2药学院,广东 广州 510515
2School of Pharmaceutical Sciences, Southern Medical University, Guangzhou, 510515, China
WEI Qiang, E-mail: weiqiang0915@163.com
Prostate cancer is the most common noncutaneous malignancy and the second leading cause of cancerassociated mortality in men in Western countries[1]. While localized prostate cancer can be treated effectively, the treatment of metastatic prostate cancer remains difficult [2] and the median survival of these patients is only 12-15 months [3]. The proliferation, migration and invasion of the tumor cells contribute critically to prostate cancer metastasis [4], and understanding of the molecular mechanisms underlying the malignant behaviors of the tumor cells is therefore of vital importance for devising an effective therapy.
Recent studies suggested that ADP-ribosylation factor 6 (Arf6), a small GTP-binding protein of the Arf family, plays a pivotal role in a wide variety of cellular events, including cell exocytosis, endocytosis, endosome membrane trafficking, phospholipid metabolism and cytoskeleton reorganization[5, 6]. These cellular processes are crucial for the cancer cells to regulate the cell morphology, invade surrounding tissues and metastasize to other organs. Accumulating evidence have shown that Arf6 activation enhances the proliferative, invasive, and migratory potentials of breast cancer, melanoma, and hepatoma cells, while silencing of Arf6 suppressed the cell proliferation, migration, and invasion of the tumor cells[7-9].
A recent study indicated that Arf6 activation mediated the phosphorylation of extracellular signalregulated kinase (ERK) in HepG2 cells and increased intracellular activity of Ras-related C3 botulinum toxin substrate 1 (Rac1), which in turn enhanced the cell motility, migration, and invasion[8]. As a member of the Mitogen-activated protein kinase (MAPK) family, ERK is the primary signaling molecule that regulates gene expression, cell differentiation, mitosis, survival, and apoptosis [10]. An elevated expression of p-ERK is associated with increased tumor cell proliferation, invasion and metastasis [11]. ERK is involved in Rac1 signaling pathway in various human tumor cells[12], and Rac1 has been shown to play an important role in multiple cellular processes[13].
Arf6 has been shown to regulate the proliferation of human glioma cells involving the serine/threonine protein kinase B (PKB/AKT) and ERK signaling pathway[9]. PKB/AKT is an important molecule in the phosphoinositide 3-kinases (PI3K) /AKT signaling pathway and is vital in a wide variety of cellular processes[12, 14]. Hyperactivity of the PI3K promotes the cell proliferation, migration and invasion via phosphorylation of the downstream target AKT [14]. Substantial evidences indicate that the activation of the PI3K/AKT or ERK pathway is critical to the proliferation of prostate cancer cells [12]. So far the mechanism of Arf6 in promoting proliferation of prostate cancer cell remains unclear, and in this study, we aimed to examine the biological function of Arf6 and possible molecular mechanisms in a human prostate cancer cell (PC-3) model with small interfering RNA (siRNA)-mediated Arf6 silencing.
MATERIALS AND METHODS Cell cultureHuman prostate cancer cell line PC-3 was purchased from American Type Culture Collection (ATCC)[15, 16] and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin and 100 μg/mL of streptomycin (Gibco-BRL Life Technologies, Grand Island, NY, USA) at 37 ℃ in a humidified incubator with 5% CO2.
siRNA transfectionThree siRNA duplexes targeting different encoding regions of human Arf6 gene (GenBank Access No.: NM_ 001663.3) were designed and synthesized (Ribobio, Guangzhou, China). The sequences of the Arf6 siRNA were as follows:
siRNA-1 (792-810),
5'-GGGACGCCAUAAUCCUCAUdTdT-3' (sense),
5'-AUGAGGAUUAUGGCGUCCCdTdT-3'
(antisense); siRNA-2 (534-552),
5'-CAACAAUCCUGUACAAGUUdTdT-3' (sense),
5'-AACUUGUACAGGAUUGUUGdTdT-3'(antisense),
and siRNA-3 (950-968),
5'-CUCACAUGGUUAACCUCUAdTdT-3' (sense),
5'-UAGAGGUUAACCAUGUGAGdTdT-3'(antisense).
The cells at approximately 60%-70% confluence were transfected with Arf6 siRNA with Lipofectamine 2000 (Invitrogen Life Technologies, Merelbeke, Belgium) according to the manufacturer's instructions. Briefly, siRNA duplexes and lipofectamine 2000 were diluted separately in 150 μL of serum-free RPMI 1640 medium, incubated for 5 min at room temperature, and then mixed thoroughly followed by further incubation for 20 min at room temperature. The mixture was then transferred into 6-well culture plates and mixed with the cell culture medium. The cells were incubated at 37 ℃ for 6 h for transfection before the medium was changed. A negative siRNA provided by Ribobio was used as a control siRNA for cell transfection under identical conditions.
Real-time PCRTotal RNA was extracted from prostate cancer PC-3 cells using Trizol (Takara, DaLian, China) according to the manufacturer's protocol. Equal amounts of RNA (1 μg) from each sample were used for cDNA synthesis using HiScriptQ RT SuperMix for qPCR (Takara). Real-time PCR with SYBR Green PCR Master Mix (Takara) was performed using Stratngene MX3005P qPCR System (Stratngene, USA). The PCR primers were designed using Premier Primer 5.0 software, and the sequences were as follows:
Arf6,
Forward 5'-ATGGGGAAGGTGCTATCCAAAATC-3',
Reverse 5'-GCAGTCCACTACGAAGATGAGACC-3';
GAPDH,
Forward 5'-GGCCTCCAAGGAGTAAGACC-3',
Reverse 5'-AGGGGAGATTCAGTGTGGTG-3'.
The primers were synthesized by Invitrogen, and the lengths of the amplification products of Arf6 and GAPDH were 270 bp and 122 bp, respectively. PCR amplification were carried out at an initial denaturing temperature of 95 ℃ for 3 min followed by 40 thermal cycles of 95 ℃ for 20 s, 60 ℃ for 20 s, and 72 ℃ for 20 s. The fluorescence data were collected at 72 ℃ step and analyzed with 2-ΔΔCt method using GAPDH gene expression as the reference if amplification of the target was detected below a background threshold (Ct≤35).
Western blottingFor Western blot analysis, the cells at about 80% confluence were lysed with RIPA lysis buffer (KeyGEN, Nanjing, China) with 1 mmol/L PMSF and 1% cocktail of protease inhibitors. Cell lysates were kept on ice for 30 min and centrifuged at 12 000 g for 10 min to obtain the total protein. The total protein concentration was measured using a BCA protein assay kit (Beyotime, Shanghai, China). Equal amounts of the proteins from each sample were separated by 12% SDS-PAGE and electrotransferred to a PVDF membrane (Millipore, Bedford, MA). The membranes were blocked with 5% skim milk for 2 h at room temperature and incubated with the primary antibody overnight at 4 ℃ . The following antibodies were used: mouse anti-Arf6 antibody (1∶600; Santa Cruz Biotechnology, CA, USA), rabbit anti-ERK1/2 antibody (1∶ 700; Bioworld Technology, MA, USA), rabbit anti-p-ERK1/2 antibody (1∶700; Bioworld), rabbit anti-AKT and anti-p-AKT antibody (1∶600; Bioworld), rabbit anti-Rac1 antibody (1 ∶600; Abclonal, USA), and rabbit GAPDH antibody (1∶ 600; Zsgb-Bio, Beijing, China). Following incubation with the primary antibodies, the membranes were incubated with species-specific horseradish peroxidase (HRP)-conjugated secondary antibodies (1∶5000; Zsgb- Bio) at room temperature for 1.5 h. Protein bands were visualized using the ECL regent (Vazyme, Nanjing, China) substrate, and the chemiluminescence signals were captured with X-ray film.
Cell proliferation assayThe effect of siRNA-3 on cell proliferation was examined using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer's instructions. Briefly, the cells were plated on 96-well plates at the density of 1000 cells/well in 200 μL medium and cultured at 37 °C in 5% CO2. At 48, 72 and 96 h after the transfection, 20 μL of MTT (5 mg/mL) in phosphate buffered saline (PBS) was added to each well. After 4 h of incubation at 37 ℃, the cell medium was carefully discarded, the crystals were dissolved by DMSO (150 μL), and the absorbance (A) was measured at 490 nm. All the assays were done in triplicate and performed at least 3 times. The cell survival rate (% ) and inhibition rate (% ) were calculated using the formulas: cell survival rate (% ) = [(Atreatment-Ablank)/(Acontrol-Ablank)]×100%; inhibition rate (%)= 1-cell survival rate (%).
Wound healing assayPC-3 cells in logarithmic growth phase were seeded at the density of 0.5 × 106 cells/well on 12-well culture plate. After growing to 60%-70% confluence, the cells were transfected with Arf6 siRNA or negative control siRNA. When the cells grew to full confluence, the cell monolayer was scratched with a 10 μL pipette tip under sterile condition. Floating cells were removed by washing with PBS. The scratched cell monolayers were then kept in medium containing 1% serum for 18 h and photographed under an inverted phase-contrast microscope (Olympus, Japan) with a 10× objective lens. The migration distance was calculated using the formula: migration distance=(Width 0 h-Widthx h)/2.
Transwell migration and invasion assayPC-3 cells at 60%-70% confluence were transfected with Arf6 siRNA or negative control siRNA for 48 h. The cells were then harvested and suspended in serum-free RPMI 1640 at the density of 5×105/mL. The cell suspension (300 μL) was plated on the top side of Transwell filter on the top chamber of the 24-multiwell insert system with 8 micron pores (BD Bioscience, San Jose, California, USA). The medium supplemented with 10% FBS as the chemoattractant was added to the bottom chamber. After 24 h of cell incubation, the cell migration was stopped by scraping the residual cells on the top chamber with a cotton swab. Migratory cells on the lower membrane surface were fixed in 4% paraformaldehyde for 15 min and stained with 1% crystal violet. The invasion assay was performed similarly except for the Matrigel-coated membrane (BD Bioscience) in the upper chamber. The cells were incubated for 48 h before fixation and staining. The membrane was photographed at 5 randomly selected fields under Olympus DP71 microscope with a 10 × objective lens.
Statistical analysisThe data reported are presented as Mean±SD. Statistical analyses were carried out using the SPSS software version 19.0. Data were analyzed by analysis of variance (ANOVA) followed by post hoc analysis or using Student's t test to compare the difference among the groups. A P value less than 0.05 was considered to indicate a statistically significant difference.
To test the efficiency of the 3 siRNA duplexes for silencing Arf6, we detected the mRNA and protein expressions of Arf6 in PC-3 cells after transfection for 48 h using RT-PCR and Western blotting, respectively. As shown in Fig. 1, the cells transfected with negative control siRNA had similar Arf6 expression levels with the control cells (P>0.05), while the cells transfected with siRNA-1, siRNA-2 and siRNA-3 showed decrements of Arf6 mRNA expressions by (34.82 ± 4.79)%, (56.85±1.52)% and (91.88±3.13)%, respectively, and their Arf6 protein expressions were decreased by (25.73±1.25)%, (67.11±1.08)% and (86.37±0.57)%, respectively. siRNA-3 (50 nmol/L) had the most efficient Arf6-silencing effect in PC-3 cells. We also noted a dose-dependent effect of siRNA-3 in silencing Arf6 (Fig. 2), and 50 nmol/L siRNA-3 stably suppressed Arf6 mRNA and protein expressions till 96 h after the transfection. We therefore used siRNA-3 (50 nmol/L) in the subsequent experiments.
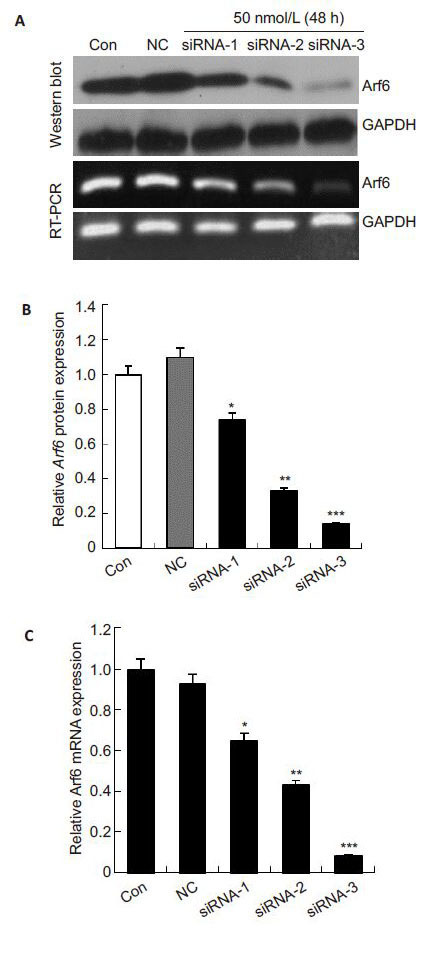
|
Fig.1 Effects of 3 different Arf6-specific siRNA on Arf6 gene in PC-3 cells. GAPDH was used as the internal control. A: Expression of Arf6 was examined by Western blotting and real-time PCR at 48 h after transfection with siRNA (50 nmol/L); B, C: Quantitative analysis of relative Arf6 protein and mRNA levels normalized to GAPDH. The levels were calculated as a percentage relative to the control group. All data are presented as the Mean±SD of 3 independent experiments. *P<0.05, **P<0.01 and ***P< 0.001 vs control. Con: Control group; NC: Negative control siRNA group. |
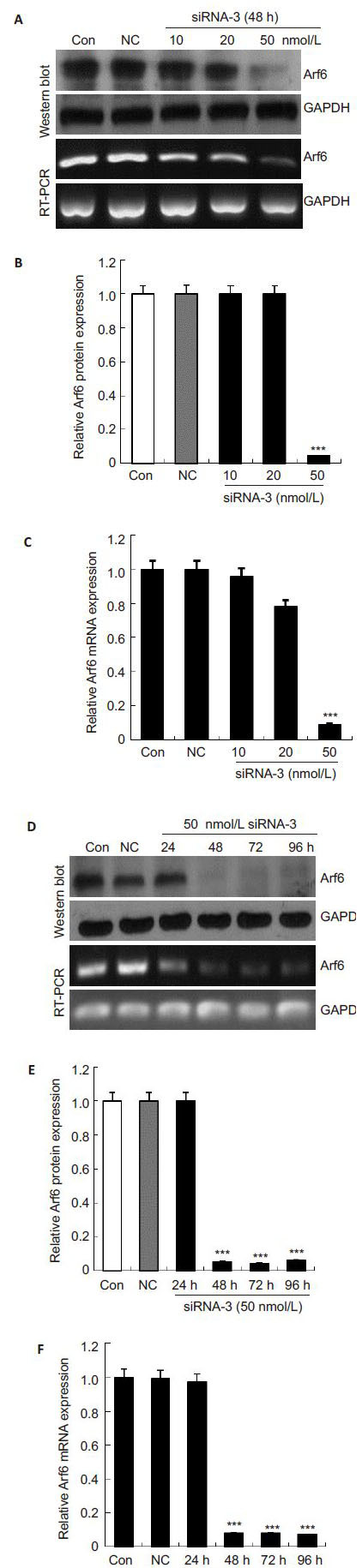
|
Fig.2 siRNA-3 suppressed Arf6 mRNA and protein expressions in PC-3 cells in a dose- and time-dependent manner. A: Effects of 10, 20, and 50 nmol/L siRNA-3 for 48 h on Arf6 mRNA and protein expression; D: Effects of the siRNA-3 (50 nmol/L) on Arf6 mRNA and protein expression at 24, 48, 72 and 96 h following transfection; B, C, E, and F: Quantitative analyisis of relative Arf6 protein and mRNA levels normalized to GAPDH. The levels were calculated as a percentage relative to the control group. All data are presented as Mean±SD of 3 independent experiments. ***P<0.001 vs control. |
To explore whether siRNA silencing affect the proliferation of PC-3 cells, the cell proliferation was examined at 48, 72 and 96 h following transfection with siRNA-3. Fig. 3 shows that at 48, 72 and 96 h following transfection with siRNA-3 (50 nmol/L), the proliferation of PC-3 cells was inhibited at the rates of (10.68 ± 0.04)%, (16.51±0.04)% and (26.35±0.03)%, respectively. These results revealed that the inhibitory effect of siRNA-3 on PC-3 cell proliferation initiated at 48 h, and was the strongest at 96 h, demonstrating a timedependent effect of siRNA-3 in silencing Arf6 in PC-3 cells.
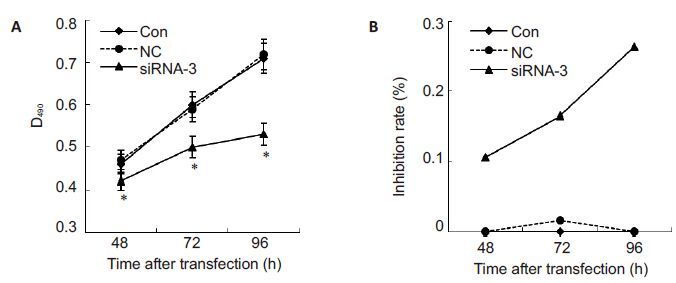
|
Fig.3 Effects of siRNA-3 on the proliferation of PC-3 cells detected using MTT assay at 48, 72 and 96 h after transfection. A: Absorbance of PC-3 cells at different time points following siRNA-3 transfection. The experiments were repeated 3 times; B: Inhibition rate of PC-3 cells at different time points following siRNA-3 transfection. All data are Mean±SD (n=3). *P<0.05 vs control. |
Wound healing assay showed that Arf6 silencing by siRNA-3 significantly reduced the migration distance of PC-3 cells at following an 18-h transfection (Fig. 4, P< 0.001). The cells transfected with siRNA-3 covered the area of only about 45% to 50% of that covered by control PC-3 cells. Transfection with negative control siRNA did not significantly affected the migration capacity of PC-3 cells (P>0.05).
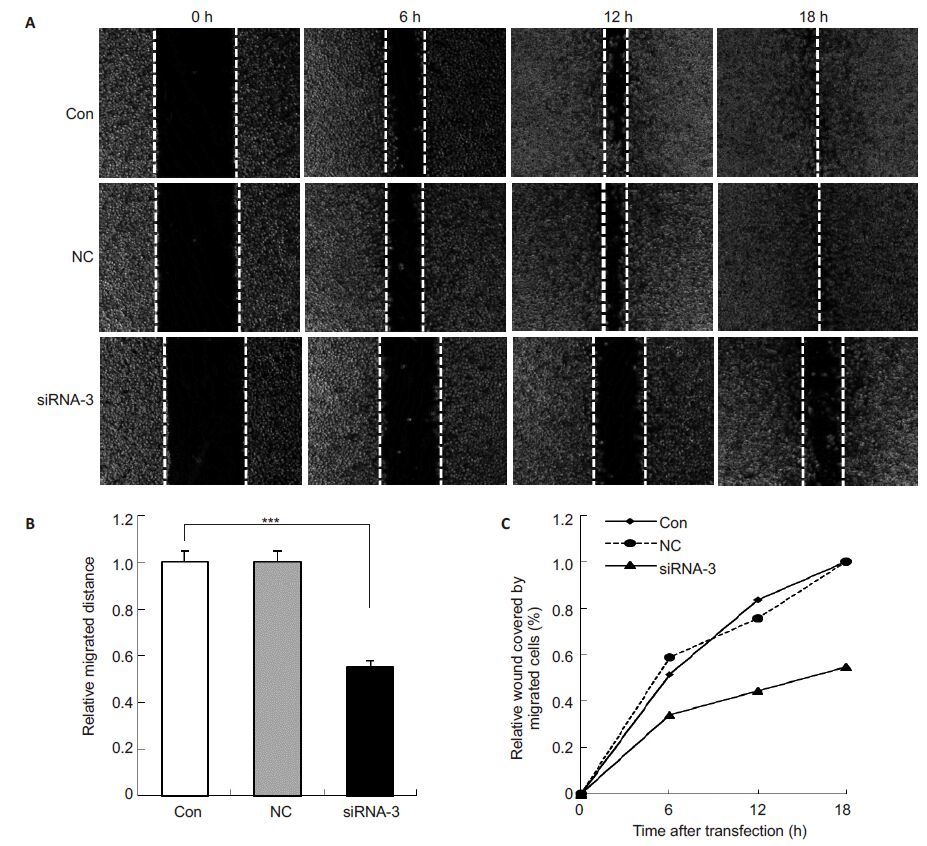
|
Fig.4 siRNA-3-induced Arf6 silencing suppressed migration of PC-3 cells. A: Representative images of control cells (Con), cells transfected with negative control siRNA (NC) and siRNA-3 at 0, 6, 12 and 18 h after monolayer wounding with a sterile 10 μL plastic pipette tip (Original magnification: ×10); B: Relative migration distance of cells in 3 groups at 18 h; C: Percentage of wound area covered by migrated cells in 3 groups at different time points. The area covered by migrated cells from 5 independent microscopic fields was quantified by Image J software. All data are Mean±SD (n=3). ***P<0.000 vs control. |
In the Transwell assay, the number of PC-3 cells transfected with siRNA-3 for 48 h that migrated across the membrane were decreased by 61.61% compared to the control cells and the cells in NC group (P<0.001, Fig. 5). siRNA-3-induced silencing of Arf6 reduced the cell invasion by 89.44% (P<0.001) compared with the control cells (Fig. 5), while no significant difference was found between the cells transfected with negative control siRNA and the control cells (P>0.05). These results demonstrate that silencing Arf6 significantly reduced the migratory and invasive capacity of PC-3 cells in vivo.
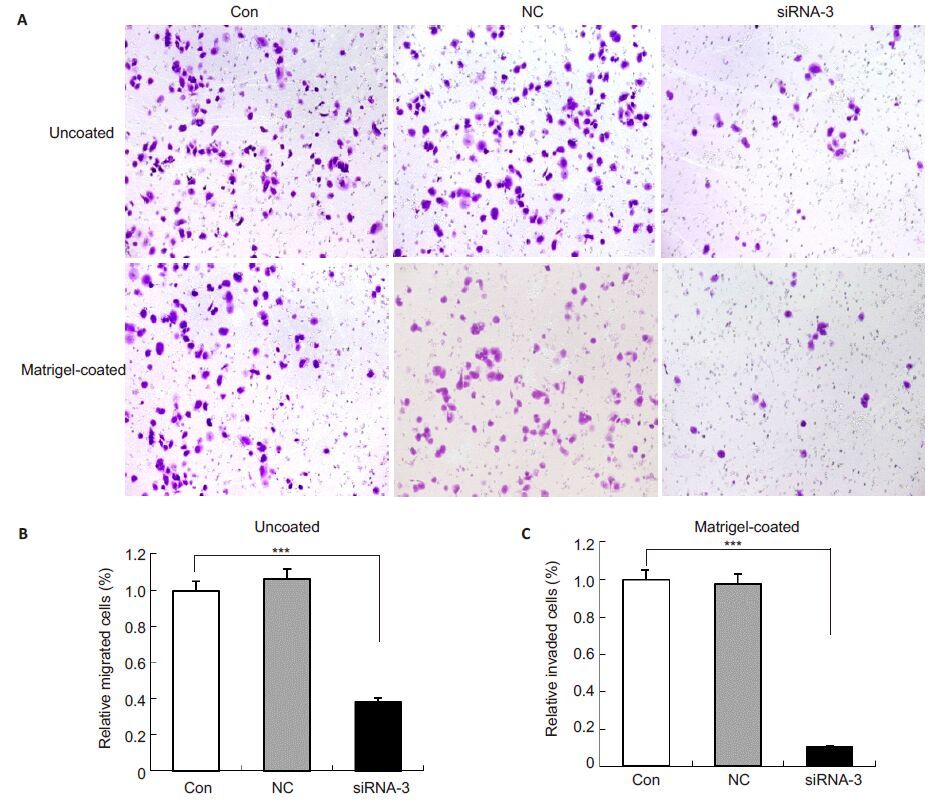
|
Fig.5 siRNA-3-induced silencing of Arf6 suppressed invasion of PC-3 cells. A: Representative images of cell invasion detected using Matrigel-uncoated and -coated Transwell chambers in the presence or absence of 10% FBS. Cells that invaded through the membrane were fixed and stained. Images were photographed at 5 random fields with Olympus DP71 (Original magnification: × 10); B: Quantification of the percentage of invading cells through uncoated membrane. The number of invaded cells was quantified by determining the area of 1% crystal violet staining using Image-Pro Plus. Values are Mean ± SD of 3 independent experiments. P values were calculated using one-way ANOVA and Dunnett's test; C: Quantification of the percentage of invading cells through Matrigel-coated membrane. ***P<0.001 vs control. |
To determine the involvement of PI3K/AKT pathway, ERK pathway, and Rac1 in proliferation, migration and invasion suppression of PC-3 cells transfected with siRNA-3, we detected the protein expression levels of AKR, p-AKT, p-ERK1/2, and Rac1 in the transfected cells. We found that the expressions of p-AKT and total AKT were comparable among all the groups, while the expressions of p-ERK1/2 and Rac1 were significantly reduced in siRNA-3-transfected cells (Fig. 6).
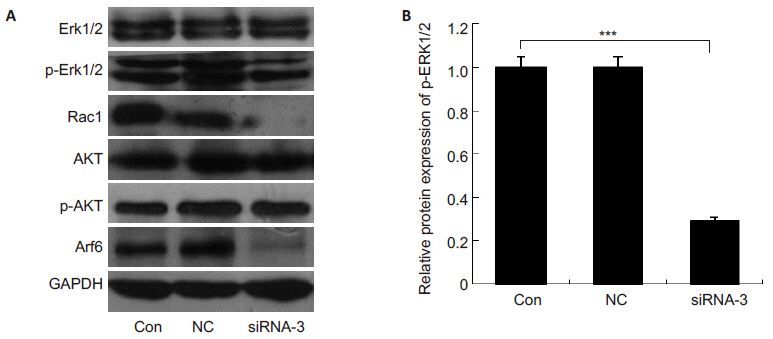
|
Fig.6 Effects of siRNA-3-induced silencing of Arf6 on total-ERK1/2, p-ERK1/2, Rac1, AKT and p-AKT expression. A: Protein levels of total-ERK1/2, p-ERK1/2, Rac1, AKT and p-AKT were detected by Western blotting. GAPDH was used as a loading control. All assays were repeated at least 3 times; B: Quantification of p-ERK1/2 protein levels. The results were calculated as percentages of the control group. ***P<0.001 vs control. |
In the present study, we demonstrated that endogenous Arf6 silencing by siRNA efficiently inhibited the proliferation, migration, and invasion of the PC-3 cell line in vitro. The possible mechanisms underlying the effect of Arf6 silencing involve the down-regulation of p-ERK1/2 and Rac1. This finding suggests that Arf6 plays an important role in the development of prostate cancer, and Arf6-specific siRNA may be of potential value for treatment of human prostate cancer.
Arf6 has been shown to correlate with the proliferation of tumor cells[17]. Li et al[9] found that Arf6 knockdown by siRNA or by expression of a dominant-negative Arf6 mutant suppressed the proliferation of glioblastoma cells. A recent study also revealed a vital role of Arf6 activation in HET-SR cell proliferation [17]. Consistent with these reports, we showed that transfection with siRNA-3 targeting Arf6 time-dependently suppressed the proliferation of PC-3 cells, suggesting the involvement of Arf6 in regulating the proliferation of PC-3 cells.
Tumor cell metastasis is the major reason for therapy failure and mortality in prostate cancer patients[18]. Previous studies showed that Arf6 was required for migration and invasion of various types of cancer cells, such as breast cancer cells, melanoma cells and glioma cells[8, 19, 20]. Silencing of Arf6 inhibited the migration and invasion of these cancer cells both in vivo and in vitro[21]. We observed similar effects of Arf6 silencing in PC-3 cells. Taken together, these results suggest that siRNA-induced Arf6 silencing decreases the migration and invasion capacities of prostate cancer cells.
PI3K/AKT signaling pathway is involved in Arf6-mediated proliferation of glioblastoma cells[9]. We found that siRNA-3-induced silencing of Arf6 inhibited the proliferation of PC-3 cells without affecting AKT activation, which is consistent with the results by Knizhnik et al, who reported that constitutively active Arf6 promoted cell proliferation and had no effect on PI3K/AKT signaling[17].
ERK has been implicated in Arf6-mediated cell proliferation, migration, and invasion [22, 23], and down-regulation of p-ERK1/2 efficiently inhibits PC-3 cell growth, migration, and invasion [11]. We found that p-ERK1/2 expression was decreased in PC-3 cells following in siRNA-3 transfection, suggesting that Arf6 silencing-induced inhibition of PC-3 cell proliferation, migration, and invasion is associated with down-regulation of p-ERK1/2.
Rac1, a member of the Rho family GTPase, leads to the formation of lamellipodia and membrane ruffles[13]. Rac1 is highly expressed in metastatic prostate cancer cells, and the suppression of Rac1 inhibits the proliferation, migration, and invasion of prostate cancer cells[13, 24, 25]. Rac1 is also a downstream effector of Arf6 in normal cells[26]. A recent study found that siRNA-mediated Arf6 silencing decreased the migratory and invasive abilities of hepatoma HepG2 cells via down-regulating p-ERK1/2 and inhibiting Rac1 activation [8]. We also found significantly decreased expressions of p-ERK1/2 and Rac1 in PC-3 cells following transfection with siRNA-3.
In addition, a previous study demonstrated that suppression of Rac1 arrested cell cycle progression at G1/S transition in PC-3 cells[25]. These results indicated that down-regulation of p-ERK1/2 and Rac1 may be the molecular mechanism for Arf6 silencing-induced inhibition of proliferation, migration, and invasion of PC-3 cells.
ConclusionIn spite of the limitation that we tested only the androgen-insensitive PC-3 cell line with a high metastatic potential in this study, we reveal that Arf6 performs a regulatory role of prostate cancer cell proliferation, migration, and invasion by downregulating the p-ERK1/2 and Rac1 expression. Whether these findings apply to other prostate cancer cell lines awaits further studies. In addition, further efforts are needed to elucidate the relationships among Arf6, ERK1/2, and Rac1 in the regulation of the proliferation, migration, and invasion of prostate cancer.
| [1] | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015[J] . CA Cancer J Clin, 2015, 65(1): 5-29. ( 1) 1) |
| [2] | Li H, Zhang Y, Zhang Y, et al. Rsf-1 overexpression in human prostate cancer, implication as a prognostic marker[J] . Tumor Biol, 2014, 35(6): 5771-6. ( 1) 1) |
| [3] | de Bono JS, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial[J] . Lancet, 2010, 376(9747): 1147-54. ( 1) 1) |
| [4] | Grant CM, Kyprianou N. Epithelial mesenchymal transition (EMT) in prostate growth and tumor progression[J] . Transl Androl Urol, 2013, 2(3): 202-11. ( 1) 1) |
| [5] | Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis[J] . Semin Cell Dev Biol, 2011, 22(1): 39-47. ( 1) 1) |
| [6] | Hongu T, Kanaho Y. Activation machinery of the small GTPase Arf6 [J] . Adv Biol Regul, 2014, 54: 59-66. ( 1) 1) |
| [7] | Hu B, Shi B, Jarzynka MJ, et al. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway [J] . Cancer Res, 2009, 69(3): 794-801. ( 1) 1) |
| [8] | Hu Z, Du J, Yang L, et al. GEP100/Arf6 is required for epidermal growth factor-induced ERK/Rac1 signaling and cell migration in human hepatoma HepG2 cells[J] . PLoS One, 2012, 7(6): e38777. ( 3) 3) |
| [9] | Li M, Wang J, Ng SS, et al. Adenosine diphosphate-ribosylation factor 6 is required for epidermal growth factor-induced glioblastoma cell proliferation[J] . Cancer, 2009, 115(21): 4959-72. ( 4) 4) |
| [10] | Mccubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/ MEK/ERK pathway in cell growth, malignant transformation and drug resistance[J] . BBA Mol Cell Res, 2007, 1773(8): 1263-84. ( 1) 1) |
| [11] | Tian Y, Guan Y, Jia Y, et al. Chloride intracellular channel 1 regulates prostate cancer cell proliferation and migration through the MAPK/ERK pathway [J] . Cancer Biother Radio, 2014, 29(8): 339-44. ( 2) 2) |
| [12] | Henderson V, Smith B, Burton LJ, et al. Snail promotes cell migration through PI3K/AKT-dependent Rac1 activation as well as PI3K/ AKT-independent pathways during prostate cancer progression[J] . Cell Adh Migr, 2015, 9(4): 255-64. ( 3) 3) |
| [13] | Kato T, Kawai K, Egami Y, et al. Rac1-dependent lamellipodial motility in prostate cancer PC-3 cells revealed by optogenetic control of Rac1 activity[J] . PLoS One, 2014, 9(5): e97749. ( 2) 2) |
| [14] | Chan CH, Jo U, Kohrman A, et al. Posttranslational regulation of Akt in human cancer[J] . Cell Biosci, 2014, 4(1): 59. ( 1) 1) |
| [15] | Wei Q, Costanzi S, Balasubramanian R, et al. A2B adenosine receptor blockade inhibits growth of prostate cancer cells [J] . Purinerg Signal, 2013, 9(2): 271-80. ( 1) 1) |
| [16] | Wei Q, Costanzi S, Liu Q, et al. Activation of the P2Y1 receptor induces apoptosis and inhibits proliferation of prostate cancer cells [J] . Biochem Pharmacol, 2011, 82(4): 418-25. ( 1) 1) |
| [17] | Knizhnik AV, Kovaleva OV, Komelkov AV, et al. Arf6 promotes cell proliferation via the PLD-mTORC1 and p38MAPK pathways[J] . J Cell Biochem, 2012, 113(1): 360-71. ( 3) 3) |
| [18] | Taichman RS, Loberg RD, Mehra R, et al. The evolving biology and treatment of prostate cancer [J] . J Clin Invest, 2007, 117(9): 2351-61. ( 1) 1) |
| [19] | Xu R, Zhang Y, Gu L, et al. Arf6 regulates EGF-induced internalization of E-cadherin in breast cancer cells[J] . Cancer Cell Int, 2015, 15(1): 11. ( 1) 1) |
| [20] | Grossmann AH, Yoo JH, Clancy J, et al. The small GTPase ARF6 stimulates β-catenin transcriptional activity during WNT5Amediated melanoma invasion and metastasis[J] . Sci Signal, 2013, 6 (265): a14. ( 1) 1) |
| [21] | Muralidharan-Chari V, Hoover H, Clancy J, et al. ADP-ribosylation factor 6 regulates tumorigenic and invasive properties in vivo[J] . Cancer Res, 2009, 69(6): 2201-9. ( 1) 1) |
| [22] | Zhang Y, Du J, Zheng J, et al. EGF-reduced Wnt5a transcription induces epithelial-mesenchymal transition via Arf6-ERK signaling in gastric cancer cells[J] . Oncotarget, 2015, 6(9): 7244-61. ( 1) 1) |
| [23] | Smyth D, Mckay CM, Gulbransen BD, et al. Interferon-gamma signals via an ERK1/2-ARF6 pathway to promote bacterial internalization by gut epithelia[J] . Cell Microbio, 2012, 14(8): 1257-70. ( 1) 1) |
| [24] | Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control[J] . Cell Commun Signal, 2010, 8: 23. ( 1) 1) |
| [25] | Kobayashi T, Inoue T, Shimizu Y, et al. Activation of Rac1 is closely related to androgen-independent cell proliferation of prostate cancer cells both in vitro and in vivo[J] . Mol Endocrinol, 2010, 24(4): 722-734. ( 2) 2) |
| [26] | Palacios F, D'Souza-Schorey C. Modulation of Rac1 and ARF6 activation during epithelial cell scattering[J] . J Biol Chem, 2003, 278(19): 17395-400.( 1) 1) |
 2016, Vol. 36
2016, Vol. 36
