肺癌死亡率位居全世界癌症之首,每年超过140万人死于肺癌[1]。尽管在早期诊断和新的治疗策略方面获得进展,但是肺癌病人5年总体生存率仍然低于15%[2]。宣威肺癌是指位于中国西南云南省宣威地区(包括宣威、富源、麒麟和沾益县等)居民所患的肺癌,女性肺癌发病率高达400/10万,高于全国肺癌平均发病率的20倍[3]。与其他地区肺癌相比,宣威地区肺癌有明显的区域聚集性等特点,为世界非吸烟女性肺癌发病率最高的地区性肺癌,其病因及发病机制复杂而尚不清楚。
微小RNA(miRNAs)是被认为参与了生命体的整个过程,包括细胞的分化、增殖及凋亡;并在疾病演变过程中起重要作用,越来越多地受到研究人员的高度重视。miRNAs表达与多种癌症相关,在肿瘤发生过程中起所起的关键作用[4]。目前研究发现不同的miRNAs作用于不同靶基因,通过不同信号通路,影响肺癌肿瘤细胞的增殖,侵袭,凋亡等生物学功能[5]。然而,miRNAs在宣威肺腺癌癌变和进展中的作用的研究少见报道。
本研究以宣威地区及非宣威地区病理确诊的肺腺癌患者手术标本为研究对象,应用基因芯片检测两者miRNAs差异表达谱,通过生物信息学的方法对其靶基因和肺癌密切相关的通路进行预测分析,为揭示miRNAs在宣威肺腺癌癌变和进展中的作用提供实验依据。
1 资料和方法 1.1 研究对象选取2014 年3 月~2014 年8 月,34例本院手术患者手术切除标本。其中24例均符合宣威肺腺癌特征:无吸烟史,3代内居住在宣威肺腺癌高发区域。另10例为系非宣威地区肺腺癌。研究对象纳入标准:无其他恶性肿瘤病史,均为第1次诊断及治疗,无放化疗治疗史,术后均得到病理证实为肺腺癌。
手术标本为肺癌组织与正常肺组织,其标本切取满足以下条件:瘤体的中央区域和正常肺组织(距离瘤体边界>5 cm),无炎症等病理学表现。所有的手术标本在切除后,均切为<0.5 cm的颗粒状,立即存放于RNALATER液(>5倍体积)冻存管中,置液氮罐临时存储,后转入置-80 ℃冰箱。所有患者均签署知情同意书,本实验流程符合伦理委员会要求。
1.2 方法 1.2.1 总RNA提取及质控研磨及匀浆:在液氮中研磨组织,并进行匀浆,使用设备:BioPulverizer,mini Beadbeater-16,Bioruptor,Branson Digital Sonifier。RNA提取:提取总RNA按照TRIzol® Reagent(Invitrogen lifetechnologies)试剂盒的要求进行。质量控制,使用Nanodrop,Agilent 2100 评估RNA 的浓度及纯度。存-80 ℃冰箱备用。
1.2.2 miRNA芯片检测芯片标记:按照miRCURYTMArray Power Labeling kit(Cat #208032-A,Exiqon)说明书进行Hy3™,Hy5™荧光标签标记。芯片杂交:使用2×Hybridization buffer,Phalanx Hyb. Assembly试剂在Agilent Hybridization Oven,MAUI Hybridization System进行杂交。简要过程:95 ℃孵育2 min,孵育过程需避光;孵育后冰上放置2 min,置95 ℃热水加温,50 ℃风干,置95 ℃热水再次加温,短时56 ℃烘干箱并置于摇床上,2 r/min过夜。
miRNAs 芯片检测和分析:使用miRCURYTMArray,Wash buffer kit(Cat #208021,Exiqon)试剂盒进行洗涤后,使用Axon GenePix 4000B进行图像扫描,采用GenePix pro V6.0进行原始数据分析。
1.3 宣威肺腺癌特异miRNAs筛选为了筛选出宣威肺腺癌特有的差异表达的miRNAs(宣威肺腺癌特异microRNAs),采用宣威肺腺癌芯片值与非宣威肺腺癌芯片值进行交集后取补集的方法对宣威特有差异miRNAs进行筛选。信号值去除背景,然后进行中值归一化。归一化后的数据计算肺癌组织与正常肺组织均值的比值,该比值>2.0且T-TEST(经过Bonferron矫正)P<0.05的被认为是miRNAs差异表达。
1.4 靶基因预测分析对预选miRNAs进行靶基因预测,GO 和Pathway分析,在mirbase,miranda和targetscan 3个数据库对预选的miRNAs进行network分析。
2 结果 2.1 宣威肺腺癌miRNAs表达宣威肺腺癌组织与相应正常肺组织的miRNAs检测结果显示:miRNAs表达量差异倍数>2倍(P<0.05),表达上调有64个,表达下调有89个(图 1、2)。
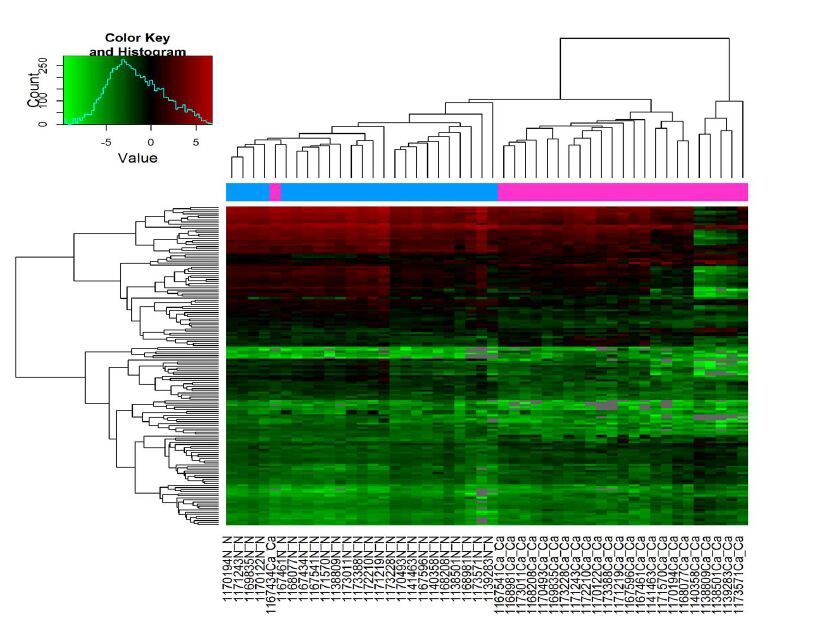
|
图 1 宣威肺腺癌差异miRNAs层次聚类分析颜色表示信号强度,绿-黑-红依次增强 Figure 1 Heat map and hierarchical clustering of miRNA profiles of lung adenocarcinoma in Xuanwei. The relative expression valuesare depicted according to the color scale. Red indicates upregulation,and green indicates down-regulation. |
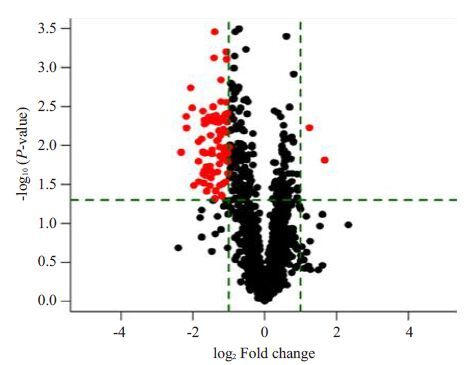
|
图 2 宣威肺腺癌芯片P值与miRNAs表达差异倍数火山图 Figure 2 Volcano plot of the differential expressed miRNAs. Reddot represents a statistically significant difference in theexpression of microRNAs(P<0.05) |
非宣威肺腺癌组织与相应正常肺组织的miRNAs检测结果显示:miRNA差异倍数>2倍(P<0.05),表达上调有2个,表达下调有81个(图 3、4)。
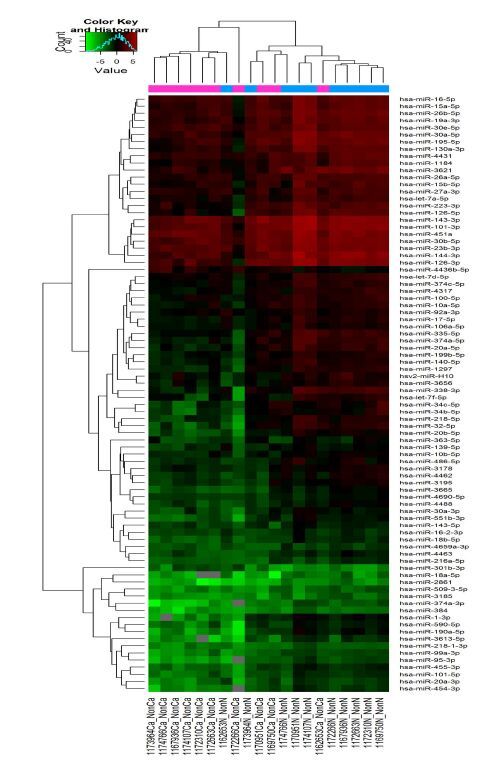
|
图 3 非宣威肺腺癌差异miRNAs层次聚类分析颜色表示信号强度,绿-黑-红依次增强 Figure 3 Heat map and hierarchical clustering of miRNA profiles oflung adenocarcinoma in regions other than Xuanwei. The relativeexpression values are depicted according to the color scale. Redindicates upregulation,and green indicates down-regulation. |
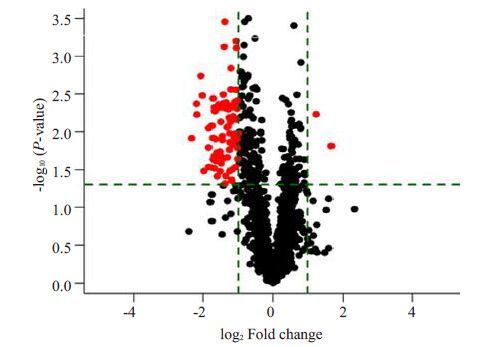
|
图 4 非宣威肺腺癌芯片P值与miRNAs表达差异倍数火山图红点:差异表达miRNAs Figure 4 Volcano plot of the differential expressed miRNA inlung adenocarcinoma in regions other than Xuanwei. Red dotrepresents a statistically significant difference in theexpression of microRNAs(P<0.05) |
宣威肺腺癌特异miRNAs显示:在宣威肺腺癌表达显著的特异34 个miRNAs(P<0.05,fold change >2)。表达上调的miRNAs 有23 个,包括hsa-miR-504-5p、hsa-miR-501-5p、hsa-miR-147a、hsa-miR-372-3p、hsa-miR-187-3p、hsa-miR-325、hsa-miR-369-3p、hsa-miR-33b-5p、hsa-miR-182-5p、hsa-miR-210-3p、hsa-miR-595、hsa-miR-9-5p、hsa-miR-224-5p、hsa-miR-558、hsa-miR-495-3p、hsa-miR-485-5p、hsa-miR-134-5p、hsa-miR-519d-3p、hsa-miR-96-5p、hsa-miR-375、hsa-miR-645、hsa-miR-938 和hsa-miR-135b-5p;表达下调的miRNAs 有11 个,包括hsa-miR-598-3p、hsa-miR-138-5p、hsa-miR-99a-5p、hsa-miR-34b-3p、hsa-miR-30d-5p、hsa-miR-140-3p、hsa-miR-34c-3p、hsa-miR-497-5p、hsa-miR-585-3p、hsa-miR-30c-5p和hsa-miR-363-3p。
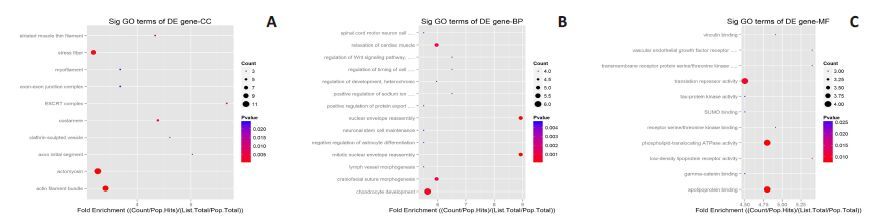
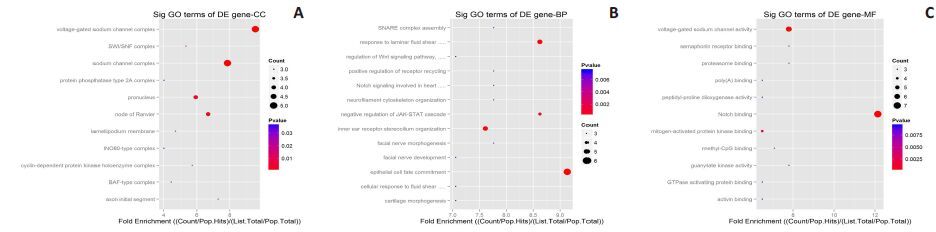
2.4 宣威肺腺癌特异表达miRNAs的靶基因AGO分析对34个宣威肺腺癌特有差异表达的miRNAs进行生物过程、细胞定位和分子功能的GO分析,并统计miRNAs可能的靶基因在每个信号通路中的富集程度(enrich p-value)分析发现:(1)表达上调的miRNAs:靶基因的生物功能主要富集在对细胞代谢调节,对细胞分化及神经系统过程也有富集;细胞定位主要在细胞核内,部分在细胞器;分子功能是与蛋白质的结合,影响蛋白激酶活性,对蛋白翻译的进行调节;(2)表达下调的miRNAs:靶基因的生物功能主要富集在对细胞生物过程的正/负调节,JAK-STAT信号级联,对细胞分化及神经系统过程也有富集;细胞定位:主要存在于细胞核内,也部分在神经突触;分子功能主要集中在与蛋白酶,激酶,蛋白质特定结合,蛋白复合物,受体信号传导蛋白等结合,调节下游蛋白的翻译(图 5、6)。
|
图 5 宣威肺腺癌特异表达上调miRNAs靶基因定位(A)、生物学过程(B)和分子功能(C)的倍数富集Dotplot分析 Figure 5 Dot plot analysis of CC,BP,and MF of the up-regulated microRNAs. |
|
图 6 宣威肺腺癌特异表达下调miRNAs靶基因定位(A)、生物学过程(B)和分子功能(C)的倍数富集Dotplot分析 Figure 6 Dot plot analysis of CC, BP, and MF of the down-regulated microRNAs. |
在对34个宣威肺腺癌特异的miRNAs进行KEGG数据库映射通路分析,一共涉及到78个信号通路:上调miRNAs涉及通路40个,包括有轴突导向通路,多巴胺能神经突触,cGMP-PKG,MAPK通路,粘附通路,癌症转录失调通路,雌激素信号通路,胆碱能突触,Rap1信号通路等;下调miRNAs涉及通路38个,包括有cAMP信号通路,神经突触长时程增强通路,神经突触长时程抑制通路,Hippo信号通路,cGMP-PKG信号通路,蛋白多糖通路,心肌细胞的β肾上腺素能信号通路,急性髓细胞样白血病通路等。本研究对肺癌相关的通路进行筛选发现:cGMP-PKG信号通路,MAPK通路,Rap1信号通路,cAMP 信号通路,PI3K/Alt 通路,mTOR 通路,WNT通路,癌症转录失调通路,notch通路,RAS及p53等通路关系密切。进一步对这11个通路中预测的靶基因分析发现:可能对肺癌生物学功能产生影响的靶基因主要集中在PI3K/Alt,WNT和MAPK通路(表 1)。
| 表 1 宣威肺腺癌特异表达的miRNAs与肺癌相关通路 Table 1 Predicted signaling pathways involving the differentially expressed microRNAs in lung adenocarcinoma in Xuanwei |
肺癌的发生发展是非常复杂的多基因事件,涉及到多个癌基因及抑癌基因的功能改变。miRNA是癌症的关键参与者,具有调节癌症相关基因表达的能力,在肿瘤发生、生长、迁移与侵袭过程中发挥了重要作用。Takamizawa等[6]首先描述miRNA表达与肺癌有关。随后,大量研究发现肺癌患者的miRNAs的表达改变或异常,miRNAs在体外或体内肺癌模型起到抑癌基因或癌基因的作用,在肺癌演进过程中发挥重要作用。
本研究选取来自宣威地区肺腺癌和非宣威地区肺腺癌患者手术切除的肺癌组织和相应正常肺组织标本,miRNAs芯片检测34对肺切除标本miRNAs差异表达谱结果发现,宣威肺腺癌与非宣威肺腺癌比较,其中宣威肺腺癌特异miRNAs 有34 个:(1)与肺癌相关的microRNA:hsa-miR-187-3p[7]、hsa-miR-140-3p[8]、hsamiR-34c-3p[9]、hsa-miR-30d-5p[10]、hsa-miR-375[11]和hsa-miR-138-5p[12];(2)与其他恶性肿瘤相关的microRNA:hsa-miR-325[13]、hsa-miR-501-5p[14]、hsa-miR-96-5p[15]、hsa-miR-182-5p[15]、hsa-miR-485-5p[16]、hsa-miR-9-5p[17]、hsa-miR-598-3p[18]、hsa-miR-30c-5p[19]、hsa-miR-645[20]、hsa-miR-938[21]、hsa-miR-135b-5p[22]、hsa-miR-363-3p[23]、hsa-miR-147a[24]、hsa-miR-224-5p[25]、hsa-miR-519d-3p[26]、hsa-miR-558[27]和hsa-miR-497-5p[28];(3)目前未在肿瘤研究的microRNA:hsa-miR-33b-5p、hsa-miR-134-5p、hsa-miR-504-5p、hsa-miR-99a-5p、hsa-miR-369-3p、hsa-miR-585-3p 与hsa-miR-495-3p。这些microRNA的前体或发夹结构的另一端所产生的microRNA可能与肿瘤关系密切,如与hsa-miR-33b-5p有相同前体而在发夹结构的3’端生成miR-33b-3p则可以通过对靶基因p21的作用增强肺癌细胞的生存能力及对顺铂的抵抗[29],而hsa-miR-134-5p、hsa-miR-99a-5p、hsa-miR-504-5p、hsa-miR-585-3p 和hsa-miR-495-3p的前体hsa-miR-134[30]、has-miR-99a[31]、has-miR-504[32],hsa-miR-585[33]与hsa-miR-495[34]的作用已在肿瘤研究中得到证实。
本研究进一步对宣威肺腺癌特有的差异表达的miRNAs进行AGO富集分析及KEGG数据库映射分析发现:这些miRNAs与肺癌密切相关的通路有关,包括cGMP-PKG信号通路、MAPK信号通路、Rap1 信号通路、cAMP信号通路、PI3K/Alt通路、mTOR通路、WNT通路、癌症转录失调通路、notch通路、RAS和p53通路;这些miRNAs可能对肺癌生物学功能产生影响的靶基因主要集中在PI3K/Alt,WNT和MAPK信号通路。我们的结果提示这些宣威肺腺癌特异性miRNAs可能通过调节PI3K/Alt,WNT和MAPK信号通路中相关靶基因而发挥肺癌生物学作用。
宣威肺腺癌特异性miRNAs与PI3K/Alt信号通路相关芯片预测的靶基因发现,预测主要集中在PI3K/Alt信号通路附近,其预测靶基因有:GF、RTK、SOS、IRS1、BCAP、CYTOKINSR、ECM、ITGB、FAK和GβY。在目前的非小细胞肺癌研究中,与PI3K-Akt 通路相关miRNAs的靶基因也主要集中在GF、RTK和PI3K,其中以RTK中IGF1R及EGFR为靶基因的miRNAs[35-38],分别是miR-122<A549、H460>,miR-30a<A549>、miR-31<A549>和miR-34a<pc9、hcc827>。研究还发现miR-126以VEGFA<A549>、p1k3R2<A549>、P-AKT<sk-kes-1>为靶基因[39]、miR-21 以IGF<A549>、PTEN<pc9>作为靶基因[40]。这说明同一个miRNA在同一信号通路可以在同一分型的肺癌组织学类型的不同细胞系有着不同的靶基因,可能宣威肺癌细胞系与其他肺癌细胞系在功能与靶基因方面存在一定的差异。
宣威肺腺癌特异性miRNAs与WNT信号通路相关芯片预测的靶基因:FRP、WNT、FRIZZLED、LRP5/6、BAMB1、CKIε、AXarn和gsk-3β。已知的与非小细胞肺癌相关的miR-708[41]、miR-3619-5P[42]和miR-29[43]其靶基因分别是:TMEM88、β-CATENIN和WIF1。研究发现miR-582-3P 可以调节DKK3,dAXIN2作用WNT通路[44]。miR-410在WNT通路的作用是通过DVL2与β-CATENIN,降低Gsk3β蛋白的表达[45]。
宣威肺腺癌特异性miRNA与MAPK通路芯片预测靶基因:CACN、BDNF、FGF、FGFR、RAS、NF1、RAP1、MEK1、MKP、MNK1/2、NFκB和P120GAF。目前已证实与非小细胞肺癌相关的miRNAs 有:miR-34a[46]、miR-7[47]和miR-34c-3p[48],其靶基因分别是:KRAS、pax6和Pac1,其聚集性与芯片预测也存在高度相似。
综上所述,通过已知miRNAs 在PI3K/Alt、WNT和MAPK信号通路的非小细胞肺癌靶基因集中趋势,结合本研究芯片预测结果,说明宣威肺癌特异性miRNAs与已报道的非小细胞肺癌miRNA靶基因聚集趋势相同。从以上3条通路的预测靶基因聚集情况可以认为,宣威肺癌与非宣威肺癌在信号通路上可能存在一致性。这些提示筛选出这些特异性miRNAs可能在宣威肺腺癌癌变和进展中起重要作用,其靶基因的作用可能与调节PI3K/Alt、WNT和MAPK信号通路有关。这为后续深入研究探讨miRNAs在宣威肺腺癌发生发展中作用及其机制奠定一定研究基础。
| [1] | Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012[J]. CA Cancer J Clin, 2015, 65 (2): 87-108. DOI: 10.3322/caac.21262. |
| [2] | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013[J]. CA Cancer J Clin, 2013, 63 (1): 11-30. DOI: 10.3322/caac.v63.1. |
| [3] | Mumford JL, He XZ, Chapman RS, et al. Lung cancer and indoor air pollution in Xuan Wei, China[J]. Science, 1987, 235 (4785): 217-20. DOI: 10.1126/science.3798109. |
| [4] | Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer[J]. Annu Rev Pathol, 2014, 9 : 287-314. DOI: 10.1146/annurev-pathol-012513-104715. |
| [5] | Joshi P, Middleton J, Jeon YJ, et al. MicroRNAs in lung cancer[J]. World J Methodol, 2014, 4 (2): 59-72. DOI: 10.5662/wjm.v4.i2.59. |
| [6] | Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 miRNAs in human lung cancers in association with shortened postoperative survival[J]. Cancer Res, 2004, 64 (11): 3753-6. DOI: 10.1158/0008-5472.CAN-04-0637. |
| [7] | Sun C, Li S, Yang C, et al. MiRNA-187-3p mitigates non-small cell lung cancer (NSCLC) development through down-regulation of BCL6[J]. Biochem Biophys Res Commun, 2016, 471 (1): 82-8. DOI: 10.1016/j.bbrc.2016.01.175. |
| [8] | Kong XM, Zhang GH, Huo YK, et al. MiRNA-140-3p inhibits proliferation, migration and invasion of lung cancer cells by targeting ATP6AP2[J]. Int J Clin Exp Pathol, 2015, 8 (10): 12845-52. |
| [9] | Zhou YL, Xu YJ, Qiao CW. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer(NSCLC)by inhibiting PAC1/MAPK pathway[J]. Int J Clin Exp Pathol, 2015, 8 (6): 6312-22. |
| [10] | Chen D, Guo W, Qiu Z, et al. MiRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer[J]. Cancer Lett, 2015, 362 (2): 208-17. DOI: 10.1016/j.canlet.2015.03.041. |
| [11] | Yoda S, Soejima K, Hamamoto J, et al. Claudin-1 is a novel target of miR-375 in non-small-cell lung cancer[J]. Lung Cancer, 2014, 85 (3): 366-72. DOI: 10.1016/j.lungcan.2014.06.009. |
| [12] | Gao Y, Fan X, Li W, et al. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124[J]. Biochem Biophys Res Commun, 2014, 446 (1): 179-86. DOI: 10.1016/j.bbrc.2014.02.073. |
| [13] | Li H, Huang W, Luo R. The microRNA-325 inhibits hepatocellular carcinoma progression by targeting high mobility group box 1[J]. Diagn Pathol, 2015, 10 : 117. DOI: 10.1186/s13000-015-0323-z. |
| [14] | Huang DH, Wang GY, Zhang JW, et al. MiR-501-5p regulates CYLD expression and promotes cell proliferation in human hepatocellular carcinoma[J]. Jpn J Clin Oncol, 2015, 45 (8): 738-44. DOI: 10.1093/jjco/hyv063. |
| [15] | Assal RA, El Tayebi HM, Hosny KA, et al. A pleiotropic effect of the single clustered hepatic metastamiRs miR-96-5p and miR-182- 5p on insulin-like growth factor II, insulin-like growth factor-1 receptor and insulin-like growth factor-binding protein-3 in hepatocellular carcinoma[J]. Mol Med Rep, 2015, 12 (1): 645-50. |
| [16] | Sun X, Liu Y, Li M, et al. Involvement of miR-485-5p in hepatocellular carcinoma progression targeting EMMPRIN[J]. Biomed Pharmacother, 2015, 72 : 58-65. DOI: 10.1016/j.biopha.2015.04.008. |
| [17] | Xie CH, Cao YM, Huang Y, et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression[J]. Tumour Biol, 2016, 37 (11): 15031-41. DOI: 10.1007/s13277-016-5391-5. |
| [18] | Fu L, Li Z, Zhu J, et al. Serum expression levels of miRNA- 3823p, -598-3p, -1246 and-184 in breast cancer patients[J]. Oncol Lett, 2016, 12 (1): 269-274. |
| [19] | Jiang L, Zang D, Yi S, et al. A miRNA-mediated decrease in eukaryotic initiation factor 2α promotes cell survival during PS-341 treatment[J]. Sci Rep, 2016, 6 : 21565. DOI: 10.1038/srep21565. |
| [20] | Feng X, Wang Y, Ma Z, et al. MiRNA-645, up-regulated in human adencarcinoma of gastric esophageal junction, inhibits apoptosis by targeting tumor suppressor IFIT2[J]. BMC cancer, 2014, 14 : 633. DOI: 10.1186/1471-2407-14-633. |
| [21] | Arisawa T, Tahara T, Shiroeda H, et al. Genetic polymorphisms of IL17A and pri-miRNA-938, targeting IL17A 3'-UTR, influence susceptibility to gastric cancer[J]. Hum Immunol, 2012, 73 (7): 747-52. DOI: 10.1016/j.humimm.2012.04.011. |
| [22] | Ozcan O, Kara M, Yumrutas O, et al. MTUS1 and its targeting miRNAs in colorectal carcinoma: significant associations[J]. Tumour Biol, 2016, 37 (5): 6637-45. DOI: 10.1007/s13277-015-4550-4. |
| [23] | Song B, Yan J, Liu C, et al. Tumor suppressor role of miR-363-3p in gastric cancer[J]. Med Sci Monit, 2015, 21 : 4074-80. DOI: 10.12659/MSM.896556. |
| [24] | Wang F, Zhang H, Xu N, et al. A novel hypoxia-induced miR-147a regulates cell proliferation through a positive feedback loop of stabilizing HIF-1α[J]. Cancer Biol Ther, 2016, 17 (8): 790-8. DOI: 10.1080/15384047.2016.1195040. |
| [25] | Zhao H, Bi T, Qu Z, et al. Expression of miR-224-5p is associated with the original cisplatin resistance of ovarian papillary serous carcinoma[J]. Oncol Rep, 2014, 32 (3): 1003-12. |
| [26] | Ding J, Huang F, Wu G, et al. MiR-519d-3p suppresses invasion and migration of trophoblast cells via targeting MMP-2[J]. PLoS One, 2015, 10 (3): e0120321. DOI: 10.1371/journal.pone.0120321. |
| [27] | Qu H, Zheng L, Pu J, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase[J]. Hum Mol Genet, 2015, 24 (9): 2539-51. DOI: 10.1093/hmg/ddv018. |
| [28] | Chen Y, Kuang D, Zhao X, et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma[J]. Oncotarget, 2016, 7 (36): 58148-61. |
| [29] | Xu S, Huang H, Chen YN, et al. DNA damage responsive miR- 33b-3p promoted lung cancer cells survival and cisplatin resistance by targeting p21WAF1/CIP1[J]. Cell Cycle, 2016, 15 (21): 2920-30. DOI: 10.1080/15384101.2016.1224043. |
| [30] | Qin Q, Wei F, Zhang J, et al. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor[J]. J Cell Mol Med, 2016, 20 (10): 1974-83. DOI: 10.1111/jcmm.12889. |
| [31] | Wang X, Li Y, Qi W, et al. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells[J]. Oncotarget, 2015, 6 (32): 32737-47. |
| [32] | Soutto M, Chen Z, Saleh MA, et al. TFF1 activates p53 through down-regulation of miR-504 in gastric cancer[J]. Oncotarget, 2014, 5 (14): 5663-73. DOI: 10.18632/oncotarget. |
| [33] | Ding X, Yang Y, Sun Y, et al. MicroRNA-585 acts as a tumor suppressor in non-small-cell lung cancer by targeting hSMG-1[J]. Clin Transl Oncol: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico, 2016 : 1-7. |
| [34] | Li Z, Cao Y, Jie Z, et al. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3[J]. Cancer Lett, 2012, 323 (1): 41-7. DOI: 10.1016/j.canlet.2012.03.029. |
| [35] | Qin H, Sha J, Jiang C, et al. mir-122 inhibits metastasis and epithelial-mesenchymal transition of non-small-cell lung cancer cells[J]. Onco Targets Ther, 2015, 8 : 3175-84. |
| [36] | Wen XP, Ma HL, Zhao LY, et al. MiR-30a suppresses non-small cell lung cancer progression through AKT signaling pathway by targeting IGF1R[J]. Cell Mol Biol(Noisy-le-grand), 2015, 61 (2): 78-85. |
| [37] | Hou C, Sun B, Jiang Y, et al. MiRNA-31 inhibits lung adenocarcinoma stem-like cells via down-regulation of MET-PI3k-Akt signaling pathway[J]. Anticancer Agents Med Chem, 2016, 16 (4): 501-18. DOI: 10.2174/1871520615666150824152353. |
| [38] | Zhou JY, Chen X, Zhao J, et al. MicroRNA-34a overcomes HGFmediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET[J]. Cancer lett, 2014, 351 (2): 265-71. DOI: 10.1016/j.canlet.2014.06.010. |
| [39] | Song L, Li D, Gu Y, et al. MiRNA-126 targeting PIK3R2 inhibits NSCLC A549 cell proliferation, migration, and invasion by regulation of PTEN/PI3K/AKT pathway[J]. Clin Lung Cancer, 2016, 17 (5): e65-e75. DOI: 10.1016/j.cllc.2016.03.012. |
| [40] | Ma Y, Xia H, Liu Y, et al. Silencing miR-21 sensitizes non-small cell lung cancer A549 cells to ionizing radiation through inhibition of PI3K/Akt[J]. Biomed Res Int, 2014, 2014 : 617868. |
| [41] | Jang JS, Jeon HS, Sun Z, et al. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers[J]. Clin Cancer Res, 2012, 18 (13): 3658-67. DOI: 10.1158/1078-0432.CCR-11-2857. |
| [42] | Niu X, Liu S, Jia L, et al. Role of MiR-3619-5p in β-catenin- mediated non-small cell lung cancer growth and invasion[J]. Cell Physiol Biochem, 2015, 37 (4): 1527-36. DOI: 10.1159/000438520. |
| [43] | Tan M, Wu J, Cai Y. Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer[J]. Biochem Biophys Res Commun, 2013, 438 (4): 673-9. DOI: 10.1016/j.bbrc.2013.07.123. |
| [44] | Fang L, Cai J, Chen B, et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/[J]. Nat Commun, 2015, 6 : 8640. DOI: 10.1038/ncomms9640. |
| [45] | Zhang X, Ke X, Pu Q, et al. MiRNA-410 acts as oncogene in NSCLC through downregulating SLC34A2 via activating Wnt/ β-catenin pathway[J]. Oncotarget, 2016, 7 (12): 14569-85. |
| [46] | Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer[J]. Proc Natl Acad Sci USA, 2014, 111 (34): E3553-61. DOI: 10.1073/pnas.1412686111. |
| [47] | Luo J, Li H, Zhang C. MiRNA-7 inhibits the malignant phenotypes of nonsmall cell lung cancer in vitro by targeting Pax6[J]. Mol Med Rep, 2015, 12 (4): 5443-8. |
| [48] | Zhou YL, Xu YJ, Qiao CW. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer(NSCLC)by inhibiting PAC1/MAPK pathway[J]. Int J Clin Exp Pathol, 2015, 8 (6): 6312-22. |
 2017, Vol. 37
2017, Vol. 37

