Journal of Southern Medical University ›› 2025, Vol. 45 ›› Issue (6): 1280-1288.doi: 10.12122/j.issn.1673-4254.2025.06.17
Huaxuan ZHAO1( ), Guichao ZHANG1, Jiarong LIU3, Futian MO3, Taoen LI3, Chengyong LEI3(
), Guichao ZHANG1, Jiarong LIU3, Futian MO3, Taoen LI3, Chengyong LEI3( ), Shidong LÜ2(
), Shidong LÜ2( )
)
Received:2025-04-06
Online:2025-06-20
Published:2025-06-27
Contact:
Chengyong LEI, Shidong Lü
E-mail:2974463417@qq.com;345041047@qq.com;lsd990@smu.edu.cn
Huaxuan ZHAO, Guichao ZHANG, Jiarong LIU, Futian MO, Taoen LI, Chengyong LEI, Shidong LÜ. Correlations of immune cell infiltration characteristics with clinicopathological parameters in patients with clear cell renal cell carcinoma[J]. Journal of Southern Medical University, 2025, 45(6): 1280-1288.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.j-smu.com/EN/10.12122/j.issn.1673-4254.2025.06.17
| Antibody | Clone | Company | Dilution ratio |
|---|---|---|---|
| CD3 | MX036 | MXB Biotechnologies | Ready to use |
| CD4 | SP35 | MXB Biotechnologies | Ready to use |
| CD8 | SP16 | Amresco | Ready to use |
| FOXP3 | 236A/E7 | Abcam | 1:250 |
| CD68 | C68/684 | Abcam | 1:100 |
| CD20 | EP459Y | Abcam | 1:300 |
| CD79 | EP3618 | Abcam | 1:250 |
| Tryptase | EPR8476 | Abcam | 1:2500 |
| Programmed Death-1 | D4W2J | Cell Signaling Technology | 1:200 |
| Programmed Death-Ligand 1 | EPR19759 | Abcam | 1:250 |
Tab.1 Antibodies used for immunohistochemistry
| Antibody | Clone | Company | Dilution ratio |
|---|---|---|---|
| CD3 | MX036 | MXB Biotechnologies | Ready to use |
| CD4 | SP35 | MXB Biotechnologies | Ready to use |
| CD8 | SP16 | Amresco | Ready to use |
| FOXP3 | 236A/E7 | Abcam | 1:250 |
| CD68 | C68/684 | Abcam | 1:100 |
| CD20 | EP459Y | Abcam | 1:300 |
| CD79 | EP3618 | Abcam | 1:250 |
| Tryptase | EPR8476 | Abcam | 1:2500 |
| Programmed Death-1 | D4W2J | Cell Signaling Technology | 1:200 |
| Programmed Death-Ligand 1 | EPR19759 | Abcam | 1:250 |
| Antibody | Coupled fluorescein | Clone | Company |
|---|---|---|---|
| CD3 | FITC | UCHT1 | BD Biosciences |
| CD4 | APC-CY7 | RPA-T4 | BD Biosciences |
| CD8 | BV605 | SK1 | BD Biosciences |
| CD278 (ICOS) | PE | DX29 | BD Biosciences |
| CD134 (OX40) | PE-CY7 | ACT35 | BD Biosciences |
| CD137 | BV421 | 4B4-1 | BD Biosciences |
| CD25 | APC | M-A251 | BD Biosciences |
Tab.2 Antibodies used for flow cytometry
| Antibody | Coupled fluorescein | Clone | Company |
|---|---|---|---|
| CD3 | FITC | UCHT1 | BD Biosciences |
| CD4 | APC-CY7 | RPA-T4 | BD Biosciences |
| CD8 | BV605 | SK1 | BD Biosciences |
| CD278 (ICOS) | PE | DX29 | BD Biosciences |
| CD134 (OX40) | PE-CY7 | ACT35 | BD Biosciences |
| CD137 | BV421 | 4B4-1 | BD Biosciences |
| CD25 | APC | M-A251 | BD Biosciences |
| Characteristic | Immunohistochemical cohort (n=154) | PDTF cohort (n=22) | P |
|---|---|---|---|
| Gender | >0.999 | ||
| Male | 106 (68.8) | 15 (68.2) | |
| Female | 48 (31.2) | 7 (31.8) | |
| Age (year) | 0.811 | ||
| <50 | 51 (33.1) | 8 (36.4) | |
| ≥50 | 103 (66.9) | 14 (63.6) | |
| Tumor size (cm) | 0.819 | ||
| <5 | 90 (58.4) | 12 (54.5) | |
| ≥5 | 64 (41.6) | 10 (45.5) | |
| Tumor stage | 0.964 | ||
| 1a | 75 (48.7) | 10 (45.4) | |
| 1b | 49 (31.8) | 8 (36.4) | |
| 2a | 16 (10.4) | 2 (9.1) | |
| 3a | 11 (7.1) | 2 (9.1) | |
| 3b | 2 (1.3) | 0 (0) | |
| 4 | 1 (0.7) | 0 (0) | |
| ISUP grade | 0.872 | ||
| 1 | 9 (5.8) | 2 (9.1) | |
| 2 | 125 (81.2) | 18 (81.8) | |
| 3 | 15 (9.7) | 2 (9.1) | |
| 4 | 5 (3.3) | 0 (0) | |
| Clinical stage | >0.999 | ||
| Ⅰ | 122 (79.2) | 18 (81.8) | |
| Ⅱ | 17 (11.0) | 2 (9.1) | |
| Ⅲ | 13 (8.5) | 2 (9.1) | |
| IV | 2 (1.3) | 0 (0) |
Tab.3 Clinical characteristics of patients with ccRCC in immunohistochemical and PDTF cohorts [n (%)]
| Characteristic | Immunohistochemical cohort (n=154) | PDTF cohort (n=22) | P |
|---|---|---|---|
| Gender | >0.999 | ||
| Male | 106 (68.8) | 15 (68.2) | |
| Female | 48 (31.2) | 7 (31.8) | |
| Age (year) | 0.811 | ||
| <50 | 51 (33.1) | 8 (36.4) | |
| ≥50 | 103 (66.9) | 14 (63.6) | |
| Tumor size (cm) | 0.819 | ||
| <5 | 90 (58.4) | 12 (54.5) | |
| ≥5 | 64 (41.6) | 10 (45.5) | |
| Tumor stage | 0.964 | ||
| 1a | 75 (48.7) | 10 (45.4) | |
| 1b | 49 (31.8) | 8 (36.4) | |
| 2a | 16 (10.4) | 2 (9.1) | |
| 3a | 11 (7.1) | 2 (9.1) | |
| 3b | 2 (1.3) | 0 (0) | |
| 4 | 1 (0.7) | 0 (0) | |
| ISUP grade | 0.872 | ||
| 1 | 9 (5.8) | 2 (9.1) | |
| 2 | 125 (81.2) | 18 (81.8) | |
| 3 | 15 (9.7) | 2 (9.1) | |
| 4 | 5 (3.3) | 0 (0) | |
| Clinical stage | >0.999 | ||
| Ⅰ | 122 (79.2) | 18 (81.8) | |
| Ⅱ | 17 (11.0) | 2 (9.1) | |
| Ⅲ | 13 (8.5) | 2 (9.1) | |
| IV | 2 (1.3) | 0 (0) |
| Immune marker | Subcellular localization | Present on |
|---|---|---|
| CD3+ | Cytomembrane | T lymphocytes |
| CD4+ | Cytomembrane | T helper lymphocytes |
| CD8+ | Cytomembrane | Cytotoxic T lymphocytes |
| FOXP3+ | Cell nucleus | Regulatory T lymphocytes |
| CD68+CD163- | Cytomembrane | M1 Macrophages |
| CD68+CD163+ | Cytomembrane | M2 Macrophages |
| CD20+CD79α- | Cytomembrane | B lymphocytes |
| CD20-CD79α+ | Cytomembrane | Plasma cells |
| Tryptase+ | Cytomembrane | Mast cells |
Tab.4 List of immune cell markers and the represented cells
| Immune marker | Subcellular localization | Present on |
|---|---|---|
| CD3+ | Cytomembrane | T lymphocytes |
| CD4+ | Cytomembrane | T helper lymphocytes |
| CD8+ | Cytomembrane | Cytotoxic T lymphocytes |
| FOXP3+ | Cell nucleus | Regulatory T lymphocytes |
| CD68+CD163- | Cytomembrane | M1 Macrophages |
| CD68+CD163+ | Cytomembrane | M2 Macrophages |
| CD20+CD79α- | Cytomembrane | B lymphocytes |
| CD20-CD79α+ | Cytomembrane | Plasma cells |
| Tryptase+ | Cytomembrane | Mast cells |
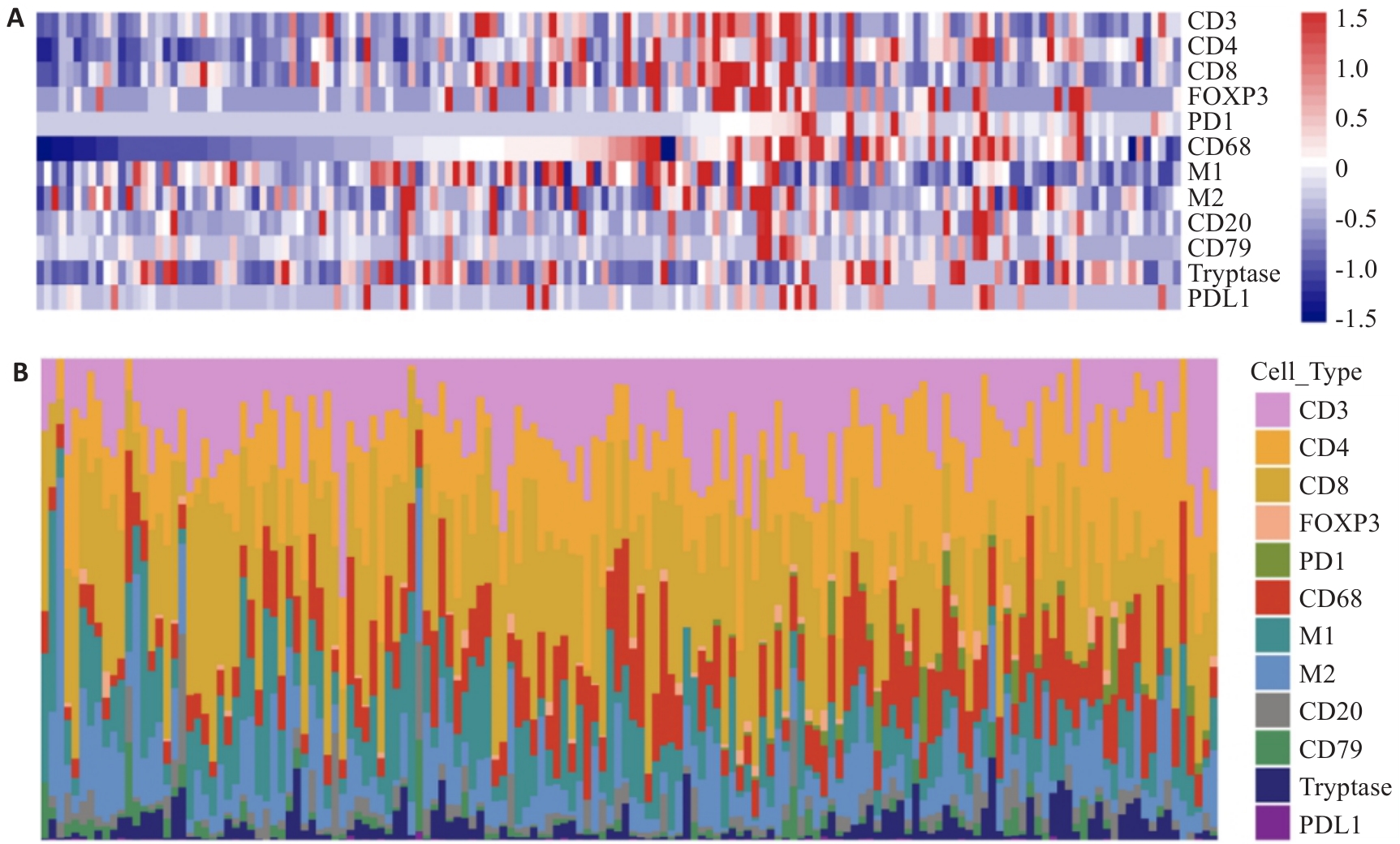
Fig.1 Landscape of immune cell infiltration in clear cell renal cell carcinomas (ccRCC). A: The overall immune cell infiltration in 154 patients with ccRCC (each row represents an immune cell feature, and each column represents an individual. B: Percentage of each immune cell from the 154 patients with ccRCC.
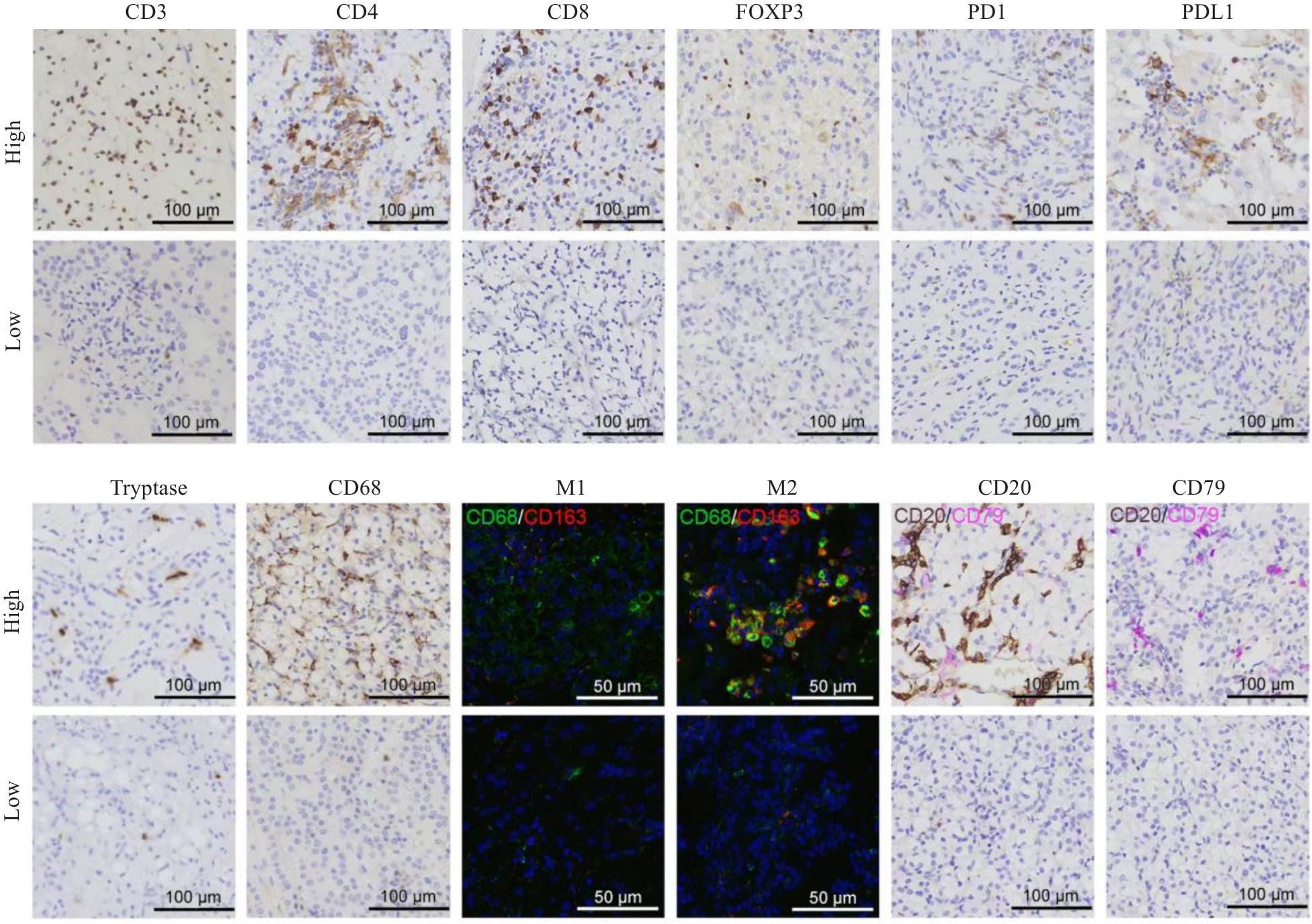
Fig.2 Immunohistochemical and immunofluorescence features of tumor-infiltrating immune cells. CD3, CD4, CD8, FOXP3, PD1, PDL1, Tryptase, CD68, CD20, and CD79 expressions were detected with immunohistochemistry, and M1 and M2 expressions were detected with immunofluorescence staining.
| Index | Gender | P | Age (year) | P | Tumor size (cm) | P | Tumor stage | P | ISUP grade | P | Clinical stage | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | <50 | ≥50 | <5 | ≥5 | 1,2 | 3,4 | 1, 2 | 3, 4 | Ⅰ-Ⅱ | Ⅲ-Ⅳ | |||||||
| CD3 | 0.241 | >0.999 | 0.004 | 0.702 | 0.023 | 0.269 | ||||||||||||
| High | 20 | 5 | 8 | 17 | 8 | 17 | 22 | 3 | 18 | 7 | 21 | 4 | ||||||
| Low | 86 | 43 | 43 | 86 | 82 | 47 | 118 | 11 | 116 | 13 | 118 | 11 | ||||||
| CD4 | 0.310 | 0.326 | 0.814 | 0.026 | 0.155 | 0.035 | ||||||||||||
| High | 89 | 44 | 42 | 91 | 77 | 56 | 124 | 9 | 118 | 15 | 123 | 10 | ||||||
| Low | 17 | 4 | 9 | 12 | 13 | 8 | 16 | 5 | 16 | 5 | 16 | 5 | ||||||
| CD8 | 0.281 | 0.860 | 0.239 | 0.256 | 0.045 | 0.173 | ||||||||||||
| High | 70 | 27 | 33 | 64 | 53 | 44 | 86 | 11 | 80 | 17 | 85 | 12 | ||||||
| Low | 36 | 21 | 18 | 39 | 37 | 20 | 54 | 3 | 54 | 3 | 54 | 3 | ||||||
| FOXP3 | 0.684 | 0.691 | 0.087 | >0.999 | 0.262 | >0.999 | ||||||||||||
| High | 27 | 10 | 11 | 26 | 17 | 20 | 34 | 3 | 30 | 7 | 34 | 3 | ||||||
| Low | 79 | 38 | 40 | 77 | 73 | 44 | 106 | 11 | 104 | 13 | 105 | 12 | ||||||
| PD1 | 0.464 | >0.999 | 0.020 | 0.441 | 0.014 | 0.243 | ||||||||||||
| High | 14 | 9 | 8 | 15 | 8 | 15 | 20 | 3 | 16 | 7 | 19 | 4 | ||||||
| Low | 92 | 39 | 43 | 88 | 82 | 49 | 120 | 11 | 118 | 13 | 120 | 11 | ||||||
| CD68 | 0.793 | 0.608 | 0.049 | >0.999 | 0.276 | 0.696 | ||||||||||||
| High | 14 | 5 | 5 | 14 | 7 | 12 | 18 | 1 | 15 | 4 | 18 | 1 | ||||||
| Low | 92 | 43 | 46 | 89 | 83 | 52 | 122 | 13 | 119 | 16 | 121 | 14 | ||||||
| M1 | 0.428 | 0.699 | 0.710 | 0.756 | 0.013 | 0.539 | ||||||||||||
| High | 76 | 38 | 39 | 75 | 68 | 46 | 104 | 10 | 104 | 10 | 104 | 10 | ||||||
| Low | 30 | 10 | 12 | 28 | 22 | 18 | 36 | 4 | 30 | 10 | 35 | 5 | ||||||
| M2 | >0.999 | 0.483 | 0.020 | 0.441 | >0.999 | 0.472 | ||||||||||||
| High | 90 | 41 | 45 | 86 | 82 | 49 | 120 | 11 | 114 | 17 | 119 | 12 | ||||||
| Low | 16 | 7 | 6 | 17 | 8 | 15 | 20 | 3 | 20 | 3 | 20 | 3 | ||||||
| CD20 | 0.793 | 0.796 | >0.999 | 0.074 | 0.020 | 0.093 | ||||||||||||
| High | 14 | 5 | 7 | 12 | 11 | 8 | 15 | 4 | 13 | 6 | 15 | 4 | ||||||
| Low | 92 | 43 | 44 | 91 | 79 | 56 | 125 | 10 | 121 | 14 | 124 | 11 | ||||||
| CD79 | >0.999 | >0.999 | 0.049 | 0.384 | 0.004 | 0.093 | ||||||||||||
| High | 13 | 6 | 6 | 13 | 7 | 12 | 16 | 3 | 12 | 7 | 15 | 4 | ||||||
| Low | 93 | 42 | 45 | 90 | 83 | 52 | 124 | 11 | 122 | 13 | 124 | 11 | ||||||
| Tryptase | 0.057 | 0.351 | 0.049 | 0.357 | 0.001 | 0.147 | ||||||||||||
| High | 69 | 39 | 33 | 75 | 69 | 39 | 100 | 8 | 101 | 7 | 100 | 8 | ||||||
| Low | 37 | 9 | 18 | 28 | 21 | 25 | 40 | 6 | 33 | 13 | 39 | 7 | ||||||
| PDL1 | 0.576 | >0.999 | 0.030 | 0.007 | 0.007 | 0.010 | ||||||||||||
| High | 10 | 6 | 5 | 11 | 5 | 11 | 11 | 5 | 10 | 6 | 11 | 5 | ||||||
| Low | 96 | 42 | 46 | 92 | 85 | 53 | 129 | 9 | 124 | 14 | 128 | 10 | ||||||
Tab.5 Correlation of the density of tumor-infiltrating lymphocytes (TILs) with the clinicopathological features of ccRCC patients
| Index | Gender | P | Age (year) | P | Tumor size (cm) | P | Tumor stage | P | ISUP grade | P | Clinical stage | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | <50 | ≥50 | <5 | ≥5 | 1,2 | 3,4 | 1, 2 | 3, 4 | Ⅰ-Ⅱ | Ⅲ-Ⅳ | |||||||
| CD3 | 0.241 | >0.999 | 0.004 | 0.702 | 0.023 | 0.269 | ||||||||||||
| High | 20 | 5 | 8 | 17 | 8 | 17 | 22 | 3 | 18 | 7 | 21 | 4 | ||||||
| Low | 86 | 43 | 43 | 86 | 82 | 47 | 118 | 11 | 116 | 13 | 118 | 11 | ||||||
| CD4 | 0.310 | 0.326 | 0.814 | 0.026 | 0.155 | 0.035 | ||||||||||||
| High | 89 | 44 | 42 | 91 | 77 | 56 | 124 | 9 | 118 | 15 | 123 | 10 | ||||||
| Low | 17 | 4 | 9 | 12 | 13 | 8 | 16 | 5 | 16 | 5 | 16 | 5 | ||||||
| CD8 | 0.281 | 0.860 | 0.239 | 0.256 | 0.045 | 0.173 | ||||||||||||
| High | 70 | 27 | 33 | 64 | 53 | 44 | 86 | 11 | 80 | 17 | 85 | 12 | ||||||
| Low | 36 | 21 | 18 | 39 | 37 | 20 | 54 | 3 | 54 | 3 | 54 | 3 | ||||||
| FOXP3 | 0.684 | 0.691 | 0.087 | >0.999 | 0.262 | >0.999 | ||||||||||||
| High | 27 | 10 | 11 | 26 | 17 | 20 | 34 | 3 | 30 | 7 | 34 | 3 | ||||||
| Low | 79 | 38 | 40 | 77 | 73 | 44 | 106 | 11 | 104 | 13 | 105 | 12 | ||||||
| PD1 | 0.464 | >0.999 | 0.020 | 0.441 | 0.014 | 0.243 | ||||||||||||
| High | 14 | 9 | 8 | 15 | 8 | 15 | 20 | 3 | 16 | 7 | 19 | 4 | ||||||
| Low | 92 | 39 | 43 | 88 | 82 | 49 | 120 | 11 | 118 | 13 | 120 | 11 | ||||||
| CD68 | 0.793 | 0.608 | 0.049 | >0.999 | 0.276 | 0.696 | ||||||||||||
| High | 14 | 5 | 5 | 14 | 7 | 12 | 18 | 1 | 15 | 4 | 18 | 1 | ||||||
| Low | 92 | 43 | 46 | 89 | 83 | 52 | 122 | 13 | 119 | 16 | 121 | 14 | ||||||
| M1 | 0.428 | 0.699 | 0.710 | 0.756 | 0.013 | 0.539 | ||||||||||||
| High | 76 | 38 | 39 | 75 | 68 | 46 | 104 | 10 | 104 | 10 | 104 | 10 | ||||||
| Low | 30 | 10 | 12 | 28 | 22 | 18 | 36 | 4 | 30 | 10 | 35 | 5 | ||||||
| M2 | >0.999 | 0.483 | 0.020 | 0.441 | >0.999 | 0.472 | ||||||||||||
| High | 90 | 41 | 45 | 86 | 82 | 49 | 120 | 11 | 114 | 17 | 119 | 12 | ||||||
| Low | 16 | 7 | 6 | 17 | 8 | 15 | 20 | 3 | 20 | 3 | 20 | 3 | ||||||
| CD20 | 0.793 | 0.796 | >0.999 | 0.074 | 0.020 | 0.093 | ||||||||||||
| High | 14 | 5 | 7 | 12 | 11 | 8 | 15 | 4 | 13 | 6 | 15 | 4 | ||||||
| Low | 92 | 43 | 44 | 91 | 79 | 56 | 125 | 10 | 121 | 14 | 124 | 11 | ||||||
| CD79 | >0.999 | >0.999 | 0.049 | 0.384 | 0.004 | 0.093 | ||||||||||||
| High | 13 | 6 | 6 | 13 | 7 | 12 | 16 | 3 | 12 | 7 | 15 | 4 | ||||||
| Low | 93 | 42 | 45 | 90 | 83 | 52 | 124 | 11 | 122 | 13 | 124 | 11 | ||||||
| Tryptase | 0.057 | 0.351 | 0.049 | 0.357 | 0.001 | 0.147 | ||||||||||||
| High | 69 | 39 | 33 | 75 | 69 | 39 | 100 | 8 | 101 | 7 | 100 | 8 | ||||||
| Low | 37 | 9 | 18 | 28 | 21 | 25 | 40 | 6 | 33 | 13 | 39 | 7 | ||||||
| PDL1 | 0.576 | >0.999 | 0.030 | 0.007 | 0.007 | 0.010 | ||||||||||||
| High | 10 | 6 | 5 | 11 | 5 | 11 | 11 | 5 | 10 | 6 | 11 | 5 | ||||||
| Low | 96 | 42 | 46 | 92 | 85 | 53 | 129 | 9 | 124 | 14 | 128 | 10 | ||||||
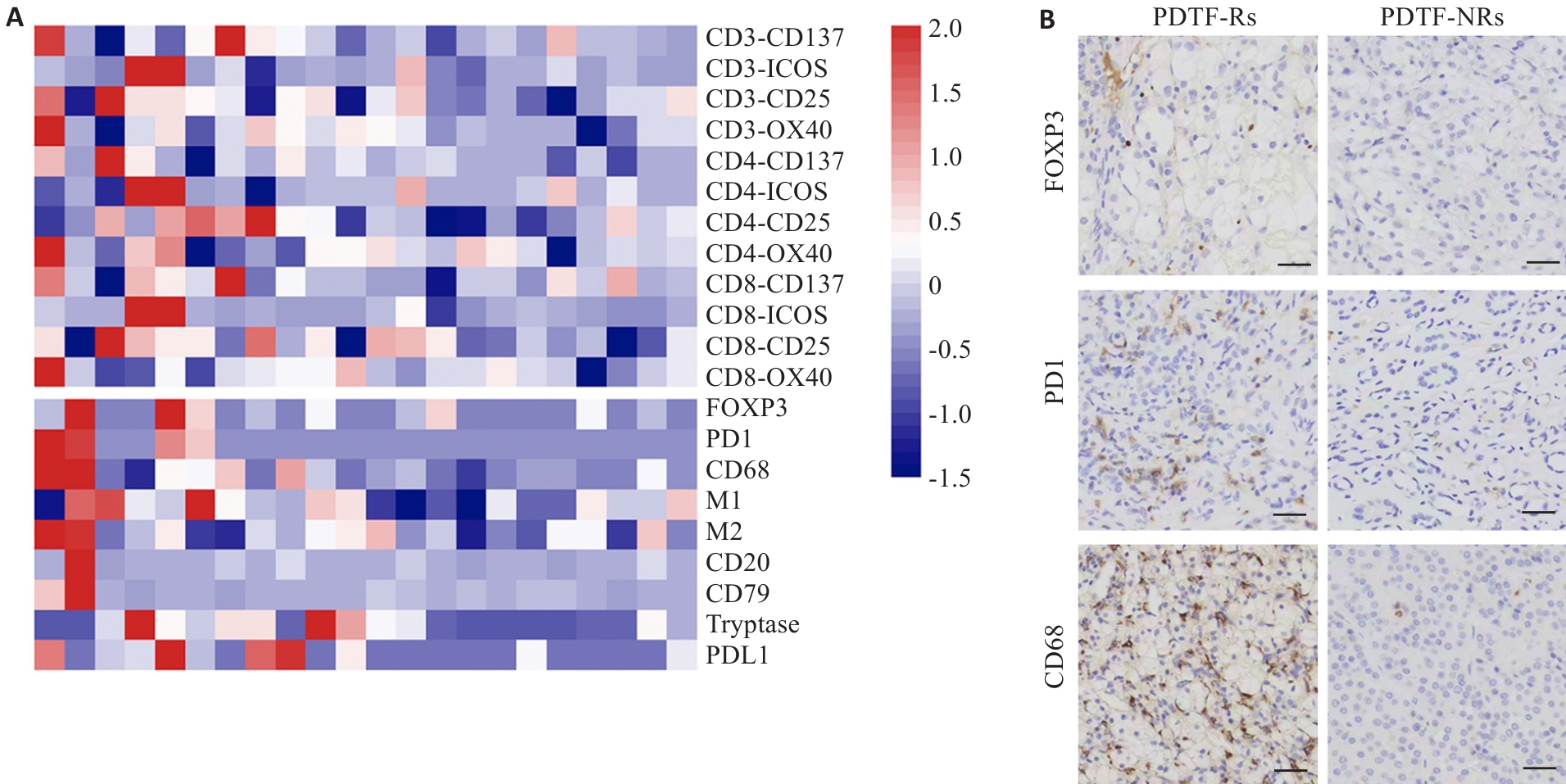
Fig.3 Immunological responses of PD-1 antibody treatment are positively correlated with immune cell infiltration in ccRCC. A: Heatmap displaying the z-score for the difference in positive T cell activation markers between PD-1 antibody treatment versus the control in the PDTF model (upper panel) and the density of immune cells (lower panel), with each column representing one individual. B: Representative immunohistochemical images of FOXP3+, PD1+, CD68+ immune cell infiltration in patients with PDTF-Rs and PDTF-NRs (Scale bar=100 μm).
| 1 | Muglia VF, Prando A. Renal cell carcinoma: histological classification and correlation with imaging findings[J]. Radiol Bras, 2015, 48(3): 166-74. doi:10.1590/0100-3984.2013.1927 |
| 2 | Schödel J, Grampp S, Maher ER, et al. Hypoxia, hypoxia-inducible transcription factors, and renal cancer[J]. Eur Urol, 2016, 69(4): 646-57. doi:10.1016/j.eururo.2015.08.007 |
| 3 | Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update[J]. Eur Urol, 2015, 67(5): 913-24. doi:10.1016/j.eururo.2015.01.005 |
| 4 | Frank I, Blute ML, Cheville JC, et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score[J]. J Urol, 2002, 168(6): 2395-400. doi:10.1016/s0022-5347(05)64153-5 |
| 5 | Patard JJ, Kim HL, Lam JS, et al. Use of the University of California Los Angeles integrated staging system to predict survival in renal cell carcinoma: an international multicenter study[J]. J Clin Oncol, 2004, 22(16): 3316-22. doi:10.1200/jco.2004.09.104 |
| 6 | Wolff I, May M, Hoschke B, et al. Do we need new high-risk criteria for surgically treated renal cancer patients to improve the outcome of future clinical trials in the adjuvant setting? Results of a comprehensive analysis based on the multicenter CORONA database[J]. Eur J Surg Oncol EJSO, 2016, 42(5): 744-50. doi:10.1016/j.ejso.2016.01.009 |
| 7 | Meskawi M, Sun M, Trinh QD, et al. A review of integrated staging systems for renal cell carcinoma[J]. Eur Urol, 2012, 62(2): 303-14. doi:10.1016/j.eururo.2012.04.049 |
| 8 | Sánchez-Gastaldo A, Kempf E, González del Alba A, et al. Systemic treatment of renal cell cancer: a comprehensive review[J]. Cancer Treat Rev, 2017, 60: 77-89. doi:10.1016/j.ctrv.2017.08.010 |
| 9 | Porta C, Cosmai L, Leibovich BC, et al. The adjuvant treatment of kidney cancer: a multidisciplinary outlook[J]. Nat Rev Nephrol, 2019, 15(7): 423-33. doi:10.1038/s41581-019-0131-x |
| 10 | Pons-Tostivint E, Latouche A, Vaflard P, et al. Comparative analysis of durable responses on immune checkpoint inhibitors versus other systemic therapies: a pooled analysis of phase III trials[J]. JCO Precis Oncol, 2019, 3: 1-10. doi:10.1200/po.18.00114 |
| 11 | Borgeaud M, Sandoval J, Obeid M, et al. Novel targets for immune-checkpoint inhibition in cancer[J]. Cancer Treat Rev, 2023, 120: 102614. doi:10.1016/j.ctrv.2023.102614 |
| 12 | Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy[J]. Nat Rev Cancer, 2020, 20(11): 662-80. doi:10.1038/s41568-020-0285-7 |
| 13 | Fortis SP, Sofopoulos M, Sotiriadou NN, et al. Differential intratumoral distributions of CD8 and CD163 immune cells as prognostic biomarkers in breast cancer[J]. J Immunother Cancer, 2017, 5: 39. doi:10.1186/s40425-017-0240-7 |
| 14 | Jiang YM, Zhang Q, Hu YF, et al. ImmunoScore signature: a prognostic and predictive tool in gastric cancer[J]. Ann Surg, 2018, 267(3): 504-13. doi:10.1097/sla.0000000000002116 |
| 15 | Sjöberg E, Frödin M, Lövrot J, et al. A minority-group of renal cell cancer patients with high infiltration of CD20+B-cells is associated with poor prognosis[J]. Br J Cancer, 2018, 119(7): 840-6. doi:10.1038/s41416-018-0266-8 |
| 16 | Zhang JW, Li SW, Liu FK, et al. Role of CD68 in tumor immunity and prognosis prediction in pan-cancer[J]. Sci Rep, 2022, 12(1): 7844. doi:10.1038/s41598-022-11503-2 |
| 17 | Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study[J]. Lancet, 2018, 391(10135): 2128-39. |
| 18 | Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome[J]. Nat Rev Cancer, 2012, 12(4): 298-306. doi:10.1038/nrc3245 |
| 19 | Cros J, Sbidian E, Posseme K, et al. Nestin expression on tumour vessels and tumour-infiltrating macrophages define a poor prognosis subgroup of Pt1 clear cell renal cell carcinoma[J]. Virchows Arch, 2016, 469(3): 331-7. doi:10.1007/s00428-016-1973-2 |
| 20 | Voabil P, de Bruijn M, Roelofsen LM, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer[J]. Nat Med, 2021, 27(7): 1250-61. doi:10.1038/s41591-021-01398-3 |
| 21 | Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer[J]. Trends Cell Biol, 2015, 25(4): 198-213. doi:10.1016/j.tcb.2014.11.006 |
| 22 | Joyce JA, Pollard JW. Microenvironmental regulation of metastasis[J]. Nat Rev Cancer, 2009, 9(4): 239-52. doi:10.1038/nrc2618 |
| 23 | Krishna C, DiNatale RG, Kuo FS, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy[J]. Cancer Cell, 2021, 39(5): 662-77.e6. doi:10.1016/j.ccell.2021.03.007 |
| 24 | Giraldo NA, Becht E, Pagès F, et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer[J]. Clin Cancer Res, 2015, 21(13): 3031-40. doi:10.1158/1078-0432.ccr-14-2926 |
| 25 | Lu ZW, Yin Y, Rao T, et al. Interaction of immune cells with renal cancer development: Mendelian randomization (MR) study[J]. BMC Cancer, 2024, 24(1): 439. doi:10.1186/s12885-024-12196-8 |
| 26 | Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment[J]. Nat Rev Clin Oncol, 2017, 14(12): 717-34. doi:10.1038/nrclinonc.2017.101 |
| 27 | Yao JX, Xi W, Zhu YJ, et al. Checkpoint molecule PD-1-assisted CD8+ T lymphocyte count in tumor microenvironment predicts overall survival of patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors[J]. Cancer Manag Res, 2018, 10: 3419-31. doi:10.2147/cmar.s172039 |
| 28 | Braun DA, Street K, Burke KP, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma[J]. Cancer Cell, 2021, 39(5): 632-48.e8. doi:10.1016/j.ccell.2021.02.013 |
| 29 | Germano G, Frapolli R, Belgiovine C, et al. Role of macrophage targeting in the antitumor activity of trabectedin[J]. Cancer Cell, 2013, 23(2): 249-62. doi:10.1016/j.ccr.2013.01.008 |
| 30 | Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation[J]. Nat Rev Immunol, 2008, 8(12): 958-69. doi:10.1038/nri2448 |
| 31 | Chen YN, Hu MR, Wang L, et al. Macrophage M1/M2 polarization[J]. Eur J Pharmacol, 2020, 877: 173090. doi:10.1016/j.ejphar.2020.173090 |
| 32 | Luo L, Zhou HY, Su H. Identification of 4-genes model in papillary renal cell tumor microenvironment based on comprehensive analysis[J]. BMC Cancer, 2021, 21(1): 553. doi:10.1186/s12885-021-08319-0 |
| 33 | Yue YW, Cai XY, Lu CH, et al. Unraveling the prognostic significance and molecular characteristics of tumor-infiltrating B lymphocytes in clear cell renal cell carcinoma through a comprehensive bioinformatics analysis[J]. Front Immunol, 2023, 14: 1238312. doi:10.3389/fimmu.2023.1238312 |
| 34 | Fu HC, Zhu Y, Wang YW, et al. Tumor infiltrating mast cells (TIMs) confers a marked survival advantage in nonmetastatic clear-cell renal cell carcinoma[J]. Ann Surg Oncol, 2017, 24(5): 1435-42. doi:10.1245/s10434-016-5702-5 |
| 35 | Oldford SA, Haidl ID, Howatt MA, et al. A critical role for mast cells and mast cell-derived IL-6 in TLR2-mediated inhibition of tumor growth[J]. J Immunol, 2010, 185(11): 7067-76. doi:10.4049/jimmunol.1001137 |
| [1] | HAO Yuwei, GAO Sheng, ZHANG Xiaoyue, CUI Mengqiu, DING Xiaohui, WANG He, YANG Dawei, YE Huiyi, WANG Haiyi. Comparison of diagnostic performance of Clear Cell Likelihood Score v1.0 and v2.0 for clear renal cell carcinoma [J]. Journal of Southern Medical University, 2023, 43(5): 800-806. |
| [2] | . Expression of miR-223 in clear cell renal cell carcinoma and its significance [J]. Journal of Southern Medical University, 2015, 35(03): 338-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||