南方医科大学学报 ›› 2024, Vol. 44 ›› Issue (6): 1188-1197.doi: 10.12122/j.issn.1673-4254.2024.06.21
彭声旺1,2( ), 王永波1,2, 边兆英1,2, 马建华1,2, 黄静1(
), 王永波1,2, 边兆英1,2, 马建华1,2, 黄静1( )
)
收稿日期:2024-02-21
出版日期:2024-06-20
发布日期:2024-07-01
通讯作者:
黄静
E-mail:swpeng24@smu.edu.cn;hjing@smu.edu.cn
作者简介:彭声旺,在读硕士研究生,E-mail: swpeng24@smu.edu.cn
基金资助:
Shengwang PENG1,2( ), Yongbo WANG1,2, Zhaoying BIAN1,2, Jianhua MA1,2, Jing HUANG1(
), Yongbo WANG1,2, Zhaoying BIAN1,2, Jianhua MA1,2, Jing HUANG1( )
)
Received:2024-02-21
Online:2024-06-20
Published:2024-07-01
Contact:
Jing HUANG
E-mail:swpeng24@smu.edu.cn;hjing@smu.edu.cn
Supported by:摘要:
目的 提出一种基于改进可微分域转换的双域锥束计算机断层扫描(CBCT)重建框架DualCBR-Net用于锥角伪影校正。 方法 所提出的双域CBCT重建框架DualCBR-Net包含3个模块:投影域预处理、可微分域转换和图像后处理。投影域预处理模块首先对投影数据进行排方向扩充,使被扫描物体能够被X射线完全覆盖。可微分域转换模块引入重建和前投影算子去完成双域网络的前向和梯度回传过程,其中几何参数对应扩大的数据维度,扩大几何在网络前向过程中提供了重要先验信息,在反向过程中保证了回传梯度的精度,使得锥角区域的数据学习更为精准。图像域后处理模块对域转换后的图像进一步微调以去除残留伪影和噪声。 结果 在Mayo公开的胸部数据集上进行的验证实验结果显示,本研究提出的DualCBR-Net在伪影去除和结构细节保持方面均优于其他竞争方法;定量上,这种DualCBR-Net方法在PSNR和SSIM上相对于最新方法分别提高了0.6479和0.0074。 结论 本研究提出的基于改进可微分域转换的双域CBCT重建框架DualCBR-Net用于锥角伪影校正方法使有效联合训练CBCT双域网络成为可能,尤其是对于大锥角区域。。
彭声旺, 王永波, 边兆英, 马建华, 黄静. 基于改进可微分域转换的双域锥束计算机断层扫描重建网络用于锥角伪影校正[J]. 南方医科大学学报, 2024, 44(6): 1188-1197.
Shengwang PENG, Yongbo WANG, Zhaoying BIAN, Jianhua MA, Jing HUANG. A dual-domain cone beam computed tomography reconstruction framework with improved differentiable domain transform for cone-angle artifact correction[J]. Journal of Southern Medical University, 2024, 44(6): 1188-1197.
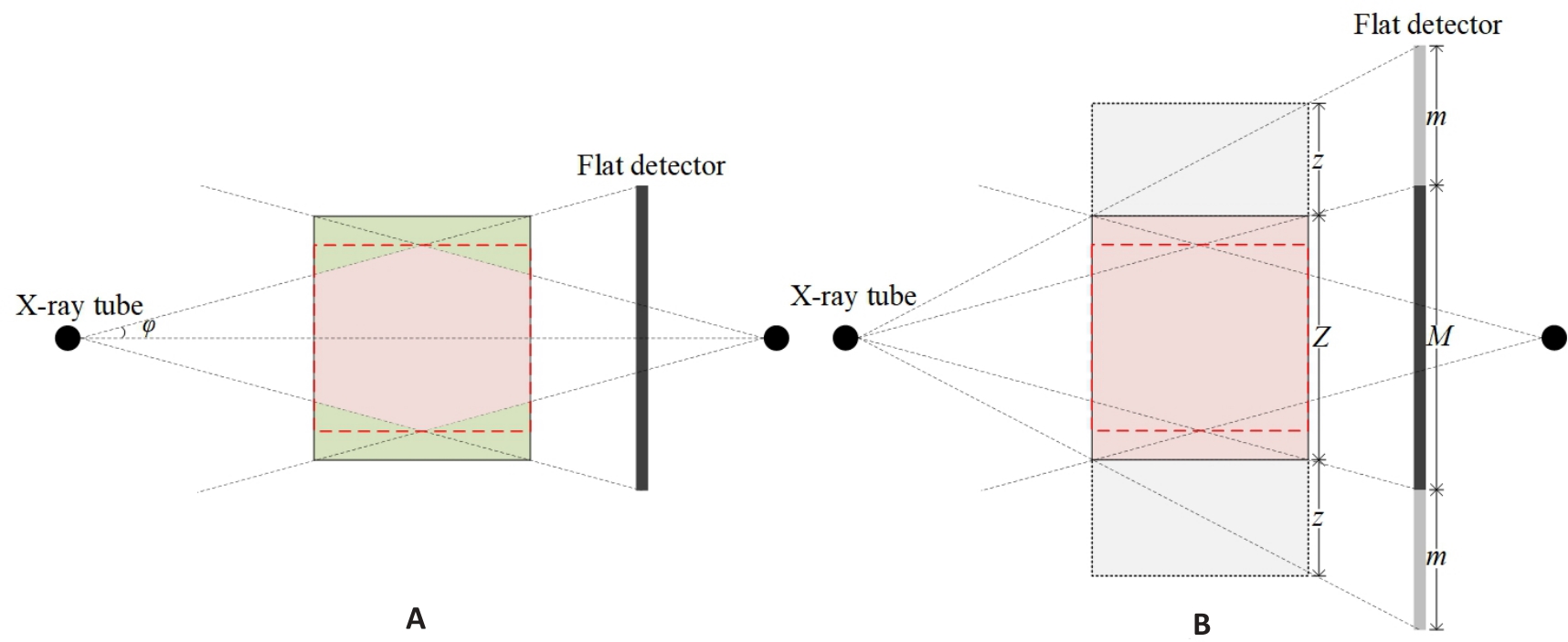
图1 CBCT初始几何和扩展几何示意图
Fig.1 Initial and extended geometry of CBCT. A: Initial geometry of CBCT. φ: cone-angle. Pink and green zones represent the effective volume and missing volume, respectively. B: Extended geometry of CBCT. M: Number of detector rows; m: Extended number of detector rows in each ends; Z: Number of reconstructed slices; z: Number of extended reconstructed slices in both ends.
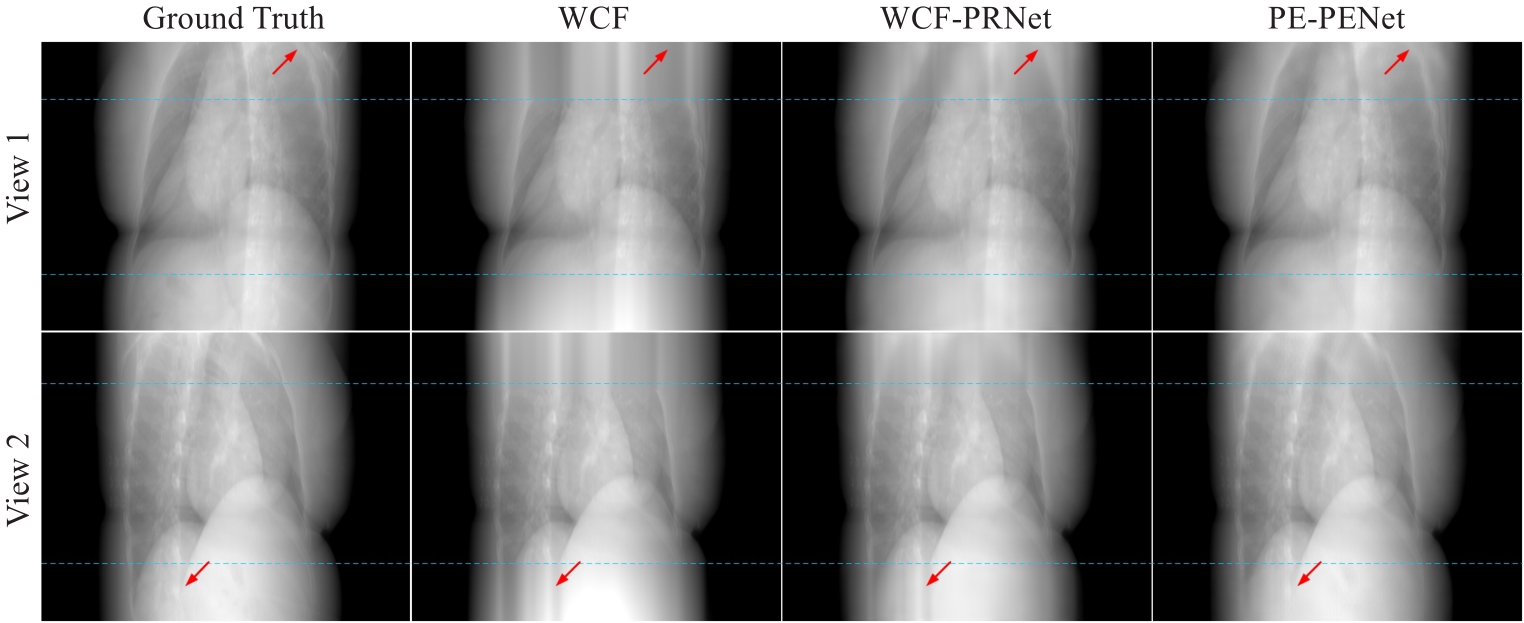
图3 两个不同角度下各个方法的投影外扩结果
Fig.3 Results of expanded projection data using different methods at two different views. The blue dashed lines at the top and bottom represent the 2 m rows of the projected data. The contrast regions are indicated by red arrows.
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| WCF | 28.8022±2.5542 | 0.9650±0.0045 | 9.6567±2.7958 |
| WCF-PRNet | 40.1381±0.7931 | 0.9818±0.0023 | 2.5204±0.2356 |
| PE-PRNet | 45.9624±0.3935 | 0.9870±0.0013 | 1.2849±0.0582 |
表1 不同方法投影外扩数据的定量比较结果
Tab.1 Quantitative comparison results of expanded projection data for different methods (Mean±SD)
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| WCF | 28.8022±2.5542 | 0.9650±0.0045 | 9.6567±2.7958 |
| WCF-PRNet | 40.1381±0.7931 | 0.9818±0.0023 | 2.5204±0.2356 |
| PE-PRNet | 45.9624±0.3935 | 0.9870±0.0013 | 1.2849±0.0582 |
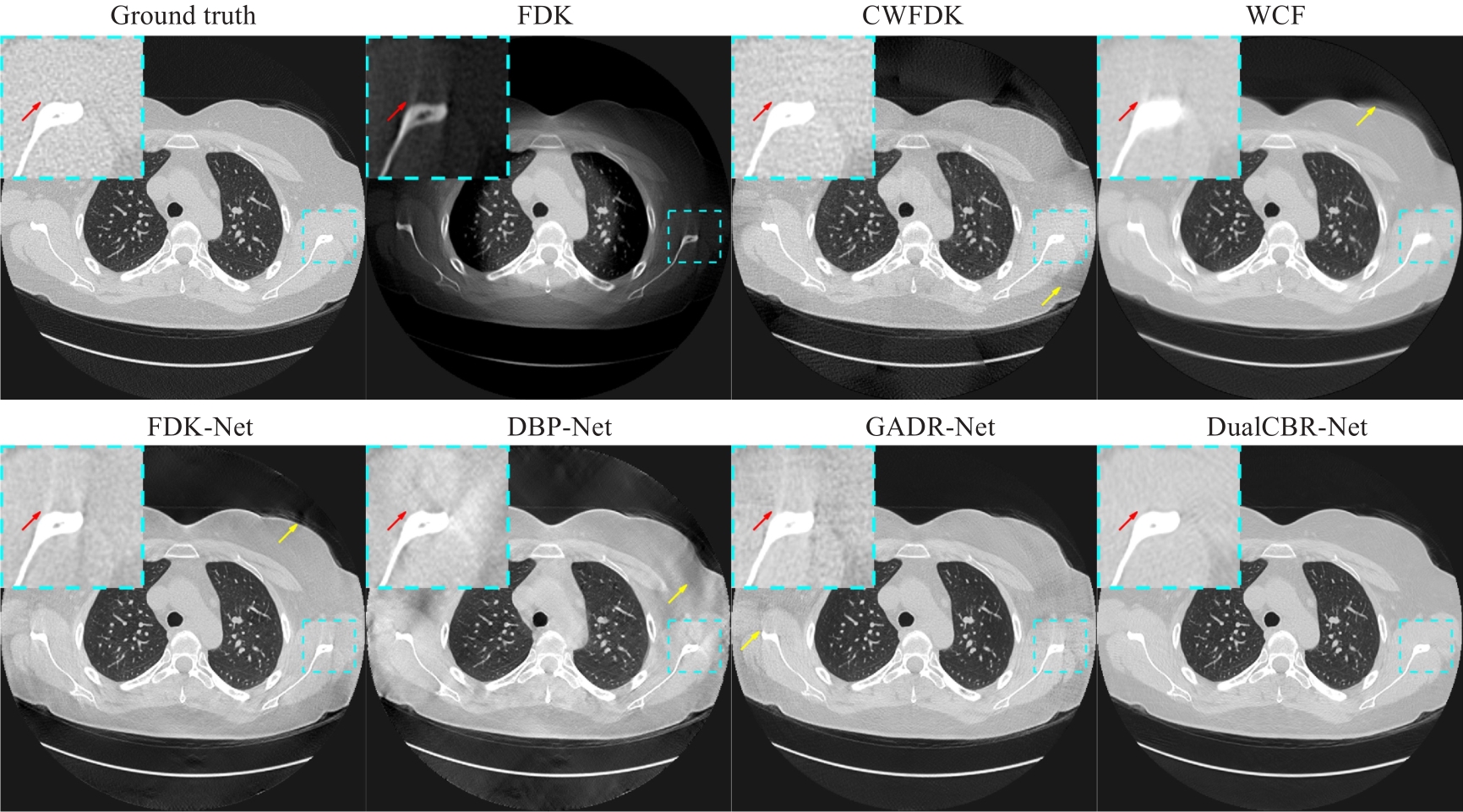
图4 轴面真值图像及各个方法的重建图像结果
Fig.4 Ground truth image and reconstruction results with different methods in the axial plane. The dashed blue boxes highlight the enlarged region of interest. The display range is (-1150, 350) Hu. The contrast regions are indicated by red arrows and the artifact regions by yellow arrows.
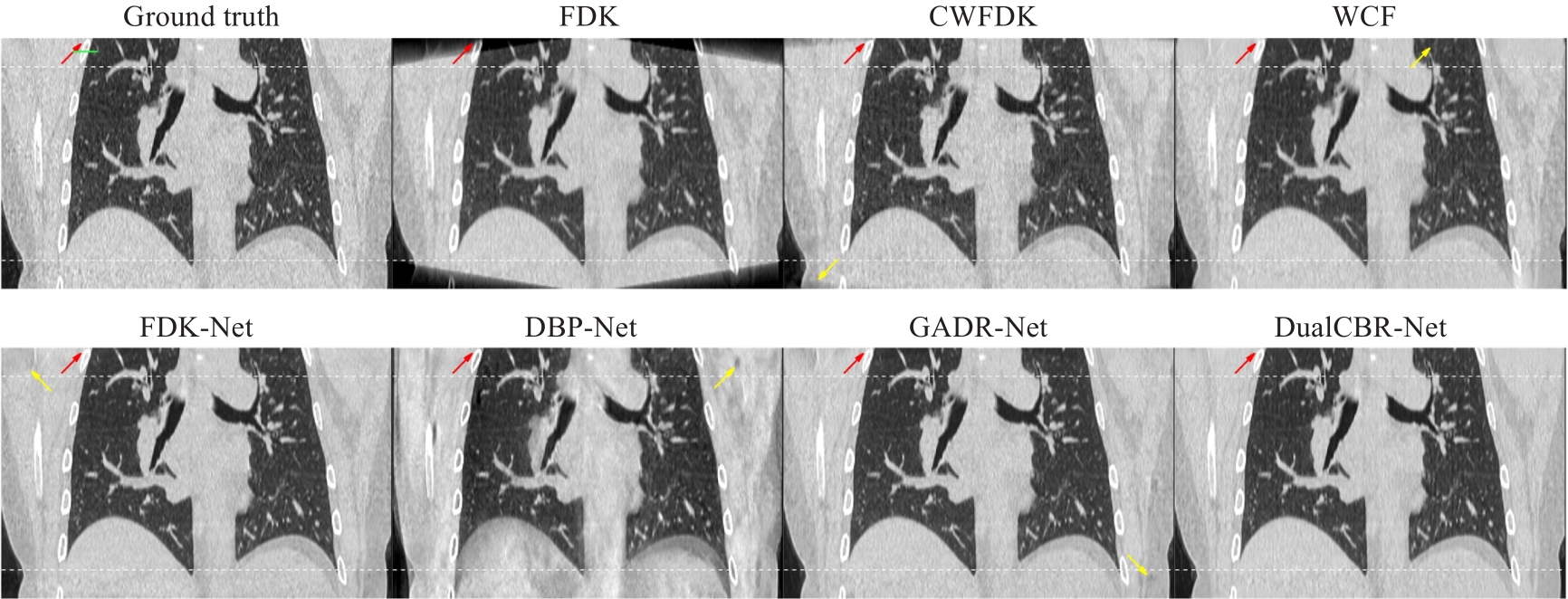
图5 冠状面真值图像及各个方法的重建图像结果
Fig.5 Ground truth image and reconstruction results with different methods in the coronal plane. The white curves at the top and bottom of the image indicate the regions affected by cone-angle artifacts. The display range is (-1150, 350) Hu. The contrast regions are indicated by red arrows and the artifact regions by yellow arrows.
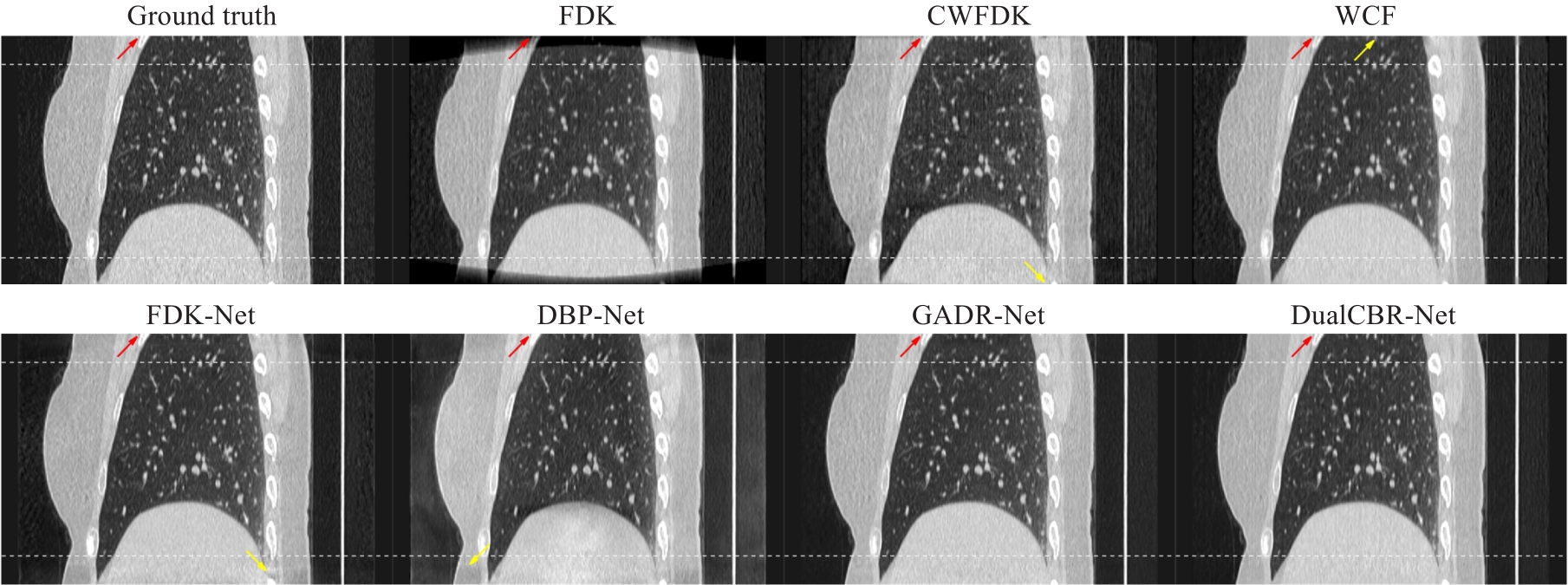
图6 矢状面真值图像及各个方法的重建图像结果
Fig.6 Ground truth image and reconstruction results with different methods in the sagittal plane. The white curves at the top and bottom of the image indicatethe regions affected by cone-angle artifacts. The display range is (-1150, 350) Hu. The contrast regions are indicated by red arrows and the artifact regions by yellow arrows.
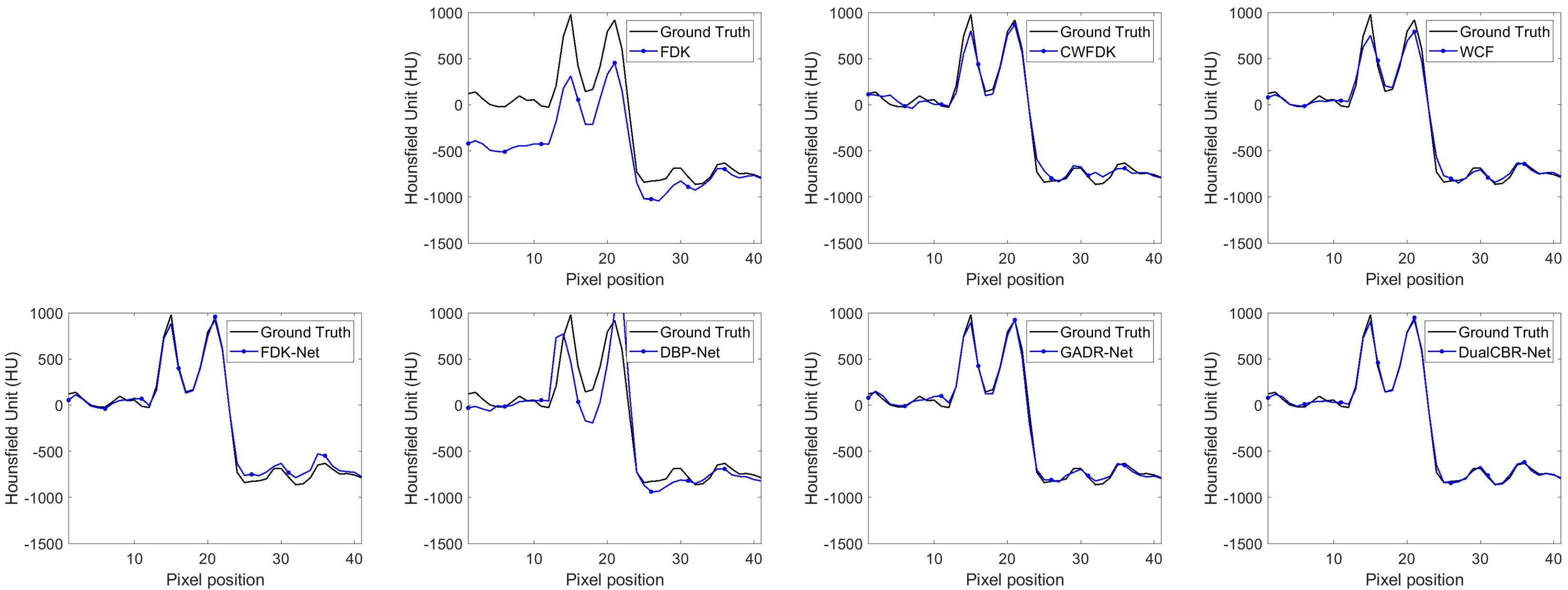
图7 真值图像和不同方法重建图像之间的轮廓图比较
Fig.7 Profile comparison between the ground truth image and different methods. The horizontal profile is indicated by green line in Fig.5.
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| FDK | 31.7062±7.1703 | 0.8049±0.1469 | 10.6439±14.0262 |
| CWFDK | 33.7062±1.8418 | 0.8417±0.0323 | 5.3882±1.2773 |
| WCF | 34.7489±1.7365 | 0.8675±0.0284 | 4.7616±0.9647 |
| FDK-Net | 35.4597±2.0657 | 0.8769±0.0286 | 4.4279±1.1314 |
| DBP-Net | 30.7556±1.6946 | 0.8471±0.0236 | 7.5277±1.3869 |
| GADR-Net | 36.3036±1.6391 | 0.8871±0.0254 | 3.9708±0.7276 |
| DualCBR-Net | 36.9515±1.6658 | 0.8945±0.0280 | 3.6882±0.6994 |
表2 不同方法锥角伪影去除的定量比较结果
Tab.2 Quantitative comparison results of cone-angle artifact removal performance for different methods (Mean±SD)
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| FDK | 31.7062±7.1703 | 0.8049±0.1469 | 10.6439±14.0262 |
| CWFDK | 33.7062±1.8418 | 0.8417±0.0323 | 5.3882±1.2773 |
| WCF | 34.7489±1.7365 | 0.8675±0.0284 | 4.7616±0.9647 |
| FDK-Net | 35.4597±2.0657 | 0.8769±0.0286 | 4.4279±1.1314 |
| DBP-Net | 30.7556±1.6946 | 0.8471±0.0236 | 7.5277±1.3869 |
| GADR-Net | 36.3036±1.6391 | 0.8871±0.0254 | 3.9708±0.7276 |
| DualCBR-Net | 36.9515±1.6658 | 0.8945±0.0280 | 3.6882±0.6994 |
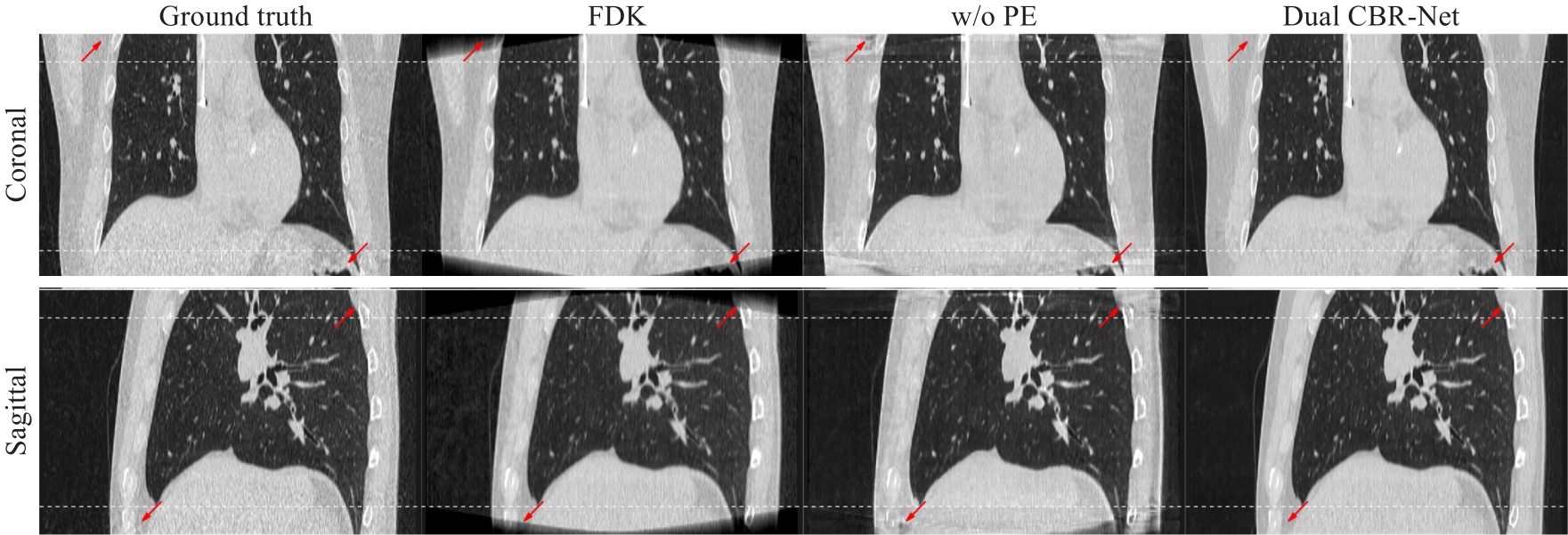
图8 投影外扩策略的验证结果
Fig.8 Validation results of the projection expansion strategy. The upper and lower rows are coronal plane and sagittal plane reconstruction images, respectively. The white curves at the top and bottom of the image indicated the regions affected by cone-angle artifacts. Projection extension strategy is not used for images in the "w/o PE" column. The display range was (-1150, 350) Hu. The contrast areas are indicated by red arrows.
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| PENet | 35.0010±1.2653 | 0.8523±0.0284 | 4.6987±0.6780 |
| DualCBR-Net (w/o IRNet) | 35.2349±1.1689 | 0.8695±0.0281 | 4.3842±0.6548 |
| DualCBR-Net (w/o AM) | 36.4023±1.5633 | 0.8825±0.0356 | 3.9054±0.5966 |
| DualCBR-Net (4 slices) | 36.5393±1.5784 | 0.8890±0.0325 | 3.8529±0.6498 |
| DualCBR-Net (6 slices) | 36.6722±1.6950 | 0.8901±0.0278 | 3.7021±0.6447 |
| DualCBR-Net (10 slices) | 36.2839±1.7725 | 0.8760±0.0252 | 4.0133±0.6561 |
| DualCBR-Net (w/o | 36.4529±1.5612 | 0.8796±0.0265 | 3.9255±0.6846 |
| DualCBR-Net | 36.9515±1.6658 | 0.8945±0.0280 | 3.6882±0.6994 |
表3 消融实验定量比较结果
Tab.3 Quantitative comparison results in ablation studies (Mean±SD)
| Methods | PSNR | SSIM | RMSE |
|---|---|---|---|
| PENet | 35.0010±1.2653 | 0.8523±0.0284 | 4.6987±0.6780 |
| DualCBR-Net (w/o IRNet) | 35.2349±1.1689 | 0.8695±0.0281 | 4.3842±0.6548 |
| DualCBR-Net (w/o AM) | 36.4023±1.5633 | 0.8825±0.0356 | 3.9054±0.5966 |
| DualCBR-Net (4 slices) | 36.5393±1.5784 | 0.8890±0.0325 | 3.8529±0.6498 |
| DualCBR-Net (6 slices) | 36.6722±1.6950 | 0.8901±0.0278 | 3.7021±0.6447 |
| DualCBR-Net (10 slices) | 36.2839±1.7725 | 0.8760±0.0252 | 4.0133±0.6561 |
| DualCBR-Net (w/o | 36.4529±1.5612 | 0.8796±0.0265 | 3.9255±0.6846 |
| DualCBR-Net | 36.9515±1.6658 | 0.8945±0.0280 | 3.6882±0.6994 |
| 1 | Jaffray DA, Siewerdsen JH, Wong JW, et al. Flat-panel cone-beam computed tomography for image-guided radiation therapy[J]. Int J Radiat Oncol Biol Phys, 2002, 53(5): 1337-49. |
| 2 | Wang G, Zhao SY, Heuscher D. A knowledge-based cone-beam X-ray CT algorithm for dynamic volumetric cardiac imaging[J]. Med Phys, 2002, 29(8): 1807-22. |
| 3 | Pauwels R, Araki K, Siewerdsen JH, et al. Technical aspects of dental CBCT: state of the art[J]. Dentomaxillofac Radiol, 2015, 44(1): 20140224. |
| 4 | Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm[J]. J Opt Soc Am A, 1984, 1(6): 612. |
| 5 | Tuy HK. An inversion formula for cone-beam reconstruction[J]. SIAM J Appl Math, 1983, 43(3): 546-52. |
| 6 | Grass M, Köhler T, Proksa R. Angular weighted hybrid cone-beam CT reconstruction for circular trajectories[J]. Phys Med Biol, 2001, 46(6): 1595-610. |
| 7 | Tang X, Hsieh J, Nilsen RA, et al. A three-dimensional-weighted cone beam filtered backprojection (CB-FBP) algorithm for image reconstruction in volumetric CT-helical scanning[J]. Phys Med Biol, 2006, 51(4): 855-74. |
| 8 | Mori S, Endo M, Komatsu S, et al. A combination-weighted Feldkamp-based reconstruction algorithm for cone-beam CT[J]. Phys Med Biol, 2006, 51(16): 3953-65. |
| 9 | Grimmer R, Oelhafen M, Elstrøm U, et al. Cone-beam CT image reconstruction with extended z range[J]. Med Phys, 2009, 36(7): 3363-70. |
| 10 | Zhu L, Starman J, Fahrig R. An efficient estimation method for reducing the axial intensity drop in circular cone-beam CT[J]. Int J Biomed Imaging, 2008: 242841. |
| 11 | Hsieh J, Chao E, Thibault J, et al. A novel reconstruction algorithm to extend the CT scan field-of-view[J]. Med Phys, 2004, 31(9): 2385-91. |
| 12 | Hsieh J. Two-pass algorithm for cone-beam reconstruction[C]. San Diego: Medical Imaging 2000 : Image Processing, 2000: 533-40. |
| 13 | Han C, Baek J. Multi-pass approach to reduce cone-beam artifacts in a circular orbit cone-beam CT system[J]. Opt Express, 2019, 27(7): 10108-26. |
| 14 | Han Y, Kim J, Ye JC. Differentiated backprojection domain deep learning for conebeam artifact removal[J]. IEEE Trans Med Imaging, 2020, 39(11): 3571-82. |
| 15 | Minnema J, van Eijnatten M, der Sarkissian H, et al. Efficient high cone-angle artifact reduction in circular cone-beam CT using deep learning with geometry-aware dimension reduction[J]. Phys Med Biol, 2021, 66(13): 135015. |
| 16 | Xia WJ, Shan HM, Wang G, et al. Physics-/ Model-Based and Data-Driven Methods for Low-Dose Computed Tomography: a survey[J]. IEEE Signal Process Mag, 2023, 40(2): 89-100. |
| 17 | He J, Wang YB, Ma JH. Radon inversion via deep learning[J]. IEEE Trans Med Imaging, 2020, 39(6): 2076-87. |
| 18 | He J, Chen SL, Zhang H, et al. Downsampled imaging geometric modeling for accurate CT reconstruction via deep learning[J]. IEEE Trans Med Imaging, 2021, 40(11): 2976-85. |
| 19 | Würfl T, Hoffmann M, Christlein V, et al. Deep learning computed tomography: learning projection-domain weights from image domain in limited angle problems[J]. IEEE Trans Med Imag, 2018, 37(6): 1454-63. |
| 20 | Hu DL, Liu J, Lv TL, et al. Hybrid-domain neural network processing for sparse-view CT reconstruction[J]. IEEE Trans Radiat Plasma Med Sci, 2021, 5(1): 88-98. |
| 21 | Zhang YK, Hu DL, Zhao QL, et al. CLEAR: comprehensive learning enabled adversarial reconstruction for subtle structure enhanced low-dose CT imaging[J]. IEEE Trans Med Imag, 2021, 40(11): 3089-101. |
| 22 | Yang QS, Yan PK, Zhang YB, et al. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss[J]. IEEE Trans Med Imaging, 2018, 37(6): 1348-57. |
| 23 | Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation[EB/OL]. [2015-05-18]. . |
| 24 | Guo MH, Xu TX, Liu JJ, et al. Attention mechanisms in computer vision: a survey[J]. Comput Vis Medium, 2022, 8(3): 331-68. |
| 25 | Moen TR, Chen BY, Holmes DR 3rd, et al. Low-dose CT image and projection dataset[J]. Med Phys, 2021, 48(2): 902-11. |
| 26 | van Aarle W, Palenstijn WJ, De Beenhouwer J, et al. The ASTRA Toolbox: a platform for advanced algorithm development in electron tomography[J]. Ultramicroscopy, 2015, 157: 35-47. |
| 27 | Kingma DP, Ba J. Adam: a method for stochastic optimization[EB/OL]. [2014-12-22]. |
| 28 | Jin KH, McCann MT, Froustey E, et al. Deep convolutional neural network for inverse problems in imaging[J]. IEEE Trans Image Process, 2017, 26(9): 4509-22. |
| 29 | Parker DL. Optimal short scan convolution reconstruction for fanbeam CT[J]. Med Phys, 1982, 9(2): 254-7. |
| 30 | Lee D, Choi S, Kim HJ. High quality imaging from sparsely sampled computed tomography data with deep learning and wavelet transform in various domains[J]. Med Phys, 2019, 46(1): 104-15. |
| 31 | Zhou B, Chen XC, Zhou SK, et al. DuDoDR-Net: dual-domain data consistent recurrent network for simultaneous sparse view and metal artifact reduction in computed tomography[J]. Med Image Anal, 2022, 75: 102289. |
| 32 | Chao LY, Wang ZW, Zhang HB, et al. Sparse-view cone beam CT reconstruction using dual CNNs in projection domain and image domain[J]. Neurocomputing, 2022, 493(7): 536-47. |
| 33 | Chen DD, Tachella J, Davies ME. Equivariant imaging: learning beyond the range space[C]. Montreal: 2021 IEEE/CVF International Conference on Computer Vision, 2021: 4359-68. |
| 34 | Chen DD, Tachella J, Davies ME. Robust Equivariant Imaging: a fully unsupervised framework for learning to image from noisy and partial measurements[C]. New Orleans, 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022: 5637-46. |
| 35 | Peng SW, Liao JY, Li DY, et al. Noise-conscious explicit weighting network for robust low-dose CT imaging[C]. San Diego: Medical Imaging 2023 : Physics of Medical Imaging, 2023: 711-8. |
| 36 | Chen J, Zhang R, Yu T, et al. Label-retrieval-augmented diffusion models for learning from noisy labels[J]. Adv Neural Inf Process Syst, 2024, 36. doi: 10.48550/arXiv.2305.19518 |
| [1] | 林宗悦, 王永波, 边兆英, 马建华. 基于深度模糊学习的牙科CBCT运动伪影校正算法[J]. 南方医科大学学报, 2024, 44(6): 1198-1208. |
| [2] | 廖静怡, 彭声旺, 王永波, 边兆英. 基于生成式投影插值的双域CBCT稀疏角度重建方法[J]. 南方医科大学学报, 2024, 44(10): 2044-2054. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||