2. Institute of Cancer Research, Southern Medical University, Guangzhou 510515, China;
3. Henan Provincial People's Hospital, Henan Eye Institute, Henan Eye Hospital, Zhengzhou 450003, China
2. 南方医科大学癌症研究所,广东 广州 510515;
3. 河南省人民医院,河南省立眼科研究所,河南 郑州 450003
Uveitis represents a spectrum of common intraocular inflammatory diseases induced by infections or noninfectious factors [1], and is a major cause of visual impairment worldwide[2, 3]. Among all the cases of uveitis, infectious uveitis accounts for 30%-50% in developing countries[4]. The pathogens of infectious uveitis includes bacteria, fungi, viruses, and parasites, and the treatment remains clinically challenging [5]. As a well-studied model for acute infectious uveitis in human, endotoxininduced uveitis (EIU) is induced by intravenous or intravitreal injection with lipopolysaccharide (LPS)[6, 7]to produce such prominent features as iris hyperemia, leukocyte infiltration and protein leakage, normally reaching their peak levels at 24 h after LPS injection[7, 8]. LPS stimulation can evoke the production of numerous inflammatory cytokines including interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), monocyte chemoattractant protein-1 (MCP-1) and intercellular adhesion molecule-1 (ICAM-1), and blocking the activity of these cytokines reduces intraocular inflammatory response [8-12], indicating the essential role of inflammatory cytokines in the pathologies of LPS-induced uveitis.
Corticosteroids, non-steroidal anti-inflammatory drugs (NSAIDs) and immunosuppressive agents have been used to treat uveitis for several decades [13, 14]. Corticosteroids currently remain the primary choice for initial treatment of uveitis. Dexamethasone (DEX) is used for treatment of a wide variety of diseases including anaphylactic diseases, immune diseases and inflammatory diseases, and is also a conventional drug prescribed for various inflammatory ocular diseases due to its strong anti-inflammatory effects[15]. Nevertheless, long-term use of DEX has been shown to lead to a series of systemic and topical side effects[15, 16]. So far studies of DEX focus primarily on specific pathways and cytokines, and an overall picture of the mechanisms for its beneficial or adverse effects is not available; integrating these molecular mechanisms into a general map provides better insights into the beneficial and side effects of DEX in uveitis treatment.
RNA sequencing (RNA-seq) is a novel approach for both mapping and quantifying transcriptomes based on high-throughput sequencing technology [17]. The quantitative RNA-seq platforms provide unbiased profiles, a snapshot of the transcriptome of cells, and the ability to identify unknown transcribed regions, and can be extremely precise if a sufficient level of coverage is obtained[18, 19]. Hence, RNA-seq can be instrumental for revealing comprehensive yet specific changes in the transcriptome. Gene expression profiling using nextgeneration sequencing (NGS)-based RNA-seq technology allows for more thorough understanding of the molecular events caused by DEX. In the present study, we aimed to obtain the transcriptome profile of the retinas from DEX-treated mice with EIU using NGS-based RNA-seq technology. Our findings may add to the current knowledge base and shed light on the mechanisms of the protective roles of DEX against ocular inflammation.
MATERIALS AND METHODS Ethics statementAll the experimental procedures were carried out in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. The protocols were approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. In all the procedures the mice were appropriately anesthetized and efforts were made to relieve the discomforts and stress[20].
Animal grouping and treatmentsFemale BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME) and housed in the Animal Care Service of Chongqing Medical University with a 12 h:12 h light/dark cycle. The mice at the age of 6 to 8 weeks were randomly assigned into 3 groups: normal control group without any treatment, LPS group with intravitreal injection of 125 ng/μL LPS (SigmaAldrich, St. Louis, MO, USA) in one eye to induce EIU, and DEX + LPS group with LPS injection followed by treatment with topical application of 0.1% DEX eyedrops every 4 h for 24 h.
Clinical Assessment of EIUThe inflammatory signs in the anterior chamber were assessed using a digital slit-lamp microscope at 24 h after LPS injection. To ensure the accuracy of assessment, the clinical severity of ocular inflammation was classified by two independent observers in line with the clinical scoring criteria as previously described[21].
HistopathologyFor histopathological assessment, the eyeballs of the mice were enucleated at 24 h after LPS injection and fixed in a mixture consisting of acetone (50 mL), glacial acetic acid (2 mL), formaldehyde (4 mL), and distilled water (3 mL) at 4 ℃ for 24 h. After embedment in paraffin, the samples were serially sliced (4 to 6 μm) through the pupillary-optic nerve axis and stained with hematoxylin and eosin (HE). Histopathologic evaluation of the eyeballs was carried out by assessing the number of infiltrating cells in the posterior and anterior chambers under an optical microscope.
Real-time PCR analysisThe total RNA of neural retina was extracted with Trizol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. For quantitative PCR, cDNA was synthesized using the PrimeScript RT reagent kit (Takara Biotechnology, Dalian, China). PCR amplification was performed using a real-time PCR system (ABI 7500 system, Applied Biosystems, Foster City, CA, USA) with SYBR Green (Vazyme Biotech Co., Ltd, Nanjing, China). The thermal cycles were carried out with initial denaturation at 95 ℃ for 10 min, followed by 40 cycles of 95 ℃ for 15 s and 60 ℃ for 1 mi. To determine the mRNA levels, GAPDH was used as the endogenous reference gene. All the samples were prepared in duplicate and the average Ct values were used for quantification. Relative quantification was achieved using the comparative 2-ΔΔCt method. The control group was used as the calibrator to calculate the relative fold change of the mRNA of the target gene X in the experimental group (EG): ΔΔCt=(Ct.XEG-Ct.GAPDHEG)/ (Ct.Xcontrol-Ct.GAPDHcontrol)[21]. The sequences of primers and GenBank accession numbers of the target genes are listed in Tab. 1.
| Table 1 Sequences of the primers for amplifying the genes using real-time RT-PCR |
RNA-seq and data analysis were conducted by Novogene (Beijing, China). Total RNA of mouse retinas were extracted using TRIzol reagent (Invitrogen) and purified with RNase-free DNase I (New England Biolabs, MA, USA) following the manufacturer's protocol. Containing 3 μg RNA for each sample, the sequencing libraries were constructed using NEB Next® UltraTM RNA Library Prep Kit for Illumina (New England Biolabs, MA, USA). The mRNA was isolated from the total RNA using Dynabeads oligo (dT) (Invitrogen Dynal) and the double-strand cDNAs were synthesized using Superscript Ⅱ reverse transcriptase (Invitrogen). The cDNAs were then fragmented by nebulization and the RNA sequencing library was established according to standard Illumina protocol. The library quality was verified using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) followed by sequencing with the Illumina Hiseq 2500, and 100 bp/50 bp singleend reads were generated. The single-end clean reads were aligned to the genome by the split read aligner TopHat (v2.0.12) with the mismatches set at 2 under default parameter settings[20].
Analysis of differentially expressed genes (DEGs)The DEGs were analyzed using the DESeq R package (version 1.10.1), which has been a routine statistical approach to determine the digital differential expressions of genes using a model assuming negative binomial distribution of the data. To control the false positive rate, the Benjamini and Hochberg's approach was used for adjusting the P values, and a P value less than 0.05 was considered to indicate a significant difference.
Functional enrichment analysisGene Ontology (GO) enrichment analysis was performed to functionally annotate the DEGs using the GOseq R package. The GO ontologies include 3 key biological domains including the biological process, cellular component and molecular function[22]. An adjusted P value < 0.05 was considered to indicate a significantly enriched GO term. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was carried out to annotate the pathway enrichment. The KEGG pathway enrichment of the DEGs was conducted using KOBAS software, and a significantly enriched KEGG pathway was defined for an adjusted P value < 0.05.
Statistical analysisAll the data of the continuous variables are presented as Mean±SE. The clinical scores and data of real-time PCR analysis were analyzed with the GraphPad Prism 5 software. The clinical scores were analyzed using Kruskal-Wallis test and the data of the inflammatory cytokines were analyzed using one-way ANOVA followed by Bonferroni correction. The validation data were analyzed using unpaired t-test. A P value less than 0.05 was considered to indicate a significant difference.
RESULTS DEX alleviates inflammation in mice with EIUCompared with the normal control group, the mice with intravitreal injection of LPS exhibited obvious iris hyperemia, exudation into the anterior chamber, and hypopyon. Treatment with DEX markedly ameliorated these inflammatory signs (Fig. 1A). Assessment of the clinical scores indicated that DEX significantly reduced the severity of ocular inflammation induced by LPS group (Fig. 1B).
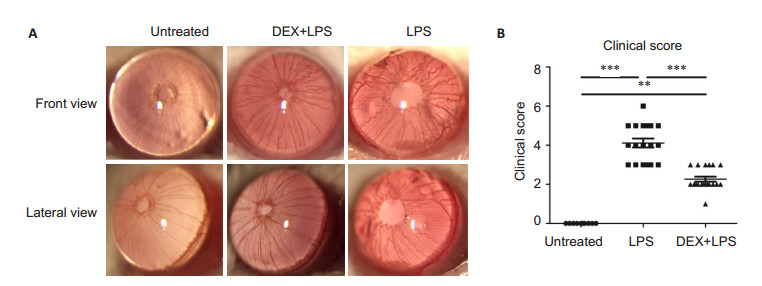
|
Figure 1 Inflammation in the anterior chamber of the mice in different groups. A: Representative images showing inflammations in the anterior chamber of the mice in the untreated (normal control), LPS and DEX+LPS groups; B: Clinical scores assessed at 24 h by two independent observers (n=10, Mean±SE). **P < 0.01, ***P < 0.001 by Kruskal-Wallis test |
Compared with the normal control group, intravitreal injection of LPS resulted in obvious inflammation in the vitreous cavity and the ciliary body, where numerous infiltrating inflammatory cells were observed 24 h after LPS injection. DEX treatment significantly reduced the number of infiltrating cells compared with the LPS group (Fig. 2).
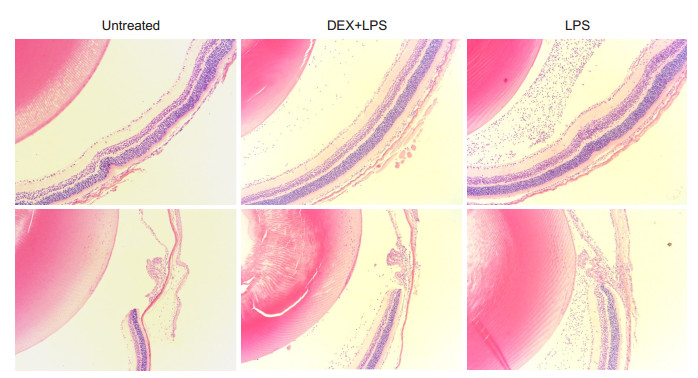
|
Figure 2 Infiltrating cells in the posterior and anterior chambers of the mice with EIU and DEX treatment at 24 h after LPS injection (HE staining, ×100) |
The inflammatory cytokines IL-6, TNF-α, MCP-1 and ICAM-1 play a vital role in the development of inflammation in mouse models of EIU [9, 11, 23, 24]. Compared with the normal control group, LPS injection induced significantly increased expressions of these cytokines, and topical application of DEX lowered the level of IL-6 by approximately 1.8 folds, TNF-α by 3.2 folds, MCP-1 by 2.1 folds and ICAM-1 by 1.9 folds (Fig. 3).
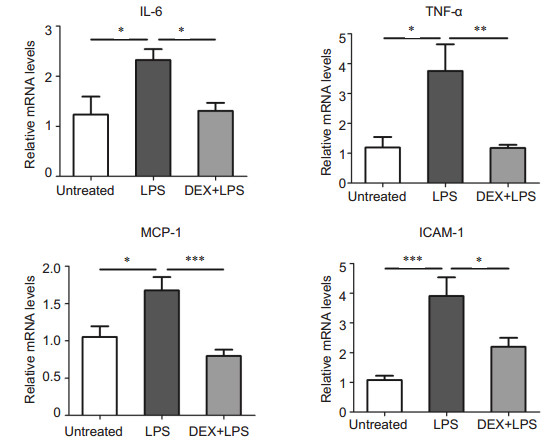
|
Figure 3 DEX decreases the over-production of inflammatory cytokines evoked by LPS. The mRNA levels of the cytokines IL-6, TNF-α, MCP-1 and ICAM-1 were detected using realtime PCR. The data are presented as Mean ± SE (n=6-8) and analyzed with one- way ANOVA followed by Bonferroni correction. ***P < 0.001, **P < 0.01, *P < 0.05 |
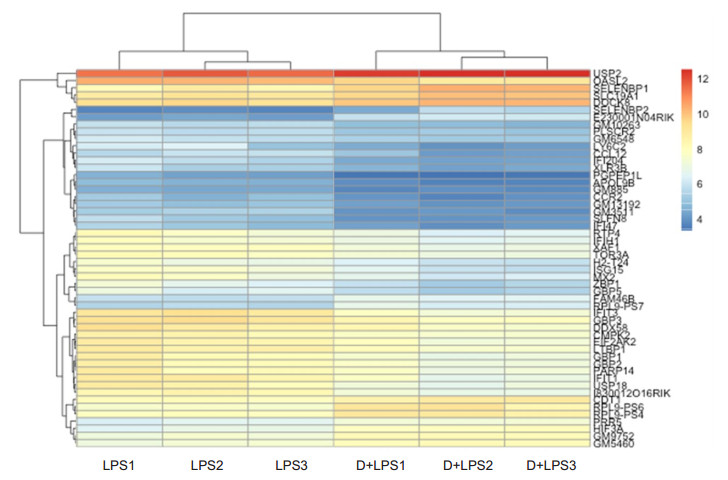
The volcano plot showed 52 DEGs (adjusted P < 0.05) in the retinas of mice following intravitreal injection of LPS with and without DEX treatment. Among these DEGs, 37 (71%) genes were up- regulated and 15 (29%) were down-regulated in the mice with EIU as compared with the mice with DEX treatment (Fig. 4). The heatmap and hierarchical clusters analyses (Fig. 5) highlighted distinct differences in the expression levels of the 52 DEGs between the two groups.
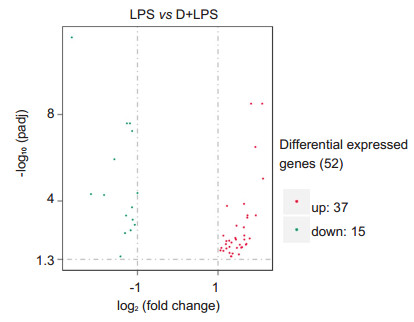
|
Figure 4 Volcano plot showing 52 DEGs (adjusted P < 0.05) in the retina of mice with LPS versus DEX+LPS treatment |
|
Figure 5 Heatmap and hierarchical clusters analysis of the DEGs in the retina between mice in LPS and DEX+LPS groups (n=3). Each small box indicates the expression status of a certain gene in one sample, red for up-regulated and blue for down-regulated expression |
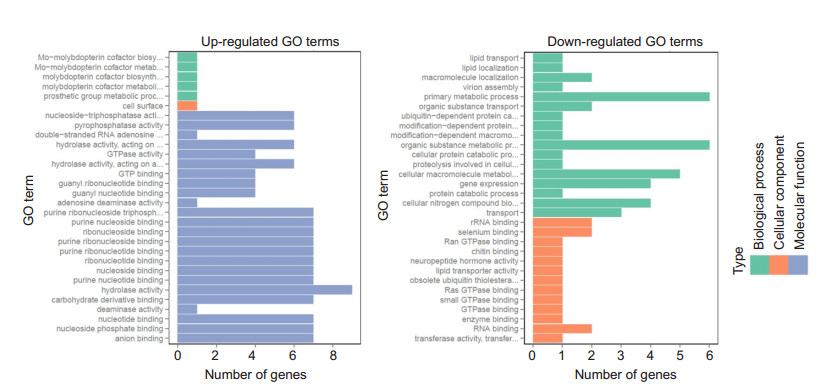
GO functional enrichment analysis was performed for the DEGs identified by RNA-seq, and the top 30 enriched up-regulated and down-regulated GO terms are shown in Fig. 6. No significantly enriched GO terms were found in either the up-regulated or the down-regulated terms.
|
Figure 6 GO enrichment analysis of the LPS versus DEX + LPS group. GO functional analysis shows the top 30 enriched up- and down-regulated GO Terms. No GO term was significantly enriched (adjusted P>0.05) |
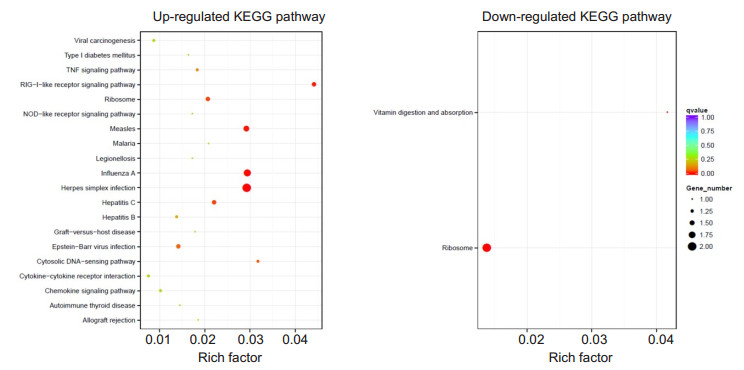
The top 20 up-regulated and down-regulated KEGG pathways with enrichment are shown in Fig. 7. Six upregulated pathways were found to be significantly enriched in mice with EIU as compared with the DEXtreated mice, involving herpes simplex infection, influenza A, measles, RIG-I-like receptor signaling pathway, hepatitis C and ribosome; 2 down-regulated pathways were significantly enriched in the mice with EIU, involving ribosome, vitamin digestion and absorption. The significantly enriched KEGG pathways are listed in Tab. 2.
|
Figure 7 KEGG enrichment analysis between LPS and DEX+LPS groups. KEGG functional analysis showed the top 20 enriched up- and down-regulated KEGG pathways, including 6 significantly up-regulated and 2 significantly down-regulated KEGG pathways (adjusted P < 0.05) |
| Table 2 Significantly enriched KEGG pathways in the retina of mice with EIU |
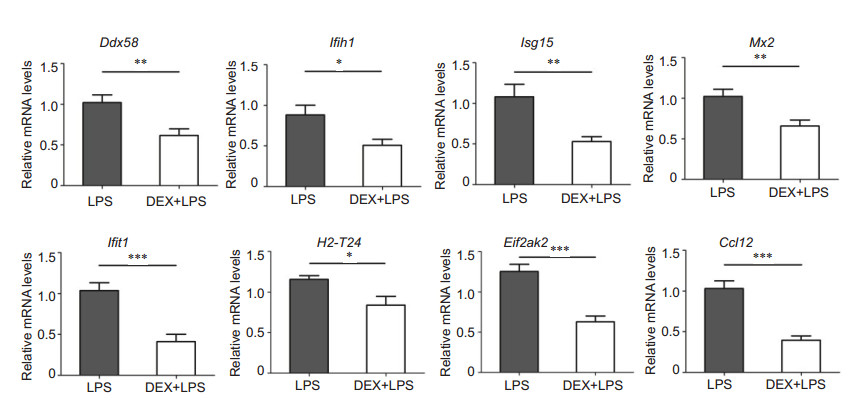
Eight genes (adjusted P < 0.05) involved in the significantly up-regulated KEGG pathways were selected for validation using real-time PCR (Tab. 3). These genes are associated with pathogen infection and innate immune response, and are divided into 3 categories: RIG-I-like receptor signaling pathway related genes (Ddx58, Ifih1, and Isg15); immune and inflammation regulation related genes (Mx2, Ifit1, H2-T24, and Eif2ak2); and inflammatory response related genes (Ccl12). The results of RNA-seq demonstrated significant up-regulation of these selected genes in mice with EIU as compared with DEX-treat mice, suggesting that DEX can significantly down-regulate these genes. The results of real-time PCR validation were consistent with the results of RNA- seq (Fig. 8).
| Table 3 Genes validated by real-time PCR |
|
Figure 8 Validation of the DEGs by real-time PCR. Eight genes from the markedly enriched KEGG pathways were selected (adjusted P < 0.05). The mRNA expressions of RIG-I-like receptor signaling pathway related genes Ddx58, Ifih1, and Isg15, immune- and inflammation-related genes Mx2, Ifit1, H2-T24, and Eif2ak2, and inflammatory response-related gene Ccl12 were significantly decreased in the DEX +LPS group compared with LPS group. All data are shown as Mean±SE. ***P < 0.001, **P < 0.01, *P < 0.05 by unpaired t-test (n=6-8) |
DEX is commonly used for treatment of many inflammatory and autoimmune ocular diseases[25]. Our results demonstrate that DEX can significantly ameliorate the inflammation in the anterior chamber and reduce the over-expression of the inflammatory cytokines IL-6, TNF-α, MCP-1 and ICAM-1 induced by LPS stimulation. The results of transcriptome analysis reveal that the protective effect of DEX is probably associated with RIG-I-like receptors (RLRs) signaling pathway and several immune- and inflammation-related genes, including Mx2, Ifit1, H2-T24 and Eif2ak2. To our knowledge, this is the first report of the specific pathway and genes that potentially mediate the therapeutic effect of DEX on uveitis; identification of this pathway and the involved genes may enrich the understanding of the mechanisms of uveitis and benefits the development of novel therapeutic strategies.
The pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), RLRs, and NOD-like receptors (NLRs), recognize the invasive pathogens and subsequently trigger an antimicrobial immune response and the production of pro-inflammatory cytokines and chemokines [26-28]. The RLRs, including retinoic acidinducible gene I (RIG-I, also known as DDX58)[29]and melanoma differentiation-associated gene 5 (MDA5, also known as IFIH1)[30], are a family of cytosolic RNA helicases that can specifically recognize viral RNA and initiate innate immunity. The RLRs signal pathway can increase the production of type I interferon and activate the antiviral genes to trigger intracellular immunity and inflammation to control virus infection[31, 32]. Anomalies in RLRs signaling is associated with several autoimmune and inflammatory diseases. Therefore, normal regulation of the RLRs signaling pathway is crucial to avoid tissue injury induced by superfluous immune response[31].
Recent studies showed that RLRs also participate in the recognition of intracellular bacteria[33, 34]. There is evidence that RLRs are capable of detecting the RNA of intracellular Salmonella enterica serovar Typhimurium and subsequently activate type I interferon to evoke the innate immunity [35]. Bacterial LPS stimulation of the endothelial cells increases the expression of RIG-I, and then selectively up-regulates the expression of inflammatory enzyme cyclooxygenase 2 (COX-2), suggesting that RIG-I plays a key role in pro-inflammatory pathways[36]. Furthermore, transmissible gastroenteritis virus (TGEV) infection is found to significantly up-regulate the RLRs (RIG-I and MDA5), whereas RLRs knockdown decreases the pro-inflammatory cytokines and the phosphorylation of NF-κB subunit p65[37]. Consistent with the previous findings, our transcriptome analysis show that DEX markedly down-regulate the RLRs in mice with EIU, suggesting that the protective effect of DEX is related to the down-regulation of RLRs, and inhibiting RIG-I may be a promising strategy for treatment of ocular inflammation.
The immune- and inflammation-related genes from the enriched KEGG pathway, including Ifit1, Mx2, H2- T24 and Eif2ak2, play a vital role in antiviral immune response and also in multiple immune and inflammatory diseases. A recent study found that Ifit1 gene was overexpressed in the peripheral blood of patients with systemic lupus erythematosus (SLE) [38], and possibly by interacting with Rho/Rac GEF [38] and subsequently activating JNK and p38MAPK, participated in the immune responses in SLE [39]. The over-expression of EIF2AK2 increases the activity of NLRP3 inflammasome, whereas silencing or suppression of EIF2AK2 expression lowers the activity of NLRP3 inflammasome to inhibit the secretion of IL-1β, IL-18 and high-mobility group box 1 (HMGB1) [40]. These evidences suggest the pro-inflammatory role of Ifit1 and Eif2ak2 in inflammatory diseases. Nevertheless, the role of Mx2 and H2-T24 in immune and inflammation regulation remains unclear. Here, we show that LPS stimulation increases the expressions of Ifit1, Mx2, H2-T24 and Eif2ak2, whereas DEX treatment down-regulates their expressions in the retina of mice, suggesting that Mx2 and H2-T24 are also involved in the immune and inflammatory responses in EIU.
Taken together, our transcriptome analysis indicate that DEX significantly down-regulates RLRs (DDX58 and IFIH1) and the immune- and inflammation-related genes including Ifit1, Mx2, H2-T24 and Eif2ak2, indicating their involvement in mediating the protective effect of DEX. Further investigations for functional verification of RLRs signaling pathway and these genes using RNA interference or pharmacological inhibition are warranted to confirm the mechanism underlying the protective effect of DEX against EIU.
| [1] | Tabbara KF. Infectious uveitis: a review[J]. Arch Soc Esp Oftalmol, 2000, 75(4): 215-9. |
| [2] | Miserocchi E, Fogliato G, Modorati G, Bandello F. Review on the worldwide epidemiology of uveitis[J]. Eur J Ophthalmol, 2013, 23(5): 705-17. DOI: 10.5301/ejo.5000278. |
| [3] | Munoz-Fernandez S, Martin-Mola E. Uveitis[J]. Best Pract Res Clin Rheumatol, 2006, 20(3): 487-505. DOI: 10.1016/j.berh.2006.03.008. |
| [4] | Rathinam SR, Namperumalsamy P. Global variation and pattern changes in epidemiology of uveitis[J]. Indian J Ophthalmol, 2007, 55(3): 173-83. DOI: 10.4103/0301-4738.31936. |
| [5] | Majumder PD, Ghosh A, Biswas J. Infectious uveitis: an enigma[J]. Middle East Afr J Ophthalmol, 2017, 24(1): 2-10. |
| [6] | Rosenbaum JT, Mcdevitt HO, Guss RB, et al. Endotoxin-induced uveitis in rats as a model for human disease[J]. Nature, 1980, 286(5773): 611-3. DOI: 10.1038/286611a0. |
| [7] | Bhattacherjee P, Williams RN, Eakins KE. An evaluation of ocular inflammation following the injection of bacterial endotoxin into the rat foot pad[J]. Invest Ophthalmol Vis Sci, 1983, 24(2): 196-202. |
| [8] | Koizumi K, Poulaki V, Doehmen S, et al. Contribution of TNF-α to leukocyte adhesion, vascular leakage, and apoptotic cell death in endotoxin-induced uveitis in vivo[J]. Invest Ophthalmol Vis Sci, 2003, 44(5): 2184-91. DOI: 10.1167/iovs.02-0589. |
| [9] | Becker MD, Garman K, Whitcup SM, et al. Inhibition of leukocyte sticking and infiltration, but not rolling, by antibodies to ICAM-1 and LFA-1 in murine endotoxin-induced uveitis[J]. Invest Ophthalmol Vis Sci, 2001, 42(11): 2563-6. |
| [10] | de Vos AF, Klaren VN, Kijlstra A. Expression of multiple cytokines and IL-1RA in the uvea and retina during endotoxin-induced uveitis in the rat[J]. Invest Ophthalmol Vis Sci, 1994, 35(11): 3873-83. |
| [11] | Hoekzema R, Murray PI, van Haren MA, et al. Analysis of interleukin-6 in endotoxin-induced uveitis[J]. Invest Ophthalmol Vis Sci, 1991, 32(1): 88-95. |
| [12] | Tuaillon N, Shen DF, Berger RB, et al. MCP-1 expression in endotoxin-induced uveitis[J]. Invest Ophthalmol Vis Sci, 2002, 43(5): 1493-8. |
| [13] | Lee FF, Foster CS. Pharmacotherapy of uveitis[J]. Expert Opin Pharmacother, 2010, 11(7): 1135-46. DOI: 10.1517/14656561003713534. |
| [14] | Read RW. Uveitis: advances in understanding of pathogenesis and treatment[J]. Curr Rheumatol Rep, 2006, 8(4): 260-6. DOI: 10.1007/s11926-006-0006-6. |
| [15] | Sherif Z, Pleyer U. Corticosteroids in ophthalmology: Past-Present - Future[J]. Ophthalmologica, 2002, 216(5): 305-15. DOI: 10.1159/000066189. |
| [16] | Saraiya NV, Goldstein DA. Dexamethasone for ocular inflammation[J]. Expert Opin Pharmacother, 2011, 12(7): 1127-31. DOI: 10.1517/14656566.2011.571209. |
| [17] | Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics[J]. Nat Rev Genet, 2009, 10(1): 57-63. DOI: 10.1038/nrg2484. |
| [18] | Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome[J]. Science, 2008, 321(5891): 956-60. DOI: 10.1126/science.1160342. |
| [19] | Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing[J]. Science, 2008, 320(5881): 1344-9. DOI: 10.1126/science.1158441. |
| [20] | Qiu Y, Yu P, Lin R, et al. Genome-wide retinal transcriptome analysis of endotoxin-induced uveitis in mice with next- generation sequencing[J]. Mol Vis, 2017, 23: 395-406. |
| [21] | Qiu Y, Shil PK, Zhu P, et al. Angiotensin-converting enzyme 2 (ACE2) activator diminazene aceturate ameliorates endotoxininduced uveitis in mice[J]. Invest Ophthalmol Vis Sci, 2014, 55(6): 3809-18. DOI: 10.1167/iovs.14-13883. |
| [22] | Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium[J]. Nat Genet, 2000, 25(1): 25-9. DOI: 10.1038/75556. |
| [23] | Mo JS, Matsukawa A, Ohkawara S, et al. Role and regulation of IL-8 and MCP-1 in LPS-induced uveitis in rabbits[J]. Exp Eye Res, 1999, 68(3): 333-40. DOI: 10.1006/exer.1998.0618. |
| [24] | Shen DF, Chang MA, Matteson DM, et al. Biphasic ocular inflammatory response to endotoxin-induced uveitis in the mouse[J]. Arch Ophthalmol, 2000, 118(4): 521-7. DOI: 10.1001/archopht.118.4.521. |
| [25] | Gordon DM. Use of dexamethasone in eye disease[J]. J Am Med Assoc, 1960, 172: 311-2. DOI: 10.1001/jama.1960.03020040009003. |
| [26] | Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-50. DOI: 10.1016/j.immuni.2011.05.006. |
| [27] | Elinav E, Strowig T, Henao-Mejia J, et al. Regulation of the antimicrobial response by NLR proteins[J]. Immunity, 2011, 34(5): 665-79. DOI: 10.1016/j.immuni.2011.05.007. |
| [28] | Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA[J]. Immunol Rev, 2011, 243(1): 91-8. DOI: 10.1111/imr.2011.243.issue-1. |
| [29] | Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIGI has an essential function in double-stranded RNA- induced innate antiviral responses[J]. Nat Immunol, 2004, 5(7): 730-7. DOI: 10.1038/ni1087. |
| [30] | Kang DC, Gopalkrishnan RV, Lin L, et al. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene[J]. Oncogene, 2004, 23(9): 1789-800. DOI: 10.1038/sj.onc.1207300. |
| [31] | Peng Y, Xu R, Zheng X. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1[J]. PLoS Pathog, 2014, 10(4): e1004041. DOI: 10.1371/journal.ppat.1004041. |
| [32] | Loo YM, Gale MJ. Immune signaling by RIG-I-like receptors[J]. Immunity, 2011, 34(5): 680-92. DOI: 10.1016/j.immuni.2011.05.003. |
| [33] | Patel JR, Garcia-Sastre A. Activation and regulation of pathogen sensor RIG-I[J]. Cytokine Growth Factor Rev, 2014, 25(5): 513-23. DOI: 10.1016/j.cytogfr.2014.08.005. |
| [34] | Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors[J]. Adv Immunol, 2013, 117: 99-125. DOI: 10.1016/B978-0-12-410524-9.00004-9. |
| [35] | Schmolke M, Patel JR, de Castro E, et al. RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection[J]. MBio, 2014, 5(2): e01006-14. |
| [36] | Imaizumi T, Aratani S, Nakajima T, et al. Retinoic acid-inducible gene-I is induced in endothelial cells by LPS and regulates expression of COX-2[J]. Biochem Biophys Res Commun, 2002, 292(1): 274-9. DOI: 10.1006/bbrc.2002.6650. |
| [37] | Ding Z, An K, Xie L, et al. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling[J]. Virology, 2017, 507: 170-8. DOI: 10.1016/j.virol.2017.04.024. |
| [38] | Ye S, Pang H, Gu YY, et al. Protein interaction for an interferoninducible systemic lupus associated gene, IFIT1[J]. Rheumatology (Oxford), 2003, 42(10): 1155-63. DOI: 10.1093/rheumatology/keg315. |
| [39] | Esteve P, Embade N, Perona R, et al. Rho-regulated signals induce apoptosis in vitro and in vivo by a p53-independent, but Bcl2 dependent pathway[J]. Oncogene, 1998, 17(14): 1855-69. DOI: 10.1038/sj.onc.1202082. |
| [40] | Lu B, Nakamura T, Inouye K, et al. Novel role of PKR in inflammasome activation and HMGB1 release[J]. Nature, 2012, 488(7413): 670-4. DOI: 10.1038/nature11290. |
 2018, Vol. 38
2018, Vol. 38

