2. Guangzhou Forensic Science Institute, Guangzhou510030, China
2. 广州市刑事科学技术研究所, 广东 广州 510030
The PowerPlex®21 System is a widely used commercial kit for short-tandem repeat (STR) genotyping[1-3] , which yields reliable results with a robust design[4] . Developed by Promega Corporation in 2012, this system has been shown to have a good inhibitor tolerance, a rapid PCR protocol and a good compatibility with direct amplification of FTA card punches and pretreated swabs[3] . This system contained 17 combined DNA index system (CODIS) loci (the original set including D3S1358, D13S317, D16S539, D18S51, CSF1PO, TH01, vWA, D21S11, D7S820, D5S818, TPOX, D8S1179 and FGA with an additional set including D1S1656, D2S1338, D12S391 and D19S433) and D6S1043, Penta E and Penta D, which contains more loci than other widely used kits such as PowerPlex 16 System (Promega, Madison, WI, USA), AmpFℓ STR Identifier System (Applied Biosystems, Foster City, CA, USA), AmpFℓ STR Sinofiler System and GoldeneyeTM DNA ID system 20A kit (Goldeneye Ltd, Beijing, China).
So far in China little population-based genetic data of the PowerPlex®21 System have been available, especially in terms of the locus D1S1656. In this study, we carried out a population investigation using this kit among the Chinese Han population living in Guangdong Province of South China. The data we obtained may enrich the international DNA database that can be used as a reference for human identification in forensic studies and genetic diversity studies.
SUBJECTS AND METHODS PopulationBloodstains and mouth swabs of 2367 unrelated Chinese Han individuals living in Guangdong Province in South China were collected from the Center of Forensic Science, Southern Medical University (Guangzhou, China). The native birthplaces of these individuals were mostly Guangdong Province (1744 individuals), and a small portion of them were born in nearby provinces and regions including Guangxi Zhuang Autonomous Region (71 individuals), Hunan Province (196 individuals), Sichuan Province (175 individuals), Jiangxi Province (83 individuals) and Hubei Province (97 individuals). Informed consent to participating in this study was obtained from all the individuals.
DNA Extraction and PCR amplificationGenomic DNA was extracted according to the Chelex-100 method and proteinase K protocol[5] . The extracted DNA was amplified using the PowerPlex®21 System (Promega) in the GeneAmp 9700 PCR System (Applied Biosystems) following the manufacturer's protocol.
Genotyping and quality controlThe PCR products were detected by capillary electrophoresis in ABI 3130xl Genetic Analyzer (Applied Biosystems) using CC5 ILS 500 size standard and reference allelic ladder provided along with the PowerPlex®21 System. Data analysis and genotyping were performed automatically using GeneMapper ID v3.2 software (ABI Company, Forster City, USA). The internal controls (negative control and the 2800 M DNA positive control) were genotyped along with each batch of samples to ensure the reproducibility and accuracy of the results.
Data analysisThe allelic frequencies, matching probability, power of discrimination, power of exclusion, polymorphism information content and typical paternity index were calculated using the PowerStats V12.xls software (http://www.promega.com/geneticidtools/). The P values of exact test of Hardy-Weinberg's equilibrium, expected heterozygosity and observed heterozygosity were calculated using Genepop software (http://genepop.curtin.edu.au/). The allele frequencies in this southern Chinese Han population were compared with the published data of other populations using Fst pairwise distance by Arlequin v3.5 software[6] . A phylogenetic tree showing the inter-population relationshipwas constructed with the neighbor-joining method and based on Fst distances with 1000 bootstrap replications by software package POPTREE2[7] according to the allele frequency data of 15 shared STR loci (D3S1358, D13S317, D16S539, D18S51, D2S1338, CSF1PO, TH01, vWA, D21S11, D7S820, D5S818, TPOX, D8S1179, D19S433 and FGA) from this present study and 13 reported populations. A principal component analysis (PCA) plot was drawn with Past 3.11 software[8] based on allelic frequencies of 15 STRs.
RESULTSTab. 1 shows the statistical parameters of the 20 STR loci in this Han population. In this population, the expected heterozygosity ranged from 0.6040 (TPOX) to 0.9120 (Penta E), and the observed heterozygosity ranged from 0.6054 (TPOX) to 0.9071 (Penta E); the and 0.9852 (Penta E), and the combined power of discrimination for the 20 STR loci was over 0.999 999 999 999 999 999 999 999 999 999. The power of exclusionvaried between 0.2974 (TPOX) and 0.8099 (PentaE), and the combined probability of excluding paternity for the 20 loci was over 0.999 999 999 999 999 999 999 999 999 999. The polymorphism information content varied between 0.5428 (TPOX) and 0.9053 (Penta E). Among all the studied loci, no significant deviations from Hardy-Weinberg expectations were observed after Bonferroni correction[9] except for the locus D5S818 (P<0.0025).
| Table 1 Allele frequencies and forensic parameters of 20 STR loci in the southern Chinese Han population (n=2367) |
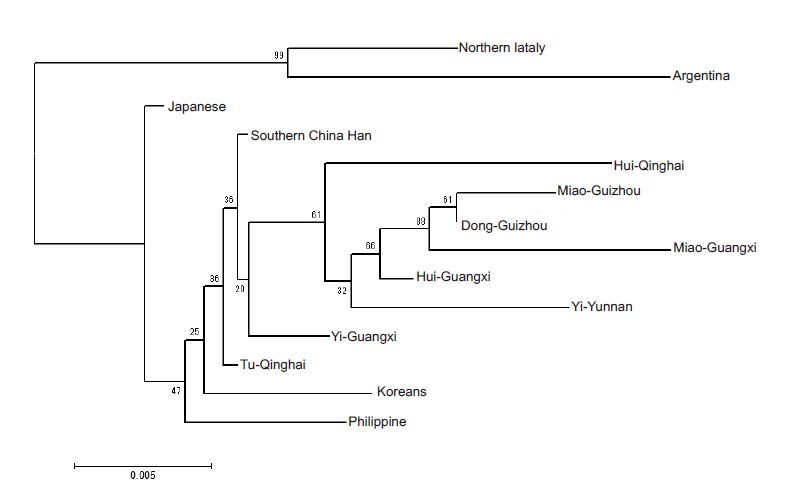
The differences between this Chinese Han population and other populations are shown in Tab. 2 and Tab. 3. Fst distance was used to compare this population and 13 other reported populations in relation to the allele frequencies of 15 autosomal STR loci. The neighbor-joining phylogenetic tree generated is shown in Fig. 1. The Northern Italian and Argentinian populations were clustered by one branch, and the Asian populations were clustered by another branch. Seven Chinese ethnic minority groups containing Hui (Qinghai), Miao (Guizhou), Dong (Guizhou), Miao (Guangxi), Hui (Guangxi), Yi (Guangxi) and Yi (Yunnan) were clustered by one small branch, and Tu (Qinghai) was far away from the other ethnic minority groups. The southern Chinese Han population in this study was near Yi (Yunnan) group.
| Table 2 Genetic distances between the southern Chinese Han population and 8 reported Chinese ethnic minority populations (Fst and P values) |
| Table 3 Genetic distances between the southern Chinese Han population and some different foreign populations (Fst and P values) |
|
Figure 1 Neighbor-joining tree based on Fst distances estimated among the 14 populations. The northern Italian and Argentinian populations were clusteredto one branch andthe other populations were clustered to another branch; Hui (Qinghai), Miao (Guizhou), Dong (Guizhou), Miao (Guangxi), Hui (Guangxi), Yi (Guangxi) and Yi (Yunnan) populations were clustered to one small branch. |
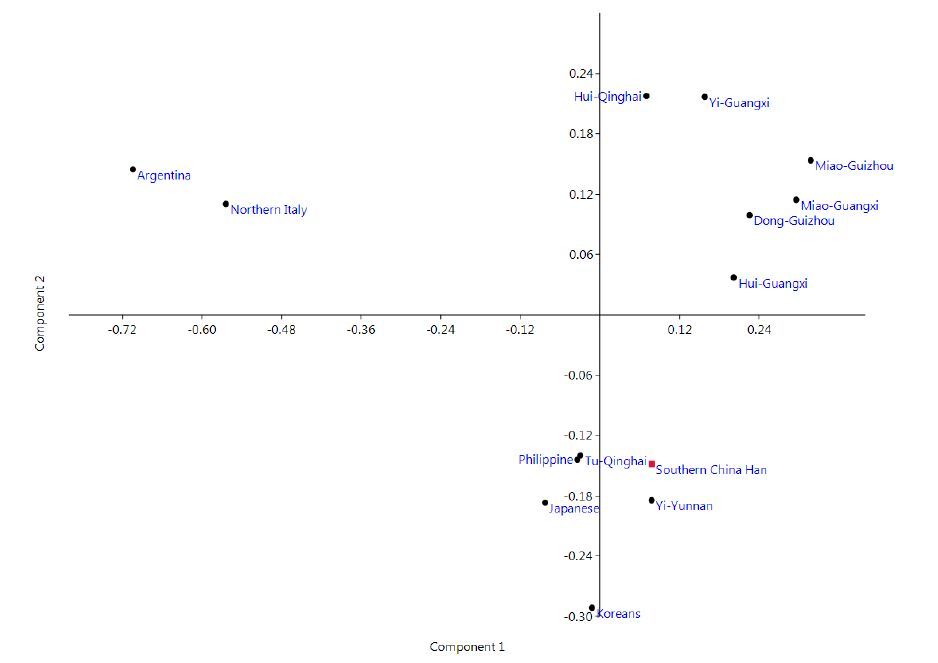
Principal component analysis of allele frequencies in this Han population under study and 13 reported populations was performed. As shown in Fig. 2, the Argentinian and northern Italian populations were on the left upper quadrant, and 6 Chinese ethnic minority populations were up on the right. The Japanese, Philippine and Korean populations were on the left lower quadrant. The southern Chinese Han population under study and Yi (Yunnan) population were on the right lower quadrant.
|
Figure 2 Principal component analysis plot based on allele frequencies of the 20 loci in the 14populations. The Argentinian and northern Italian populations were clusteredon the left upper quadrant; The Chinese Hui (Qinghai), Miao (Guizhou), Dong (Guizhou), Miao (Guangxi), Hui (Guangxi), and Yi (Guangxi) populations were clustered up on the right; The Japanese, Philippine and Korean populations were clustered on the left lower quadrant. The southern Chinese Han population and the Yi (Yunnan) population were clustered on the right lower quadrant. |
In this study, we evaluated the performance of PowerPlex®21 System and presented allele frequencies and forensically relevant statistical parameters of 20 STR loci in a Chinese Han population in South China.
A STR locus can be considered highly polymorphic when its PD value is over 0.80 or/and its PE value is over 0.50[10] . The results of this study showed that most of the examined loci were highly polymorphic. The combined discrimination power and the probability of excluding paternity of the 20 STR loci were both over 0.999 999 999 999 999 999 999 999 999 999, suggesting that PowerPlex®21 System is suitable for forensic personal identification and paternity testing.
We found that after Bonferroni correction (P<0.05/ 20 [0.0025] ), the P value of exact test for Hardy-Weinberg equilibrium was less than 0.0025 (P= 0.0013) for the locus D5S818. This divergence from Hardy-Weinberg equilibrium may be due to an excess of homozygotes. In contrast, the discrepancies reported in this study can be easily identified by using two commercial kits from different manufacturers. We previously showed the existence of a silent allele of D5S818, caused by mutations at primer-binding sites, in 6 members of 3 paternity cases[11] . The primers for D5S818 in this system need to be optimized to better adapt to southern Chinese Han population.
In this southern Chinese Han population, we found significant differences from Yi (Guangxi) group[12] at 8 STR loci from, from Hui (Guangxi) group[12] at 3 STR loci, from Miao (Guangxi) group[12] at 3 STR loci, from Dong (Guizhou) group[13] at 5 STR loci, from Miao (Guizhou) group[14] at 10 STR loci, from Tujia (Qinghai) group[15] at 2 STR loci (vWA and D7S820), from Yi (Yunnan) group[16] at 2 STR loci (D3S1358 and D21S11), from Hui (Qinghai) group [17] at 10 STR loci, from Philippine group[18] at 9 STR loci, and from Korean group[19] at 8 STR loci. Significant differences were found in this Chinese Han population at all the loci except for D18S51 and D5S818 from the Japanese population [20] and at all these 20 STR loci from the northern Italian population [21] . Only two STR loci (D16S539 and CSF1PO) showed no significant differences between this southern Chinese Han population and the Argentinian population [22] . The southern Chinese Han population showed significant differences at the locus D1S1656 from 5 foreign populations and also from the Han populations in Beijing (P<0.0001)[23] , Zhejiang Province (P<0.0001)[24] and Fujian Province of China (P<0.0001)[25] . These results indicate that D1S1656 hasa highly ethnic diversity and is suitable for Chinese population.
The neighbor-joining phylogenetic tree showed the clustering of all the Asian populations as one group and of the northern Italian and Argentinian populations as another. The southern Chinese Han population showed the nearest affinities to Yi (Yunnan) population. The cluster branch of the Chinese populations showed the nearest affinity between Guangxi and Guizhou minority populations, while the Tu (Qinghai) population branched away from the other Chinese populations. The phylogenetic tree showed that the populations were clustered basically consistent with their distributions on the continental plate. The result of principal component analysis was in agreement with the phylogenetic tree and indicated a clear pattern of regional distribution.
In conclusion, our results demonstrate that these 20 STR loci can provide highly informative polymorphic data for paternity testing, individual identification and genetic population studies. PowerPlex®21 System can serve as an efficient tool in forensic science and in anthropology of southern Chinese Han population.
| [1] | Zhang X, Hu L, Du L, et al. Genetic polymorphisms of 20 autosomal STR loci in the Vietnamese population from Yunnan Province, Southwest China[J] . Int J Legal Med, 2016.[Epub ahead of print] |
| [2] | Farhan MM, Hadi S, Iyengar A, et al. Population genetic data for 20 autosomal STR loci in an Iraqi Arab population: application to the identification of human remains[J]. Forensic Sci Int Genet,2016, 25 : e10-e1. DOI: 10.1016/j.fsigen.2016.07.017. |
| [3] | Gray K, Crowle D, Scott P. Direct amplification of casework bloodstains using the Promega PowerPlex (R) 21 PCR Amplification System[J]. Forensic Sci Int Genet,2014, 12 : 86-92. DOI: 10.1016/j.fsigen.2014.05.003. |
| [4] | Ensenberger MG, Hill CR, McLaren RS, et al. Developmental validation of the PowerPlex (R) 21 System[J]. Forensic Sci Int Genet,2014, 9 : 169-78. DOI: 10.1016/j.fsigen.2013.12.005. |
| [5] | Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material[J]. Biotechniques,1991, 10 (4) : 506-13. |
| [6] | Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour,2010, 10 (3) : 567-7. |
| [7] | Takezaki N, Nei M, Tamura K. POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface[J]. Mol Biol Evol,2010, 27 (4) : 747-52. DOI: 10.1093/molbev/msp312. |
| [8] | Hammer ?, Harper D, Ryan P. PAST: Paleontological Statistics Software Package for education and data analysis[J]. Palaeontolia Electronica,2001, 4 (1) : 1-9. |
| [9] | Bland JM, Altman DG. Multiple significance tests: the Bonferroni method[J]. BMJ,1995, 310 (6973) : 170. DOI: 10.1136/bmj.310.6973.170. |
| [10] | Shriver MD, Jin L, Boerwinkle E, et al. A novel measure of genetic distance for highly polymorphic tandem repeat loci[J]. Mol Biol Evol,1995, 12 : 914-20. |
| [11] | Chen L, Tai Y, Qiu P, et al. A silent allele in the locus D5S818 contained within the PowerPlex?21 PCR Amplification Kit[J]. Leg Med (Tokyo),2015, 17 (6) : 509-11. DOI: 10.1016/j.legalmed.2015.10.012. |
| [12] | Liu C, Liu C, Wang H. STR data for the 15 loci from three minority populations in Guangxi municipality in South China[J]. Forensic Sci Int,2006, 162 (1-3) : 49-52. DOI: 10.1016/j.forsciint.2006.06.015. |
| [13] | Zhang L. Population data for 15 autosomal STR loci in the Dong ethnic minority from Guizhou Province, Southwest China[J]. Forensic Sci Int Genet,2015, 16 : 237-8. DOI: 10.1016/j.fsigen.2015.02.005. |
| [14] | Zhang L, Zhao Y, Guo F, et al. Population data for 15 autosomal STR loci in the Miao ethnic minority from Guizhou Province, Southwest China[J]. Forensic Sci Int Genet,2015, 16 : e3-4. DOI: 10.1016/j.fsigen.2014.11.002. |
| [15] | Zhu B, Yan J, Shen C, et al. Population genetic analysis of 15 STR loci of Chinese Tu ethnic minority group[J]. Forensic Sci Int,2008, 174 (2-3) : 255-8. DOI: 10.1016/j.forsciint.2007.06.013. |
| [16] | Zhu BF, Shen CM, Wu QJ, et al. Population data of 15 STR loci of Chinese Yi ethnic minority group[J]. Leg Med (Tokyo),2008, 10 (4) : 220-4. DOI: 10.1016/j.legalmed.2007.12.004. |
| [17] | Deng YJ, Zhu BF, Shen CM, et al. Genetic polymorphism analysis of 15 STR loci in Chinese Hui ethnic group residing in Qinghai province of China[J]. Mol Biol Rep,2011, 38 (4) : 2315-22. DOI: 10.1007/s11033-010-0364-z. |
| [18] | Rodriguez JJ, Salvador JM, Calacal GC, et al. Allele frequencies of 23 autosomal short tandem repeat loci in the Philippine population[J]. Leg Med (Tokyo),2015, 17 (4) : 295-7. DOI: 10.1016/j.legalmed.2015.02.005. |
| [19] | Shin CH, Jang P, Hong KM, et al. Allele frequencies of 10 STR loci in Koreans[J]. Forensic Sci Int,2004, 140 (1) : 133-5. DOI: 10.1016/j.forsciint.2003.11.027. |
| [20] | Fujii K, Iwashima Y, Kitayama T, et al. Allele frequencies for 22 autosomal short tandem repeat loci obtained by PowerPlex Fusion in a sample of 1501 individuals from the Japanese population[J]. Leg Med (Tokyo),2014, 16 (4) : 234-7. DOI: 10.1016/j.legalmed.2014.03.007. |
| [21] | Turrina S, Ferrian M, Caratti S, et al. Evaluation of genetic parameters of 22 autosomal STR loci (PowerPlex? Fusion System) in a population sample from Northern Italy[J]. Int J Legal Med,2014, 128 (2) : 281-3. DOI: 10.1007/s00414-013-0934-4. |
| [22] | Parolin ML, Real LE, Martinazzo LB, et al. Population genetic analyses of the Powerplex? Fusion kit in a cosmopolitan sample of Chubut Province (Patagonia Argentina)[J]. Forensic Sci Int Genet,2015, 19 : 221-2. DOI: 10.1016/j.fsigen.2015.07.020. |
| [23] | Zhang QX, Yang J, Liu YC, et al. Genetic polymorphisms of 16 non-CODIS STR loci in Beijing Han population[J]. Fa Yi Xue Za Zhi,2013, 29 (3) : 206-8. |
| [24] | HAO Honglei, WU Weiwei, LV Dejian, et al. Genetic polymorphism of D1S1656, SE33 and D2S1338 STR loci of Han population in Zhejiang[J]. Forensic Sci Tech,2012, 2 : 53-4. |
| [25] | Zhang W, Yang K, Zhan YY, et al. Genetic polymorphism of 3 STR loci of Han population in Fujian[J]. Forensic Sci Tech,2015, 9 : 158-9. |
 2017, Vol. 37
2017, Vol. 37
