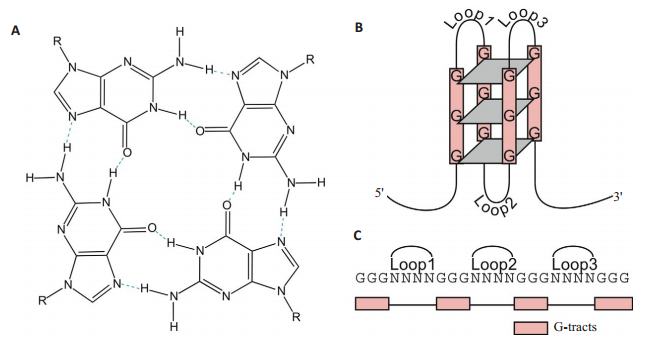
端粒在细胞的生理病理过程中起到至关重要的作用[1]。端粒中富含鸟嘌呤G的重复序列在单价离子作用下可形成高度稳定的二级结构-G四链体[2-4](图 1)[5]。G四链体结构的形成可以维持基因组的稳定性,在衰老和癌症治疗方面起到了重要的调控作用,因此G四链体成为了一个重要的抗肿瘤治疗靶点[6-8]。许多诱导该结构形成并稳定该结构的小分子配体被设计合成并用作抗癌药物[9-15]。G四链体稳定性及端粒活性是评价端粒G四链体小分子配体作用效果最常用的参数标准[16-18]。因此评定G四链体与小分子配体间的作用强度是筛选有效药物的关键。
|
图 1 G四链体结构[5] Figure 1 Structure of a G- quadruplex[5]. A: Structure of a G-quartet. The planar ring of four hydrogenbonded guanines is formed by guanines from different G-tracts, which are separated by intervening loop regions in the intra-molecular G quadruplex DNA structure; B: Schematic illustration of an intramolecular G quadruplex DNAstructure consisting of 3 G-quartets. Inter-molecular G quadruplex DNA structures can also form from 2 or 4 strands; C: The G quadruplex DNA motif sequence used in this study with 4 G-tracts of 3 guanines separated by loop regions. |
基于原子力显微镜的单分子力谱技术(AFM-SMFS)兼具高分辨成像和力学测定两种优势,在分子对相互作用强度测定方面具有很大的潜力[19-21]。AFM-SMFS在空间操纵尺度上可以跨越6个数量级,从细胞操纵(100 μm)到RNA聚合酶推进,再到单碱基对测量(0.34 nm);在力学操纵尺度上可跨越4个数量级,从共价键断裂机理分析(nN)到核酸折叠动力学分析(pN)[22]。目前已经能够对单分子间相互作用、构象变化及动力学过程进行直接、实时的检测,为解决生命科学中长期存在的许多问题提供了精准定量的解决途径。鉴于单分子力谱技术显著的技术优势,本文主要综述基于原子力显微镜单分子力谱技术在G四链体与其小分子配体相互作用中的研究现状,为人们利用单分子力谱技术开发筛选靶向G四链体抗癌药物提供参考。
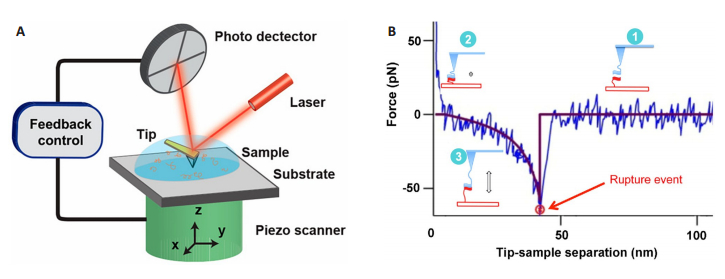
1 基于原子力显微镜的单分子力谱(AFM-SMFS)技术原子力显微镜单分子力谱技术(AFM-SMFS)仪器组成和原子力显微镜系统的唯一区别在于探针针尖是否被特异性修饰。AFM-SMFS系统主要包括4个模块:微探针、压电陶瓷扫描管、检测系统和控制系统(图 2A)[23]。当针尖靠近样品表面进行水平扫描时,可以得到样品的形貌信息;当将特异性修饰的针尖相对于固定在衬底上的样品在竖直方向来回运动,会获得相互作用分子对间的力学信息(图 2B)[24]。通过对力学信息分析可得到单分子的构象变化及其相互作用的动力学过程。
|
图 2 原子力显微镜和AFM力谱示意图[23-24] Figure 2 Schematic illustration of atomic force microscopy (AFM) andAFM force spectroscopy[23-24]. A: Main components of AFM-microprobe, piezoelectric scanner, detection system and control system; B: The experimental force-distance curve is shown with blue. Red line is the fit of the experimental data withWorm-Like Chain (WLC) model. Cartoon (1) shows the approach stage at which tethered molecules are far from each other. Cartoon (2) illustrates the complex formation, which occurs at small tip-sample distance. The retraction of the tip as shown in cartoon (3) is accompanied by stretching of the tether and this entropy driven process is approximated with the WLC fit (red curve). After full stretching of the tether, the rupture occurs at the position indicated with a red arrow. After the complex rupture, there is no interaction between the molecules as evidenced by no change of the force on the tip sample distance (horizontal red line). |
自从Lee等[25]利用链霉亲和素-生物素作用体系证实了AFM-SMFS在研究单对分子间相互作用力的可行性后,得到了广泛的发展和应用[26-28]。虽然AFM-SMFS具有超高的时空分辨率和超敏锐的力学灵敏度[29-30],但其应用受到了针尖和衬底的单分子修饰以及无法实现高通量平行测定的限制。今后AFM-SMFS的发展将会突破低通量的限制[31],这样研究人员就可以从单分子水平上评估靶向药物与特异性结合位点的作用强度,为药物筛选提供新工具。
2 AFM-SMFS在G四链体及其与小分子相互作用中的应用G四链体结构存在于染色体端粒序列和许多基因的启动子区域,包括癌基因启动子[32]。在致癌基因启动子区域内形成的分子内G四链体结构可以有效地抑制致癌基因的转录和表达[33-37]。因此,合理设计小分子配体以选择性地相互作用,稳定或切割G四链体结构是开发对癌细胞具有选择性毒性的有效抗癌药物的有希望的策略[38-40],所以许多能稳定该结构的小分子化合物被设计和合成[41-43]。研究小分子配体与G四链体间的相互作用对有效药物筛选十分必要。鉴于AFM单分子力谱在测定分子间相互作用力方面的优势,本文综述了AFM-SMFS在研究G四链体及其小分子配体相互作用中的应用,为人们利用单分子力谱技术开发筛选靶向G四链体抗癌药物提供参考。
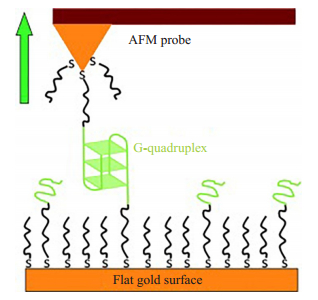
2.1 AFM-SMFS首次用于研究分子间G四链体系统2009年,Kumar Sinniah小组首次把基于原子力显微镜的单分子力谱技术用于研究双分子G四链体系统。如图 3所示[44],他们用单分子力谱研究了3G和4G四链体的展开动力学和热力学性质。研究表明在富含G碱基的重复序列中,G碱基的个数对G四链体的热力学稳定性以及展开速率有很大影响,而存在3个G碱基和4个G碱基的重复序列所形成的G四链体在抵抗机械力诱导G四链体结构展开的能力是非常相似的。此外,首次揭示了G-四链体的机械强度取决于K+浓度。此研究表明AFM SM-FS在探测G-四联体系统的结构,动力学,机械和热力学性质方面是非常有价值的。
|
图 3 AFM-SMFS研究分子间G四链体的形成[44] Figure 3 Schematic diagram of the AFM SMFS study of G-quadruplex formation[44]. Gold-coated AFM probe and surface are coated with a SAM of a thiolated 3G or 4G DNA, where each contains two G-rich domains (colored in green) and an A12 spacer sequence (black), diluted by the spacer-DNA (A10) at 1:5 molar ratio. An inter-surface quadruplex is formed between the probe and surface. |
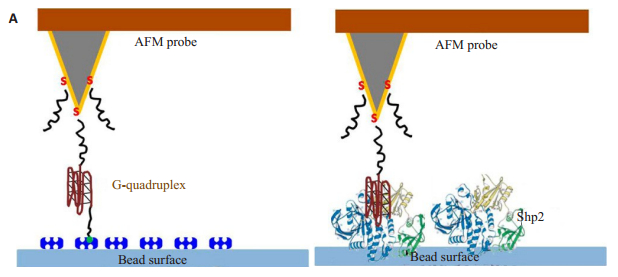
关于G-四链体构象稳定性的研究主要是将单分子G-四联体分为两部分,然后分别固定到针尖尖端和衬底表面上,如此测量的断裂力不能反映分子G-四链体真正的分子内相互作用,因为原始G-四联体结构可能不能够通过固定在针尖末端和衬底表面的两个单链部分的重组恢复。2012年毛秉伟小组报道了一种改进的G-四链体双标记方法,即在富G序列的两端分别修饰生物素和巯基,此方法允许对G四链体分子内相互作用及其与蛋白质的分子间相互作用进行研究如图 4所示。Shp2是蛋白质酪氨酸磷酸酶(PTPs)的成员,对细胞过程中的很多信号事件有调节作用。Shp2功能障碍与癌症,代谢综合征和自身免疫性疾病密切相关,而能与Shp2结合的活性适配体构象中存在一个分子内G四链体结构[45]。这项工作中开发和演示的方法为基于AFM-SMFS技术研究其他单分子G四链体结构的稳定性及其与靶蛋白的相互作用提供了新的思路。
|
图 4 AFM-SMFS研究G四链体分子内相互作用及其与蛋白质的分子间相互作用[45] Figure 4 Schematic illustration of AFM-based single molecule force spectroscopic study of intramolecular interaction in G-quadruplex DNAand its intermolecular interaction with protein[45]. A: Intramolecular interaction of G quadruplex; B: Intermolecular interaction between G quadruplex and Shp2 protein. The G-quadruplex DNAhaving 4 G-rich domains (brown) is dual-labeled (5'-biotin, 3'- thiol) through two linker DNA molecules (T20 sequence, black). Gold-coated AFM probes are modified with a SAM of the dual-labeled G-quadruplex DNAthrough the thiolated 3′ end in dilution by thiolated spacer-DNA(also T20 sequence) at 1:5 molar ratio. |
2006年ItamarWillner小组使用AFM-SMFS研究了G四链体与凝血酶间的相互作用[46]。作者使用金硫键把G四链体和凝血酶分别修饰在镀金探针和金衬底上如图 5所示,用AFM-SMFS研究两者间的相互作用力。结果发现,结合到凝血酶上的G四链体的断裂力在4.45 pN左右,这个非常低的断裂力是由于G四链体中的氢键断裂造成,同时氢键的断裂破坏了G四链体的稳定性,最终导致了G四链体与凝血酶复合体的解离。
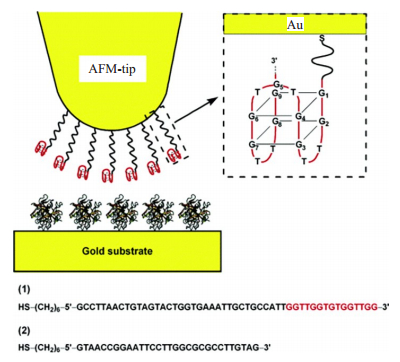
|
图 5 研究凝血酶与其适配体之间相互作用系统的示意图[46] Figure 5 Schematic illustration of the system for studying force interactions between thrombin and its aptamer[46]. The Au-coated tip is modified with the thiolated aptamer, and the gold slide is modified with thrombin. The in-set shows the G-quadruplex structure for the aptamer 1 in binding buffer solution. |
适配体(aptamer)是一段能以极高亲和力和特异性与靶分子结合的寡核苷酸序列。在过去的几十年中,已经报道了两种凝血酶适配体(15apt和27apt)。1992年,Bock等研究人员就发现了一种寡核苷酸序列(GTTGGTGTGGTTGG)适配体能抑制凝块结合的凝血酶并减少动脉血栓的形成[47]。但在凝血酶与适配体间相互作用时,其结合强度和稳定性受到了空间位阻的影响。2011年Andreas Ebner小组发表了一篇有关AFM-SMFS表征单价和二价G四链体适配体与凝血酶结合的文章[48]。在研究中,他们用立体改进的方法开发了可用于高亲和力结合的凝血酶单价适配体BFA和二价适配体BFF,如图 6A所示。实验中,他们通过改变力距循环中的提拉速度,所形成的凝血酶-适配体复合物就会在不同的加载速度下断裂。结果显示,与BFF相比,BFA适配体在所研究的加载速率下显示出更高的结合力和更低的解离速率常数,这充分证明了适配体与凝血酶的结合会受到空间位阻的影响。而对于二价适配体来说,其二价结合效率明显高于一价结合效率,证明了两个单价适配体能形成二价适配体复合物的潜力。该项研究有助于增强AFM-SMFS技术在筛选G四链体高效适配体中的应用潜力。同年,方晓红课题组也发表了基于AFM-SMFS探究二价适配体与凝血酶相互作用的文章[49]。他们使用一个含八亚磷酸酰胺(Bi-8S)的linker把15aptmer和27aptmer连接起来构成了一个二价适配体,如图 6B所示。在SMFS中,他们观察到二价适配体与凝血酶的解绑是按顺序进行的。此外,动力学光谱数据显示27apt与凝血酶的结合在很大程度上不受Bi-8S内八个间隔亚磷酰胺的影响,八间隔亚磷酰胺反而稳定了15apt与凝血酶的结合。
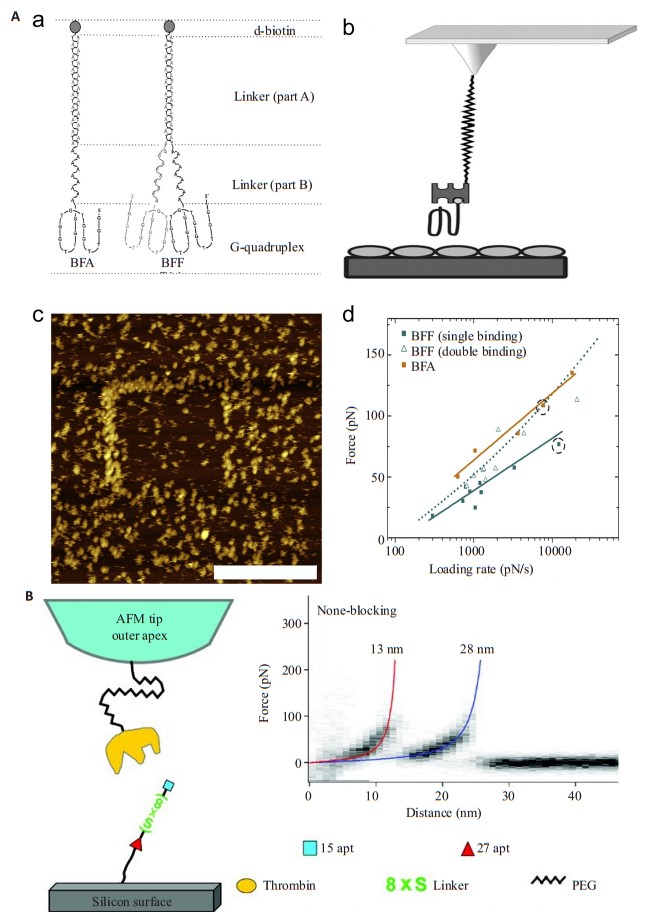
|
图 6 单价和二价适配体与凝血酶相互作用[48-49] Figure 6 A monovalent and bivalent aptamer interacting with thrombin[48-49]. A: (a) Schematic representation of thrombin aptamers (BFAand BFF). (b) The heterobifunctional cross-linker NHS-PEG18-acetal (or NHS-PEG18- aldehyde) was covalently bound to the aminopropyltrioxysilane-coated AFM tip through its NHS-ester function. Streptavidin was attached to the free aldehyde residue (after deprotection of the acetal group). Finally, the biotinylated thrombin-aptamer BFA or BFF was coupled to streptavidin. (c) A representative image of a surface with lower thrombin molecule density that was used to evaluate BFF binding characteristics; scale bar of 300 nm. (d) Comparison of the dynamic force spectra of BFA and BFF. The most probable unbinding forces were plotted against the logarithm of the loading rate.; B: α-thrombinfunctionalized AFM tip and the aptamer Bi-8S is covalently bound to the silicon surface. Superimposition of force-distance (F-D) curves recorded at a pulling speed of 300 nm/s in buffer solution. Every reproducibly occurring force peakwas then fitted using the WLC model (Materials and Methods) to determine the rupture force as well as the contour length of the thrombin-aptamer binding. The contour length is labeled at the end of eachWLCfit, approximately at 13 nm (red) and 28 nm (blue). |
AFM-SMFS研究适配体与凝血酶过程中,凝血酶通过化学试剂固定于衬底上,改变了凝血酶原有的结构和活性,使其与适配体的相互作用受到影响。为解决此问题,2013年何品刚研究组报道了一种不需要固定凝血酶,用AFM-SMFS技术直接研究凝血酶与两种适配体(15apt和27apt)之间特异性相互作用的新方法[50]如图 7所示。实验中他们将两种配体分别修饰在探针和硅衬底上,液相条件下,将修饰的探针慢慢接触衬底,由于时间设定在10 s左右,所以溶液中游离的凝血酶有足够的时间和两个配体形成配体-凝血酶-配体复合物,然后提拉探针测得配体与凝血酶间的相互作用力。他们还通过测定不同加载速率下适配体和凝血酶间的断裂力,得到了适配体/凝血酶的解离动力学。结果表明,尽管15apt和27apt在缓冲溶液中结构不同,15apt与凝血酶间的断裂力大于27apt与凝血酶间的断裂力,但是凝血酶与其两个适配体的潜在相互作用机制是相似的。这是第一次不通过接头将凝血酶固定在尖端或底物上,使用AFM直接研究凝血酶与其两种适配体之间的特异性相互作用,此研究中虽然保留了凝血酶在溶液中的结构和活性,但由于没有通过化学方法固定凝血酶,低浓度的凝血酶游离在缓冲液中,通过随机碰撞与适配体结合,从而导致了适配体与凝血酶的结合概率非常低。
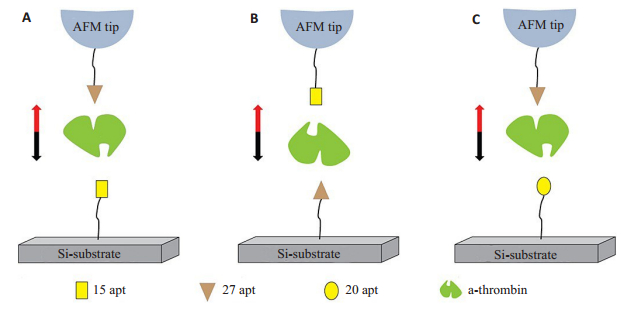
|
图 7 适配体-凝血酶-适配体复合物的机械解绑示意图[50] Figure 7 Schematic illustration for mechanical unbinding of aptamer- thrombin- aptamer complex[50]. Aptamers are covalently bound to the AFM tip and the Si substrate; thrombin is coupled to the immobilized aptamers. Single molecule force spectroscopy was performed under various modified conditions in buffer solutions. A: 15apt was coupled to the Si substrate, and 27apt was immobilized on the AFM tip; B: 27apt was coupled to the Si substrate, and 15apt was immobilized on the AFM tip; C: 20aptwas coupled to the Si substrate, and 27aptwas immobilized on the AFM tip. |
鉴于G四链体是癌症治疗的非常有吸引力的靶标,这可能使得AFM-SMFS成为筛选潜在癌症治疗试剂的新工具。尽管先前已经报道了基于AFM-SMFS的关于G-四链体与靶蛋白的分子内和分子间相互作用以及G-四链体DNA本身的稳定性的研究,但是还没有关于通过端粒酶抑制剂稳定G-四链体DNA的任何报道。2011年Chikashi Nakamura小组利用AFM-SMFS通过力学分析发现大环六唑(macrocyclic hexazole 6OTD是一种端粒酶抑制素的衍生物)单体及其二聚体与端粒DNA结合模式存在差异[51]。研究发现6OTD二聚体与G四链体间的结合是呈三明治结构,如图 8所示,其结合力比6OTD单体与G四链体间的结合力强很多。该研究使用6OTD功能化的AFM针尖提拉固定在固体基质上的染色体的端粒使表面上的染色体延长,这将有助于发展一种新的基于AFM的染色体操作技术。
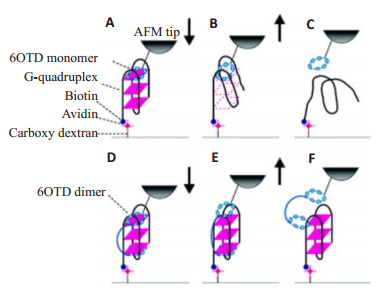
|
图 8 6OTD单体(A~C)和二聚体(D~F)与G-四联体的结合和解结合模式的示意图[51] Figure 8 Schematic model of the assumed binding and unbinding modes of the 6OTD monomer (A to C) and dimer (D to F) to and from the G-quadruplex[51]. The 6OTD monomer immobilized on an AFM tip binds one terminal of a guanine tetrad in the Gquadruplex when the monomer-immobilized AFM tip was approached to the G-quadruplex (A). When the AFM tip was retracted from the surface, the G-quadruplex was pulled and the structure was distorted (B) and then destroyed (C). The dimer on theAFM tip binds the outermost guanine tetrads at both termini of the G-quadruplex in a sandwiching manner (D). When the dimerimmobilized AFM tip was retracted from the G-quadrupleximmobilized surface, firstly the monomer unit immobilized nearby on theAFM tip might separate from the G-quadruplex. The structure of the G-quadruplex was retained because the other monomer unit maintained the structure from the bottom (E). When theAFM tip was further retracted, the dimer separated from the G-quadruplex without deforming the structure (F). |
在众多的端粒酶抑制剂中,TMpyP[5, 10, 15, 20-tetrakis(N-methylpyridinium-4-yl)porphyrin tetrakis(p-toluenesulfonate)]与端粒间具有静电作用及夹层效应而表现出非常强的端粒酶抑制性。2014年Keiichi Kimura小组采用AFM-SMFS研究了G四链体与端粒酶抑制剂(TMPyP)间的相互作用。如图 9所示[52],他们采用末端生物素化的G四链体修饰探针,用链酶素抗生物素修饰金衬底,探针和衬底间因为生物素和链酶素抗生物素间相互作用形成一个桥连,当提拉探针的时候,G四链体DNA会被完全拉伸。力曲线中突变峰力的大小和突变峰的轮廓长度可以反应DNA的拉伸,使用蠕虫状链模型分析所选择的曲线,并且使用一个拟合参数--持久长度(Lp)作为G四链体结构稳定化的指标。在可形成G-四链体结构的实验条件下,通过添加TMPyP,Lp值显著增加。此研究基于AFM-SMFS技术开发了利用轮廓长度这一参数半定量研究G四链体结构稳定性的新方法。由于端粒DNA持续长度的增加与G-四链体结构的稳定性密切相关,通过此SMFS研究获得的持久长度可作为评估端粒酶抑制剂特异性强度的重要指标。此外,这项工作中的方法也可用于评估其他端粒酶抑制剂如溴化乙锭和对二甲苯-双(溴化吡啶盐)对G-四链体DNA的稳定性。
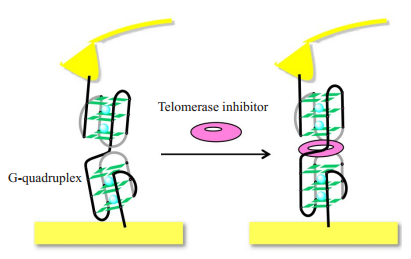
|
图 9 AFM-SMFS研究G四链体与端粒酶抑制剂间相互作用示意图[52] Figure 9 Schematic diagram of AFM- SMFS study the interaction between G quadruplex and telomerase inhibitor[52]. The telomere DNA is thiolated at its 5' position end for covalently anchoring to a gold-coated AFM tip and biotinylated at the 3' position end for binding with the SA-modified substrate. The interaction between the G quadruplex and the telomerase inhibitor can be measured by comparing the rupture forces before and after the addition of the telomerase inhibitor. |
目前以核酸为靶点的抗癌药物应该是最具发展前景的抗癌药物,因为此类药物可以从根本上抑制癌细胞的产生、发展和增殖。G-四链体DNA作为有重要生物功能的特殊二级结构,此结构的稳定性在正常体细胞中可以在细胞分裂过程中保护细胞防止过早凋亡,在癌细胞中又可以抑制端粒酶的活性,防止癌细胞永生化。因此以其为靶点的抗癌药物的设计具有很高的合理性与优越性。而且越来越多的能够诱导富含G碱基的DNA序列形成G四链体结构并使之稳定的小分子配体被开发出来,这导致筛选高效的小分子配体成为一项重要的研究工作。随着基于原子力显微镜的单分子力谱技术的不断改进和单分子修饰水平的不断完善,以及AFM-SMFS在G四链体结构稳定性及其与小分子配体间相互作用的研究有望使得AFM-SMFS成为筛选潜在癌症治疗试剂的新工具的。相信有朝一日,我们可以通过改进现有的单分子力谱技术,实现对G四链体以及药物间的相互作用的高通量测定,提高检测效率,为筛选出高效的治疗药物做出贡献。
| [1] |
Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology[J].
Nat Rev Mol Cell Biol, 2014, 15(7): 482.
DOI: 10.1038/nrm3823. |
| [2] |
Rivera T, Haggblom C, Cosconati S, et al. A balance between elongation and trimming regulates telomere stability in stem cells[J].
Nat Struct Mol Biol, 2017, 24(1): 30-9.
DOI: 10.1038/nsmb.3335. |
| [3] |
Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA Gquadruplexes in the human genome: detection, functions and therapeutic potential[J].
Nat Rev Mol Cell Biol, 2017, 18(5): 279-84.
DOI: 10.1038/nrm.2017.3. |
| [4] |
Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures[J].
Nat Rev Genet, 2012, 13(11): 770-80.
DOI: 10.1038/nrg3296. |
| [5] |
Capra JA, Paeschke K, Singh M, et al. G-quadruplex DNAsequences are evolutionarily conserved and associated with distinct genomic features in saccharomyces cerevisiae[J].
PLoS Comput Biol, 2010, 6(7): e1000861.
DOI: 10.1371/journal.pcbi.1000861. |
| [6] |
Shay JW. Role of telomeres and telomerase in aging and cancer[J].
Cancer Discov, 2016, 6(6): 584-93.
DOI: 10.1158/2159-8290.CD-16-0062. |
| [7] |
Jafri MA, Ansari SA, Alqahtani MH. Roles of telomeres and telomerase in cancer, and advances in telomerase- targeted therapies[J].
Genome Med, 2016, 8(1): 69.
DOI: 10.1186/s13073-016-0324-x. |
| [8] |
Chen BJ, Wu YL, Tanaka Y, et al. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics[J].
Int J Biol Sci, 2014, 10(10): 1084-96.
DOI: 10.7150/ijbs.10190. |
| [9] |
Das RN, Chevret E, Desplat V, et al. Design, synthesis and biological evaluation of new substituted diquinolinyl-pyridine ligands as anticancer agents by targeting G-quadruplex[J].
Molecules, 2018, 23(1): 81.
|
| [10] |
Kim MY, Duan WH, Gleason-Guzman M, et al. Design, synthesis, and biological evaluation of a series of fluoroquinoanthroxazines with contrasting dual mechanisms of action against topoisomerase Ⅱ and G-quadruplexes[J].
J Med Chem, 2003, 46(4): 571-83.
DOI: 10.1021/jm0203377. |
| [11] |
Tripathi S, Barthwal R. NMR based structure reveals groove binding of mitoxantrone to two sites of[d-(TTAGGGT)]4 having human telomeric DNA sequence leading to thermal stabilization of Gquadruplex[J].
Int J Biol Macromol, 2018, 111: 326-41.
DOI: 10.1016/j.ijbiomac.2017.12.134. |
| [12] |
Perez-Arnaiz C, Busto N, Santolaya JA, et al. Kinetic evidence for interaction of TMPyP4 with two different G-quadruplex conformations of human telomeric DNA[J].
Biochim Biophy Acta, 2018, 1862(3): 522-31.
DOI: 10.1016/j.bbagen.2017.10.020. |
| [13] |
Romera C, Bombarde O, Bonnet R, et al. Improvement of porphyrins for G-quadruplex DNAtargeting[J].
Biochimie, 2011, 93(8): 1310-7.
DOI: 10.1016/j.biochi.2011.06.008. |
| [14] |
Franceschin M, Lombardo CM, Pascucci E, et al. The number and distances of positive charges of polyamine side chains in a series of perylene diimides significantly influence their ability to induce Gquadruplex structures and inhibit human telomerase[J].
Bioorg Med Chem, 2008, 16(5): 2292-304.
DOI: 10.1016/j.bmc.2007.11.065. |
| [15] |
Campbell NH, Abd Karim NH, Parkinson GN, et al. Molecular basis of structure-activity relationships between salphen metal complexes and human telomeric DNA quadruplexes[J].
J Med Chem, 2012, 55(1): 209-22.
DOI: 10.1021/jm201140v. |
| [16] |
Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to Gquadruplex ligands[J].
Org Biomol Chem, 2008, 6(4): 627-36.
DOI: 10.1039/B714772B. |
| [17] |
Murat P, Singh Y, Defrancq E. Methods for investigating Gquadruplex DNA/ligand interactions[J].
Chem Soc Rev, 2011, 40(11): 5293-307.
DOI: 10.1039/c1cs15117g. |
| [18] |
Micco M, Collie GW, Dale AG, et al. Structure- Based design and evaluation of naphthalene diimide G-Quadruplex ligands as telomere targeting agents in pancreatic cancer cells[J].
J Med Chem, 2013, 56(7): 2959-74.
DOI: 10.1021/jm301899y. |
| [19] |
Müller DJ, Dufrene YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology[J].
Nat Nanotechnol, 2008, 3(5): 261-9.
DOI: 10.1038/nnano.2008.100. |
| [20] |
Neuman KC, Lionnet T, Allemand JF. Single- molecule micromanipulation techniques[J].
Annu Rev Mater Res, 2007, 37: 33-67.
DOI: 10.1146/annurev.matsci.37.052506.084336. |
| [21] |
Miller H, Zhou ZK, Shepherd J, et al. Single-molecule techniques in biophysics: a review of the progress in methods and applications[J].
Rep Prog Phys, 2018, 81(2): 024601.
DOI: 10.1088/1361-6633/aa8a02. |
| [22] |
于婵婵, 姚立. 生物单分子力谱技术的研究进展[J].
化学通报, 2016, 79(4): 292-8.
|
| [23] |
Zeng C, Vitale-Sullivan C, Ma X. In situ atomic force microscopy studies on nucleation and self-assembly of biogenic and bio-inspired materials[J].
Minerals, 2017, 7(9): 158.
DOI: 10.3390/min7090158. |
| [24] |
Lyubchenko YL. Amyloid misfolding, aggregation, and the early onset of protein deposition diseases: insights from AFM experiments and computational analyses[J].
AIMS Mol Sci, 2015, 2(3): 190-210.
DOI: 10.3934/molsci.2015.3.190. |
| [25] |
Gu LE, Kidwelland DA, Colton RJ. Sensing discrete streptavidinbiotin interactions with atomic force microscopy[J].
Langmuir, 1994, 10(2): 354-7.
DOI: 10.1021/la00014a003. |
| [26] |
Ott W, Jobst MA, Schoeler C, et al. Single-molecule force spectroscopy on polyproteins and receptor-ligand complexes: The current toolbox[J].
J Struct Biol, 2017, 197(1): 3-12.
DOI: 10.1016/j.jsb.2016.02.011. |
| [27] |
Li Q, Zhang T, Pan YA, et al. AFM-based force spectroscopy for bioimaging and biosensing[J].
RSCAdv, 2016, 6(16): 12893-912.
|
| [28] |
Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy[J].
Nat Methods, 2008, 5(6): 491-505.
DOI: 10.1038/nmeth.1218. |
| [29] |
Edwards DT, Perkins TT. Optimizing force spectroscopy by modifying commercial cantilevers: improved stability, precision, and temporal resolution[J].
J Struct Biol, 2017, 197(1): 13-25.
DOI: 10.1016/j.jsb.2016.01.009. |
| [30] |
张文科, 王驰, 张希. 单分子力谱[J].
科学通报, 2003, 48(11): 1113-26.
DOI: 10.3321/j.issn:0023-074X.2003.11.002. |
| [31] |
Alsteens D, Tay S, Mueller DJ. Toward high-throughput biomechanical phenotyping of single molecules[J].
Nat Methods, 2015, 12(1): 45-6.
DOI: 10.1038/nmeth.3216. |
| [32] |
Cao Q, Li Y, Freisinger E, et al. G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs[J].
Inogran Chem Front, 2017, 4(1): 10-32.
DOI: 10.1039/C6QI00300A. |
| [33] |
Neidle S. Quadruplex nucleic acids as novel therapeutic targets[J].
J Med Chem, 2016, 59(13): 5987-6011.
DOI: 10.1021/acs.jmedchem.5b01835. |
| [34] |
Zhang S, Wu Y, Zhang W. G-quadruplex structures and their interaction diversity with ligands[J].
Chem Med Chem, 2014, 9(5): 899-911.
DOI: 10.1002/cmdc.v9.5. |
| [35] |
Kaulage MH, Maji B, Pasadi S, et al. Targeting G-quadruplex DNA structures in the telomere and oncogene[romoter regions by benzimidazole-carbazole ligands[J].
Eur J Med Chem, 2018, 148: 178-94.
DOI: 10.1016/j.ejmech.2018.01.091. |
| [36] |
Diveshkumar KV, Sakrikar S, Rosu FA, et al. Specific stabilization of c-MYC and c-KIT G-Quadruplex DNA structures by indolylmethyleneindanone scaffolds[J].
Biochemistry, 2016, 55(25): 3571-85.
DOI: 10.1021/acs.biochem.6b00120. |
| [37] |
Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription[J].
Nucleic Acids Res, 2006, 34(9): 2536-49.
DOI: 10.1093/nar/gkl286. |
| [38] |
Neidle S, Parkinson GN. Quadruplex DNA crystal structures and drug design[J].
Biochimie, 2008, 90(8): 1184-96.
DOI: 10.1016/j.biochi.2008.03.003. |
| [39] |
Islam MK, Jackson PJ, Rahman KM. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies[J].
Future Med Chem, 2016, 8(11): 1259-90.
DOI: 10.4155/fmc-2015-0017. |
| [40] |
Onel B, Lin C, Yang DZ. DNA G-quadruplex and its potential as anticancer drug target[J].
Sci China Chem, 2014, 57(12): 1605-14.
DOI: 10.1007/s11426-014-5235-3. |
| [41] |
Maji B, Bhattacharya S. Advances in the molecular design of potential anticancer agents via targeting of human telomeric DNA[J].
Chem Commun, 2014, 50(49): 6422-38.
DOI: 10.1039/C4CC00611A. |
| [42] |
Ilyinsky NS, Varizhuk AM, Beniaminov AD, et al. G-quadruplex ligands: Mechanisms of anticancer action and target binding[J].
Mol Biol, 2014, 48(6): 778-94.
DOI: 10.1134/S0026893314060077. |
| [43] |
郑小辉, 穆舸, 谭彩萍, 等. G-四链体DNA稳定剂的研究进展[J].
中国科学:化学, 2014, 44(4): 484-94.
|
| [44] |
Lynch S, Baker H, Byker SG, et al. Single molecule force spectroscopy on G-quadruplex DNA[J].
Chemistry, 2009, 15(33): 8113-6.
DOI: 10.1002/chem.v15:33. |
| [45] |
Zhao XQ, Wu J, Liang JH, et al. Single-molecule force spectroscopic studies on intra- and intermolecular interactions of G-quadruplex aptamer with target Shp2 protein[J].
J Phys Chem B, 2012, 116(37): 11397-404.
DOI: 10.1021/jp303518b. |
| [46] |
Basnar B, Elnathan R, Willner I. Following aptamer-thrombin binding by force measurements[J].
Anal Chem, 2006, 78(11): 3638-42.
DOI: 10.1021/ac052289e. |
| [47] |
Bock LC, Griffin LC, Latham JA, et al. Selection of single-stranded DNA molecules that bind and inhibit human thrombin[J].
Nature, 1992, 355(6360): 564-6.
DOI: 10.1038/355564a0. |
| [48] |
Neundlinger I, Poturnayova A, Karpisova IA, et al. Characterization of enhanced monovalent and bivalent thrombin DNA aptamer binding using single molecule force spectroscopy[J].
Biophys J, 2011, 101(7): 1781-7.
DOI: 10.1016/j.bpj.2011.07.054. |
| [49] |
Ge L, Jin G, Fang XH. Investigation of the interaction between a bivalent aptamer and thrombin by AFM[J].
Langmuir, 2012, 28(1): 707-13.
DOI: 10.1021/la203954x. |
| [50] |
Jiao F, Fan HJ, Yang GD, et al. Directly investigating the interaction between aptamers and thrombin by atomic force microscopy[J].
J Mol Recognit, 2013, 26(12): 672-8.
DOI: 10.1002/jmr.2312. |
| [51] |
Amemiya Y, Furunaga Y, Iida K, et al. Analysis of the unbinding force between telomestatin derivatives and human telomeric G-quadruplex by atomic force microscopy[J].
Chem Commun, 2011, 47(26): 7485-7.
DOI: 10.1039/c0cc05781a. |
| [52] |
Funayama R, Nakahara Y, Kado S, et al. A single-molecule forcespectroscopic study on stabilization of G-quadruplex DNA by a telomerase inhibitor[J].
Analyst, 2014, 139(16): 4037-43.
DOI: 10.1039/C4AN00439F. |
 2018, Vol. 38
2018, Vol. 38

