2. Department of Oncology, First People's Hospital of Qinhuangdao,Qinhuangdao 066004, China ;
3. Department of InternalMedicine, YanshanUniversity Hospital3, Yanshan University, Qinhuangdao 066004, China
2. 秦皇岛市第一人民医院肿瘤科,河北秦皇岛066004 ;
3. 燕山大学校医院内科,河北秦皇岛066004
HIV infectionis known to stimulate the production of antibodies againstsuch antigens as envelope proteins (gp120 and gp41) and core protein(P24) of HIV [1, 2]. Typically,HIV can be detected two or three weeks after its infection,and the antigensin the blood can serve as serum markersduring the windowperiod of AIDS to allow early diagnosis and treatment [3, 4]. Currently the two main strategies for recognition and detection of the antigens rely on the detection of the interactions between antibodyand antigen and between DNA aptamers and antigen [5]. Compared with antibody/ antigen interaction,DNA aptamers have many advantages including a higher stability due to theirlow degradationrate [6, 7],a strongerand specific binding affinity to the targetmolecule [8, 9],a shorterscreening cycle (one or two months)[10, 11],a low immunogenicity,potential of repeatable application due to their quick denaturation and renaturation,absenceof the function of effector,a better sensitivity and facile chemical modification[12, 13],and a lower cost. So far a varietyof DNA aptamers with specific bindingaffinity to gp 120 and P24 have been screened for detecting the antigens for early diagnosis of HIV infection[14, 15],but no such a DNA aptamerhas been reportedwith a highly specific affinity to the antigengp 41,which plays an important role in the process of virus packaging and maturation of HIV[16, 17].
Herein we report,for the first time,the screeningof 4 DNA aptamerswith specific bindingaffinity to gp41 antigen throughsubtractive SELEX screening with commercialcarboxyl-enriched agar magnetic beads as the separation medium and subsequentconventional quantitativePCR (qPCR) techniqueand gel electrophoresis. These aptamers provide a basis for efficient,rapid and accurate detection of gp41 antigen for early diagnosis of HIV[18, 19] and may greatly facilitate the development of future high-throughput screening of HIV with a clear molecular mechanism[20].
MATERIALS AND METHODS MaterialsThe single-stranded DNA (ssDNA; 5'-CTATAGCAATG GTACGGTACTTCC[40N]-CAAAAGTGCACGCTACTT-
TGCTAA-3'),P6 primer (5'-TAATACGACTCACTATAG CAATGGTACGGTACTTCC-3'),P9 primer (5'-TTAGC AAAGTAGCGTGCACTTTTG-3'),biotin-P9 primer,universal primerM13-47 (5'-CGCCAGGGTTTTCCCAG TCACGAC-3'),and PCR mix were purchased from TaKaRa. pMDTM 18-Tvector vector and the Gel Extraction Kit were obtained from KGI NanjingBiological TechnologyDevelopment Co.,Ltd.
Buffer solutionsPBS (0.01 mol/L,pH 7.4) was purchased from Jiangsu Bollywood Biotechnology Co.,Ltd,which contained 137 mmol/L of NaCl,2.7 mmol/L of KCl,6.5 mmol/L of Na2HPO4,1.8 mmol/L of NaH2PO4,2.5 mmol/L of MgCl2,1 mmol/Lof CaCl2,and 1% (mass ratio) bovineserumalbumin (BSA) and used as the 1×screening buffer. The 1×binding buffer contained PBS (0.01 mol/L,pH 7.4),1 mmol/L of CaCl2,and 1 mg/mL of BSA. The blockingsolution contained 0.01 mol/L of PBS,1 mg/mL of BSA,and 1 mg/mL of salmon spermDNA.
Aptamer selectionssDNA librarypretreatment ssDNA library was centri⁃ fuged at 10 000 r/min for 5 min followedby addition of600 μL binding buffer,and the mixturewas well mixed.
Two roundsof positive screening with HIV gp41 antigen In the first roundof positive screening,0.3 mL of magnetic beads and 300 μL of HIV gp41 antigen (diluted at 1∶100 according to the instruction) were added in a 1.5-mL EP tube and incubated for 2 h at 37 ℃ . The productwas magnetically separated and rinsed with the screeningbuffer for 3 times,followed by the additionof 500 μL of the blockingsolution. After incubation at 37 ℃ for 1 h,the productwas separated and washedwith the screeningbuffer twice (for 1 min each time)and with the binding bufferonce for 1 min before the addition of the primaryssDNA library. The above mixture was treated at 95 ℃ for 5 min,4 ℃ for 10 min,and 37 ℃ for 5 min,and then incubated for 1 hat 37 ℃. The composites of magnetic beads and ssDNA library were separated and washed for 3 times with the screening buffer (1 min each time); 200 μL deionized water was then added and the mixture was centrifuged at 95 ℃ for 10 min. The supernatant was collected for use as the secondary libraryfor the next round of screening. The secondround of positivescreening was carriedout following the identicalprocedure in the first round of screening except that the amount of magnetic beads was 0.15 mL.
Amplification of ssDNA into dsDNA by qPCR The PCR template (160 μL) was added in half of the above solution and mixed thoroughly. The homogeneous mixture was then dividedinto 8 equal portions (30 μL) for qPCR until the amplification fluorescence reached the highest pointof the S-curve.
Preparation of the secondary ssDNA library Streptavidin-biotin magnetic beads (0.1 mL) and the amplified double-stranded DNA (dsDNA) were added into a 1.5-mL EP tube and incubatedfor 30 min at 37 ℃. The mixture of the recovered beads and 500 μL of 1% (mass ratio) TP BS was incubated at 37 ℃ for 4 min,and the procedure was repeated for 3 times. The recovered beads were washed with water followedby the addition of 200 μL of the binding buffer. After incubation at 95 ℃ for 5 min,the recoveredsupernatant was added into a counter-screening tube and incubated at 37 ℃ for 45 min. The recovered supernatant was subsequently added into a positive-screening tube and incubated for 1 h to remove unbound ssDNA library. All the recovered beads after positive-screening and counter-screening were washed with the screening buffer for 3 times (3 min each time),after which 200 μL of deionized water was added. After incubation at 95 ℃ for 5 min,the supernatant was recovered for use as the secondary libraries in the next round of screening.
Enrichment effectmonitoring PCR template (10 μL)was added into 20 μL of recovered supernatant after positive-screening and counter-screening,respectively. The corresponding homogeneous solutions were amplified by qPCR,with 20 μL of PCR template in 10 μL of deionizedwater as the control sample.
SELEX screeningwith HIV P24 antigenas counter-screening target molecule SELEX screening with HIV P24 antigenas the counter-screening targetmolecule was conducted followingidentical screening procedures described above except for the replacement of gp41 antibody by HIV P24 antigen in the third round of screening. The conditions in each round of SELEX screening were listed in Tab. 1.
Cloning and sequencing of selected aptamersdsDNA obtainedthrough secondary library The recovered supernatant in the positive-screening tube after 6 rounds of selection was used as the templateto form 300 μL of PCR reaction system,which consisted of 30 μL of 10×buffer containing Mg2 + ,45 μL of template,2.4 μL of dNTP,3 μL of P6 primer,3 μL of P9 primer,2 μL of rTaq enzyme,and 214.6 μL of deionized water. The homogeneous PCR reaction was divided into 6 equal portions and amplified by routine PCR for 26 thermal cycles of 95 ℃ for 5 min,94 ℃ for 30 s,55 ℃ for 30 s,and 72 ℃ for 30 s. The dsDNA fragment was recovered throughgel electrophoresis from the thermally-cycled solution.
Connection of dsDNA fragmentwith pMDTM18-T vector The recovered dsDNA was dissolvedin deionized water and connected with pMDTM18-T vector to prepare the connection solution,which consisted of 0.5 μL of pMDTM18-Tvector,2 μL of inserted fragment,5 μL of solution and 383.5 μL of deionized water. The connection solutionwas reacted at 16 ℃ for 30 min to obtain the secondary library dsDNA recombinant plasmid.
E. coli transformation and selection of positive clones The originalstrain of E. coli DH5α was cultured and shaken till logarithmic growth (with absorbance at 600 nm of 0.6). Competent bacterial cells (200 μL) were prepared by treatment with 0.1 mol/L CaCl2 and transferred into a 1.5-mL EP tube for storageat-80 ℃ before use. For E. coli transformation,10 μL of recombinant DNA or 10 μL of deionizedwater were added into the thawed competent cells,which were coated on the plate and culturedon a solid LB medium containing ampicillin to select the positive clones.
| Table 1 Conditions of 6 roundsof SELEX screening |
Stagger extensionprocess (StEP) to identify positive clones The monoclonal bacteria preserved at-80 ℃ was used as a template system,and the blank group was set. Staggered PCR system of the universal primers >M13-47 and P6/P9 consisted of 10 μL of 10×PCR buffer containing Mg2 + ,2 μL of template,8 μL of dNTP,1 μL of primer P6/P9,1.5 μL of rTaq enzyme and 80.7 μL of deionized water. The homogeneous PCR reaction was divided into 10 equal portions and amplified by routine PCRfor 30 thermal cycles (95 ℃ for 5 min,94 ℃ for 30 s,55 ℃ for 30 s,and 72 ℃ 30 s). The direction of the target fragment inserted into the plasmid was determined by non-denaturing polyacrylamide gel electrophoresis.
Identification of specific affinity of HIV gp41 antigen aptamer to positive clones The positive clones identified by StEP technique were used as the templateto synthesize ssDNA using asymmetric PCR method. Agar beads (0.2 mL) were packed in 4 tubes labeled with "+","1-","2-","3-",which were coupled with HIV gp41 antigen,anti-HBsAg antibodies,HIV P24 antigen,and BSA,respectively. The specific affinities of the aptamers to these proteins were determined with qPCR analysis.
Flow cytometry analysis The specific affinities of the four aptamersto HIV gp41 antigen were measured to identify the aptamer with the strongestspecificity. To monitor the enrichment of the aptamersafter selection,1×105 beads binding to HIV gp41 antigenand 1×105beads binding to HIV P24 antigen were incubated with FITC-labeled ssDNA in the binding buffer (Cf=200 nmol/L, Vf=200 μL) at 37 ℃ for 45 min. The fluorescence intensity of the beads was monitoredwith a FACS Aria cytometer by counting 10 000 events.The FITC-labeled unselected ssDNA library servedas the negative control. The binding affinities of the aptamers were evaluatedby flowing cytometry,with the initialFITC-labeled library as the negative controlto determine the background binding. All the binding assays were performed 3 times. The Kd of the interaction between the aptamerand theHIV gp41 antigen was determined using the equation Y=Bmax X∕(Kd+X).
The process of aptamer selection by SELEX was illustrated in Fig. 1.
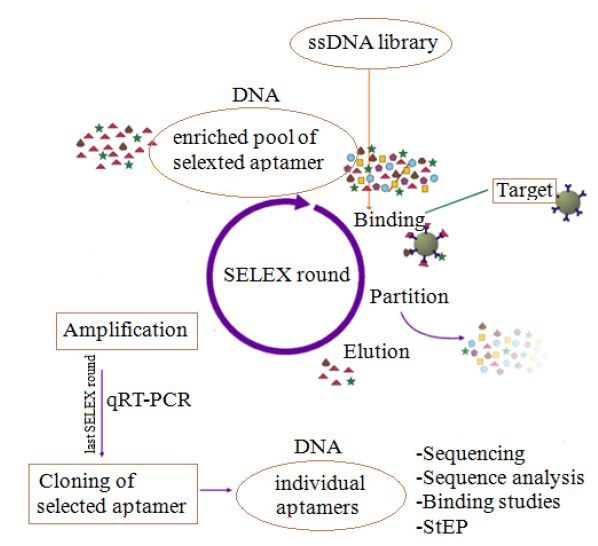
|
Figure 1 Processof aptamer selectionby SELEX. |
Through two rounds of positive screeningand 4 rounds of negative screening,4 highly specific aptamers of HIV gp41 antigenwere obtained. The qPCR resultsof positive selection and negative selection in the third-round counter-selection (Fig. 2) showed enrichment of the selected aptamersin the positive-selection tube. Although the specific aptamersbinding with the fixed target graduallyincreased in ssDNA library,aptamer enrichment did not reach saturation. Furtherscreening with an increased screening pressure was needed to obtain the highly specific aptamers binding with HIV gp41 antigen.
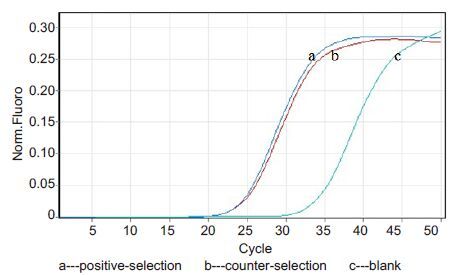
|
Figure 2 qPCR detection of the third round of negative selection. |
As shown in Fig. 3,qPCR results of positive selection and counter selectionin the fourth round of counter selection showed that the enrichment of the specific aptamersbinding with HIV gp41 antigen reached saturation in the positive-selection tube,suggesting that the non-specific ssDNA could be quickly eliminated by magnetic bead-based screening to increase the enrichment of the specificaptamers.
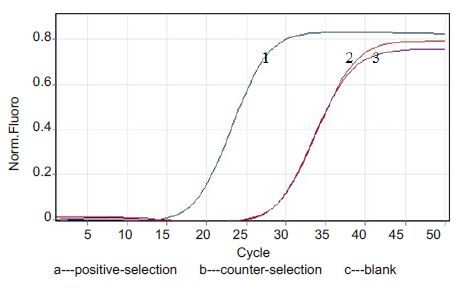
|
Figure 3 qPCR detection after the fourth and fifth rounds of counter-selection. |
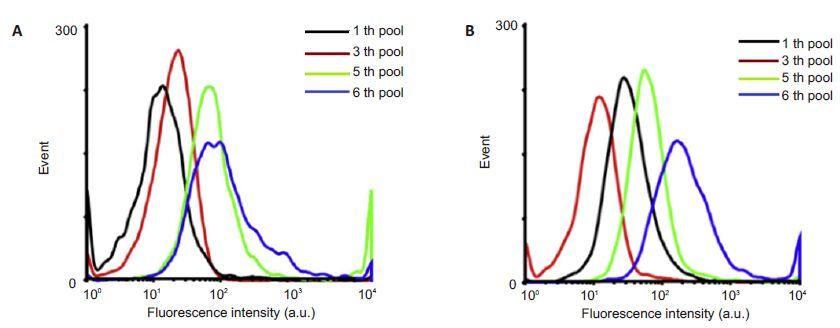
The productsof the sixth round of screening were cloned and sequencedto obtain the sequences of the 4 aptamers (Tab. 2).The secondary structures of the 4 aptamers of HIV gp41 antigen were shown in Fig. 4. Fig. 5 shows the enrichment of the DNA sequences that bind to HIV P24 and gp41 antigens.
| Table 2 Sequences of the 4 aptamersbinding with HIV gp41 antigen |
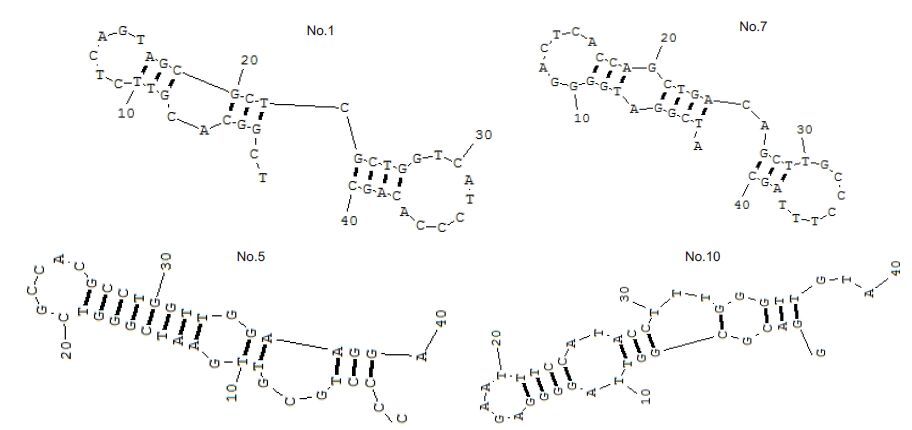
|
Figure 4 Stem-loop structures of the 4 selected aptamersbinding with HIV gp41 antigen as predictedby RNA Structure 3.7 software. |
|
Figure 5 Flow cytometry to monitor the specific affinity of the beads to HIV antigens P24 (A) and gp41 (B). |
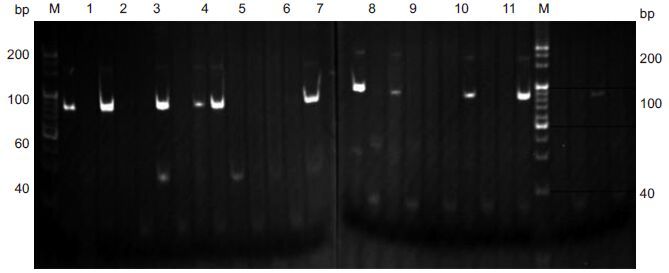
The positiveclones were identified by StEP technique. A clearcombined band of 170-180 bp appearedin some clones but not in the other clones,demonstrating successful transformation by the recombinant plasmids. The selected combination of primers M13-47 and P7x with nucleicacid bands at 170-180 bp demonstrated that the fragment was inserted forwardlyinto the vector,and the selected combination of the primersM13-47 and P11x with bands at 170-180 bp indicateda reverse insertion of the fragmentin the vector; no such bands occurred in the blank group following amplification with the two primers.A total of 9 positivemonoclones were identified,including4 with positive insertion and 5 with reverse insertionas shown by electrophoresis analysis (Fig. 6).
|
Figure 6 Electrophoresis of 9 positive monoclines identified using StEP technique.Lanes 6,11: Blank controlgroup. M: Marker. |
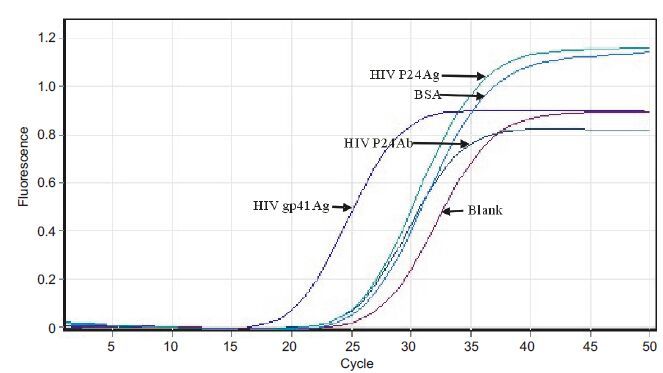
Fig. 7 showssuccessful conversion of the positiveclones with specific binding with HIV gp41. The No.15 positive clone had a highly specific affinityto HIV gp41 without adsorption to other non-specific proteins.
|
Figure 7 Identification of the specificaptamer of HIV gp41. |
SELEX techniqueis the first requirement for designing and synthesizing random ssDNA librariesof suitable length (60-100 bp). The central segment of the library is a randomsequence containing 20-60 base pairsflanked by a primerannealing sequence as the lateralsegments. Theoretically,a random sequencecontains 20 bases of random ssDNA library,in which at least 1×1015 different sequences could be formed so that the storage capacity of the library could reach at least 1×1015.
Magnetic beads can be used as a good separation medium due to their affinity to a variety of target proteins. Ligandsbound to the beads could be rapidly separated from unbound ssDNA library by washing the beads with the screening buffer. As biotin and streptavidin have the strongestinteraction known in nature,commercial streptavidin-biotin magnetic beads were utilizedto screen HIV gp41 antigen aptamers.
The library enrichment is an importantindicator of SELEX screening.The aptamers with affinity to HIV gp41 antigenwere successfully obtainedafter two rounds of positive-screening and 4 rounds of counter-screening. Analysis of positive-screening and counter-screening using qPCR in the third round of counter-screening showed that the screenedaptamer began to be enriched in positive-screening tube. Although the specific aptamersbinding with the fixed target graduallyincreased in ssDNA library,aptamer enrichment did not reach saturation. After the fourth round of counter-selection,the specific aptamers binding with HIV gp41 antigen reachedsaturation in the positive-selection tube. Subsequent StEP and Kd value measurement indicatedthat the aptamersobtained by magnetic beads-based SELEX technique possessed a high specificity to HIV gp41 antigen.The utilization of qPCR,gel electrophoresis and other conventional techniques simplifies the procedures for screening the aptamers and shortens screening time. The aptamers obtained may facilitate future high-throughput screening and futuredevelopment of rapid test kits of HIV with a low cost.
In this study,the aptamerswith a highly specific affinity to HIV gp41 antigen were successfully obtained after 6 rounds of screening with magnetic beads.To reduce the non-specific adsorption of ssDNA as much as possible to obtain highly specific aptamers,the coupled targets were replaced in the counter-selection tube in the third round of screening. The specific aptamer obtained provides an efficient,rapid and accurateprobe for early diagnosis of HIV by detecting the gp41 antigen.
| [1] | Wang CY, Wu CY, Hung TC. Sequence-constructive SELEX: a newstrategy for screening DNA aptamer binding to Globo H[J]. BiochemBioph Res Commun,2014, 452 (3) : 484-9. DOI: 10.1016/j.bbrc.2014.08.086. |
| [2] | Shao NS, LI SH, Huang YP, et al. Advances in the SELEX techniqueand aptamer[J]. Prog Biochem Biophys,2006, 33 (4) : 329-35. |
| [3] | Ghamar SD, Mohammad JR, Ali ML, et al. Selection of DNA aptamersagainst human cardiac troponin I for colorimetric sensor based dotblot application[J]. J Biotechnol,2015, 208 : 80-6. DOI: 10.1016/j.jbiotec.2015.05.002. |
| [4] | Reda E, Mohamed S, Mohammed Z, et al. DNA aptamers selectionand characterization for development of label-free impedimetricaptasensor for neurotoxin anatoxin-a[J]. Biosens Bioelectron,2015, 68 : 295-302. DOI: 10.1016/j.bios.2015.01.002. |
| [5] | Nina M, Marko V, Jan M, et al. Optimization of SELEX: comparisonof different methods for monitoring the progress of in vitro selectionof aptamers[J]. J Pharmaceut Biomed,2014, 91 : 151-9. DOI: 10.1016/j.jpba.2013.12.031. |
| [6] | Moon J, Kim G, Lee S, et al. Identification of SalmonellaTyphimurium-specific DNAaptamers developed using whole-cellSELEX and FACS analysis[J]. J Microbiol Meth,2013, 95 (2) : 162-6. DOI: 10.1016/j.mimet.2013.08.005. |
| [7] | Xin H, Zhou ZX, Yuan L, et al. Selective collection and detection ofMCF-7 breast cancer cells using aptamer-functionalized magneticbeads and quantum dots based nano-bio-probes[J]. Anal Chim Acta,2013, 788 (14) : 135-40. |
| [8] | Elyse D, Maria C, et al. Development of a bead-based aptamer/antibody detection system for C-reactive protein[J]. Anal Biochem,2015, 472 : 67-74. DOI: 10.1016/j.ab.2014.11.017. |
| [9] | Gulshan S, Poornima V, Neetika R, et al. Bio-capture of S.Typhimurium from surface water by aptamer for culture-freequantification[J]. Ecotoxicol Environ Saf, 2012, 78(78): 320-6. |
| [10] | Wu SJ, Zhang H, Shi Z, et al. Aptamer-based fluorescence biosensorfor chloramphenicol determination using upconversion nanoparticles[J]. Food Control,2015, 50 : 597-604. DOI: 10.1016/j.foodcont.2014.10.003. |
| [11] | Georg M, Ruth M, Bilha S, et al. Multimerization of ERBB2/HER2specific aptamer leads to improved receptor binding[J]. BiochemBioph Res Commun,2015, 465 (2) : 218-24. DOI: 10.1016/j.bbrc.2015.07.157. |
| [12] | Heo K, Min SW, Sung HJ. An aptamer-antibody complex (oligobody)as a novel delivery platform for targeted cancer therapies[J]. JControl Release,2016, 229 : 1-9. DOI: 10.1016/j.jconrel.2016.03.006. |
| [13] | Eric O, Eric T, Karen C, et al. A simple method for eliminatingfixed-region interference of aptamer binding during SELEX[J]. Biotechnol Bioeng,2014, 11 : 2265-79. |
| [14] | Li X, Deng JC, Yan SH, et al. Fabrication of endothelial progenitorcell capture surface via DNA aptamer modifying dopamine/polyethyleneimine copolymer film[J]. Appl Surf Sci,2016, 386 : 138-150. DOI: 10.1016/j.apsusc.2016.06.015. |
| [15] | Reda E, Mohamed S, Mohammed Z, et al. In vitro selection,characterization, and biosensing application of high-affinitycylindrospermopsin-targeting aptamers[J]. Anal Chem,2014, 86 (18) : 9196-203. DOI: 10.1021/ac502157g. |
| [16] | Camille LH, Peng HY, Wang ZX, et al. An improved SELEXtechnique for selection of DNA aptamers binding to M-type 11 ofStreptococcus pyogenes[J]. Methods,2016, 97 : 51-7. DOI: 10.1016/j.ymeth.2015.12.005. |
| [17] | Mariia D, Silvie R, Helena G, et al. Current approaches in SELEX:An update to aptamer election technology[J]. Biotechnol Adv,2015, 33 : 1141-61. DOI: 10.1016/j.biotechadv.2015.02.008. |
| [18] | Ozer A, Pagano JM, Lis JT. New technologies provide quantumchanges in the scale, speed, and success of SELEX methods andaptamer characterization[J]. Mol Ther Nucleic Acids,2014, 3 (8) : e183. |
| [19] | Mona A, Mohammad R, Khalil A, et al. In vitro and in vivo evaluationof therapy targeting epithelial-cell adhesion-molecule aptamers fornon-small cell lung cancer[J]. J Control Release,2015, 209 : 88-100. DOI: 10.1016/j.jconrel.2015.04.026. |
| [20] | Nasa S, Jonathan N, Koichi A, et al. Selection of DNA aptamersagainst uropathogenic Escherichia coli NSM59 by quantitative PCRcontrolled Cell-SELEX[J]. J Microbiol Meth,2014, 104 : 94-100. DOI: 10.1016/j.mimet.2014.06.016. |
 2016, Vol. 36
2016, Vol. 36
