南方医科大学学报 ›› 2024, Vol. 44 ›› Issue (5): 920-929.doi: 10.12122/j.issn.1673-4254.2024.05.14
左志威1( ), 孟庆良1, 崔家康1, 郭克磊2, 卞华1,2(
), 孟庆良1, 崔家康1, 郭克磊2, 卞华1,2( )
)
收稿日期:2023-11-20
出版日期:2024-05-20
发布日期:2024-06-06
通讯作者:
卞华
E-mail:15737264121@163.com;biancrown@163.com
作者简介:左志威,硕士,E-mail: 15737264121@163.com
基金资助:
Zhiwei ZUO1( ), Qingliang MENG1, Jiakang CUI1, Kelei GUO2, Hua BIAN1,2(
), Qingliang MENG1, Jiakang CUI1, Kelei GUO2, Hua BIAN1,2( )
)
Received:2023-11-20
Online:2024-05-20
Published:2024-06-06
Contact:
Hua BIAN
E-mail:15737264121@163.com;biancrown@163.com
Supported by:摘要:
目的 建立基 于GEO数据库硬皮病线粒体相关基因的机器学习和人工神经网络联合诊断模型并评价其效果。 方法 通过GEO数据库获取3份硬皮病芯片。其中GSE95065及GSE59785合并作为实验数据集并提取线粒体相关基因表达量,使用随机森林、LASSO回归和SVM算法筛选硬皮病线粒体相关特征基因,并用特征基因构建人工神经网络模型,用10折交叉验证模型准确性。来用验证数据集GSE76807对模型进一步验证,利用ROC曲线下面积值评估模型准确性。用RT-qPCR实验验证关键基因mRNA相对表达量。最后用CIBERSORT算法预估硬皮病与筛选出的潜在生物标志物的生物信息学关联。 结果 共获取差异基因24个,其中上调基因11个,下调基因13个。通过3种机器学习算法筛选到最相关的7个线粒体相关特征基因(POLB、GSR、KRAS、NT5DC2、NOX4、IGF1、TGM2),并构建人工神经网络诊断模型。使用该模型绘制了实验组和验证组诊断的ROC曲线,AUC值为0.984。验证组AUC为0.740。10折交叉验证AUC平均值大于0.980。RT-qPCR结果显示,与对照组相比,硬皮病中POLB(P=0.004)、GSR(P=0.029)、KRAS(P=0.007)、NOX4(P=0.019)、IGF1(P=0.008)、TGM2(P<0.0001)表达量明显上调,而NT5DC2(P=0.001)表达量在硬皮病组中明显下调。免疫细胞浸润显示,特征基因与滤泡辅助T细胞、幼稚B细胞、静息树突状细胞、记忆激活CD4+T细胞、巨噬细胞M0、单核细胞、记忆静息CD4+T细胞和肥大细胞激活等相关。 结论 构建了硬皮病特征基因的人工神经网络诊断模型,为探索硬皮病发病机制提供了一个新视角。
左志威, 孟庆良, 崔家康, 郭克磊, 卞华. 基于硬皮病线粒体相关基因的人工神经网络模型的构建[J]. 南方医科大学学报, 2024, 44(5): 920-929.
Zhiwei ZUO, Qingliang MENG, Jiakang CUI, Kelei GUO, Hua BIAN. An artificial neural network diagnostic model for scleroderma and immune cell infiltration analysis based on mitochondria-associated genes[J]. Journal of Southern Medical University, 2024, 44(5): 920-929.
| DATA | Sample size | Normal sample | SSc sample | Organization type | Data type |
|---|---|---|---|---|---|
| GSE95065 | 33 | 15 | 18 | Homo sapiens | Expression profiling by array |
| GSE59785 | 82 | 2 | 80 | Homo sapiens | Expression profiling by array |
| GSE76807 | 15 | 5 | 10 | Homo sapiens | Expression profiling by array |
表1 GEO数据库芯片数据集
Tab.1 GEO database chip data set
| DATA | Sample size | Normal sample | SSc sample | Organization type | Data type |
|---|---|---|---|---|---|
| GSE95065 | 33 | 15 | 18 | Homo sapiens | Expression profiling by array |
| GSE59785 | 82 | 2 | 80 | Homo sapiens | Expression profiling by array |
| GSE76807 | 15 | 5 | 10 | Homo sapiens | Expression profiling by array |
| Primer | Sequence 5'-3' |
|---|---|
| POLB | F: CTTCACTGGGAGTGACATCTTT R: CAGCGACTCCAGTGACC |
| GSR | F: GAGCTCCAAGTGGTGACTTC R: CAGGCCCTTAGAATTTGGGT |
| KRAS | F: GTGGATGAGTATGACCCTACG R GACCTGCTGTGTCGAGAATATC |
| NT5DC2 | F: ACGTCGTCATCGTCCAG R: TCTCTAGGCGAGTGATACGG |
| NOX4 | F: AGACTCTACACATCACATGTGG R: AAAGTTGAGGGCATTCACCA |
| IGF1 | F: CCCACTGAAGCCTACAAA R: TTTCTTGTTTGTCGATAGGGA |
| TGM2 | F: TGTCTGACAATGTGGAGGAG R: GCTGTAGCGAGAGGACATT |
| β-actin | F: TGCTGTCCCTGTATGCCTCTG R: TGATGTCACGCACGATTTCC |
表2 目的基因引物序列
Tab.2 Primer sequence of the target genes
| Primer | Sequence 5'-3' |
|---|---|
| POLB | F: CTTCACTGGGAGTGACATCTTT R: CAGCGACTCCAGTGACC |
| GSR | F: GAGCTCCAAGTGGTGACTTC R: CAGGCCCTTAGAATTTGGGT |
| KRAS | F: GTGGATGAGTATGACCCTACG R GACCTGCTGTGTCGAGAATATC |
| NT5DC2 | F: ACGTCGTCATCGTCCAG R: TCTCTAGGCGAGTGATACGG |
| NOX4 | F: AGACTCTACACATCACATGTGG R: AAAGTTGAGGGCATTCACCA |
| IGF1 | F: CCCACTGAAGCCTACAAA R: TTTCTTGTTTGTCGATAGGGA |
| TGM2 | F: TGTCTGACAATGTGGAGGAG R: GCTGTAGCGAGAGGACATT |
| β-actin | F: TGCTGTCCCTGTATGCCTCTG R: TGATGTCACGCACGATTTCC |
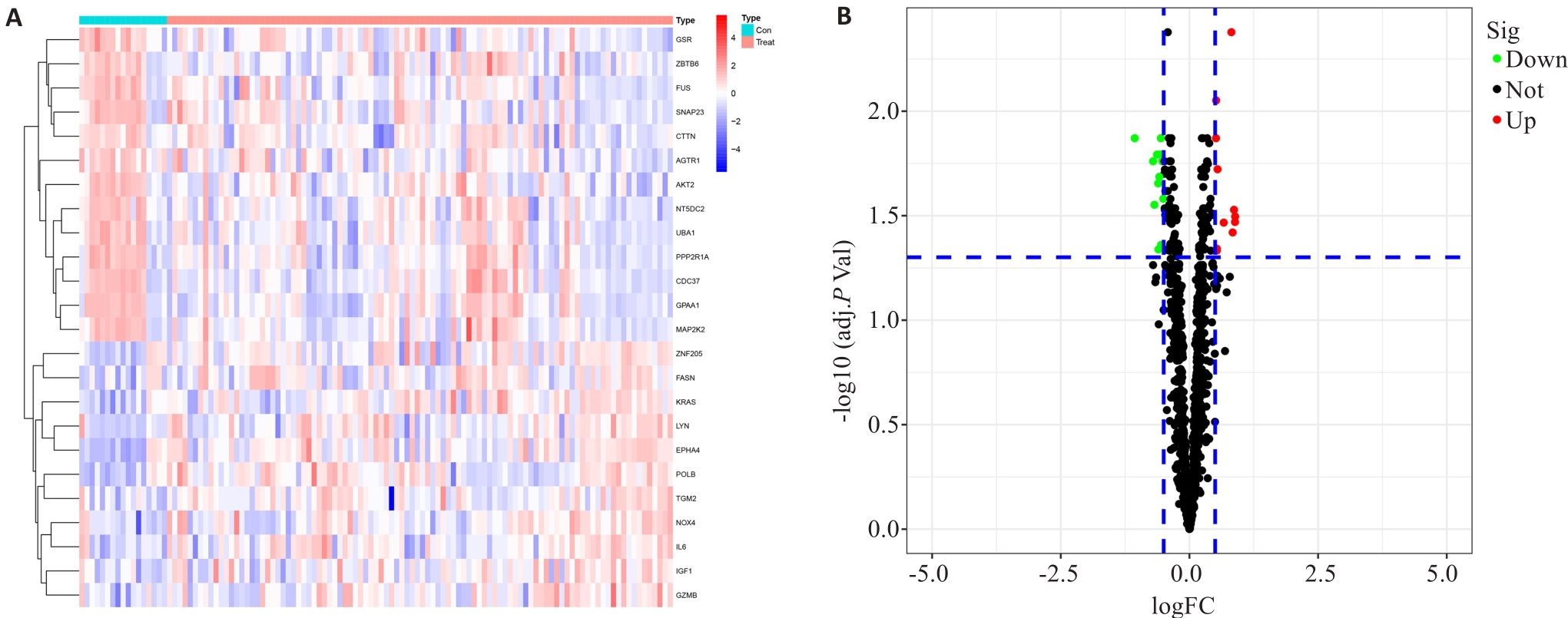
图2 硬皮病中线粒体相关差异基因表达分析
Fig.2 Analysis of the differential expressions of the differential mitochondria-related gene in scleroderma. A: DEGs heatmap (red for up-regulated and blue for down-regulated genes). B: DEGs volcano map (red for up-regulated and blue for down-regulated genes).
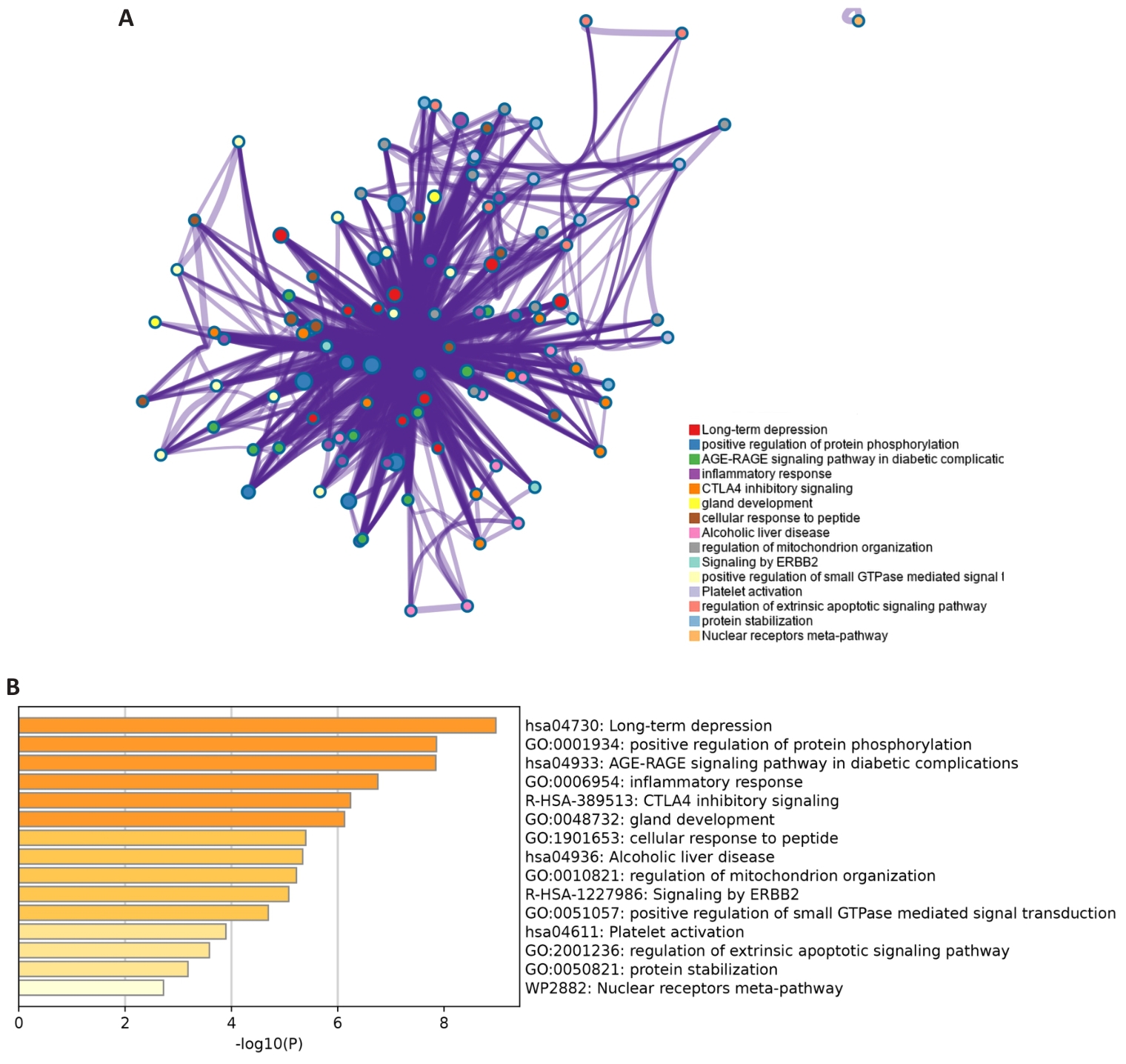
图3 差异基因的Metascape分析
Fig.3 Metascape analysis of the DEGs in scleroderma. A: DEG enrichment pathway and process network. B: Histogram of DEGs enrichment pathways and processes.
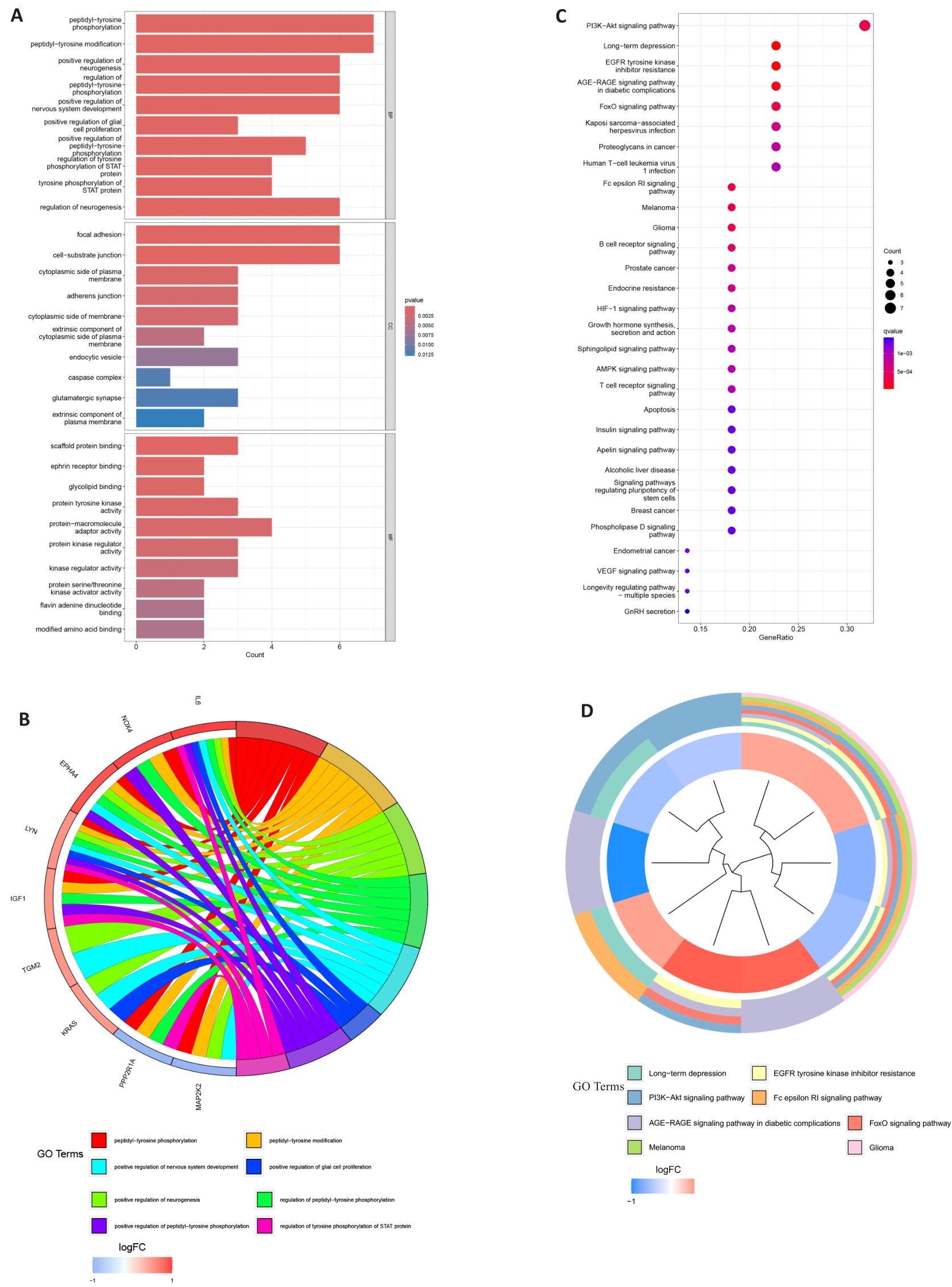
图4 差异基因的GO和KEGG富集分析
Fig.4 GO and KEGG enrichment analysis of DEGs in scleroderma. A: GO bar chart. B: GO circle. C: KEGG bubble diagram. D: KEGG cluster graph.
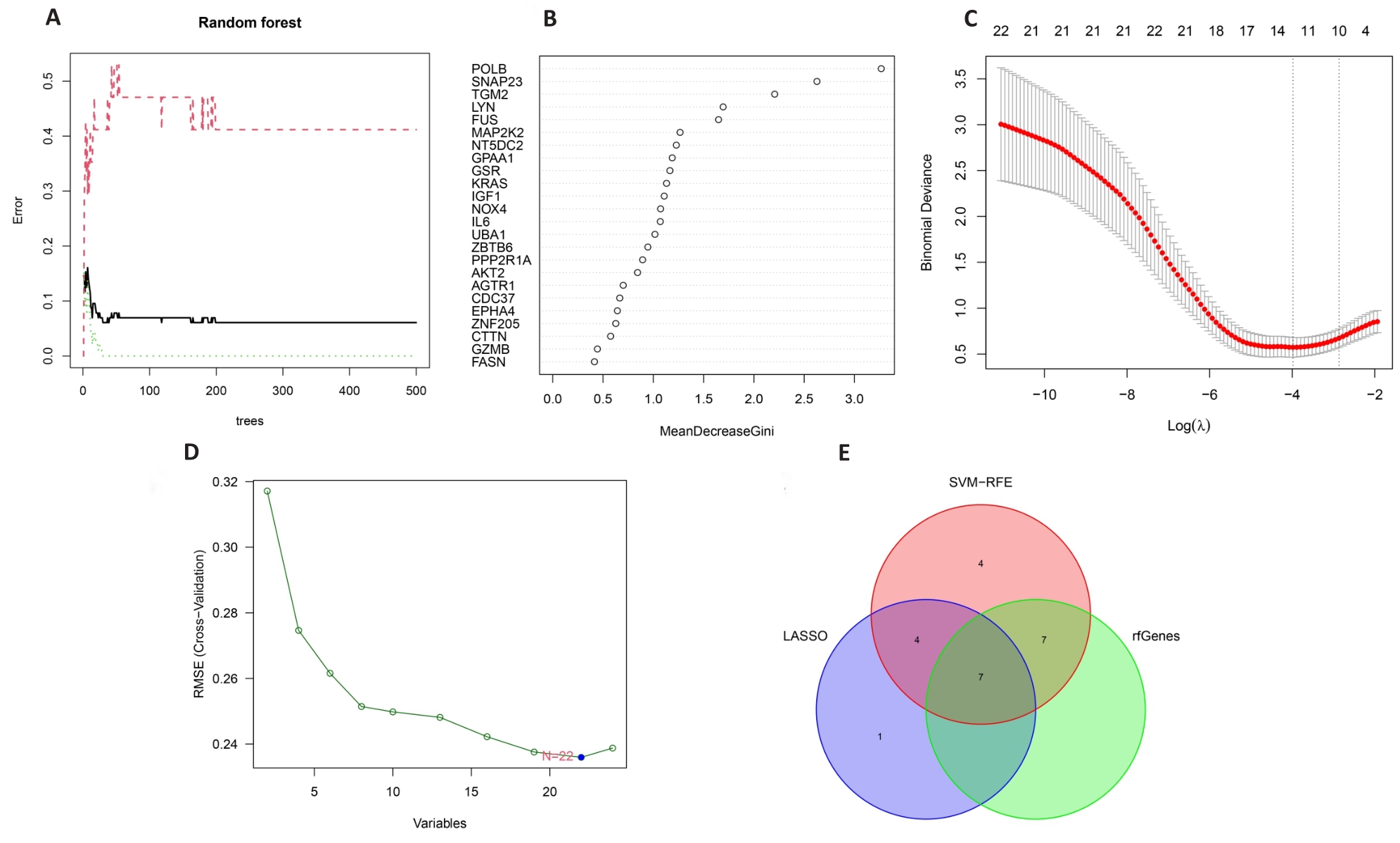
图5 机器学习算法筛选出的关键基因结果
Fig.5 Selection of the key genes using 3 machine learning algorithms. A: Correlation between the number of random forest trees and the model error. B: Result of Gini coefficient method in random forest classifier. C: Characteristic genes selected by LASSO regression algorithm. D: Feature genes screened by SVM algorithm. E: Venn diagram of the intersected genes of the 3 algorithms.
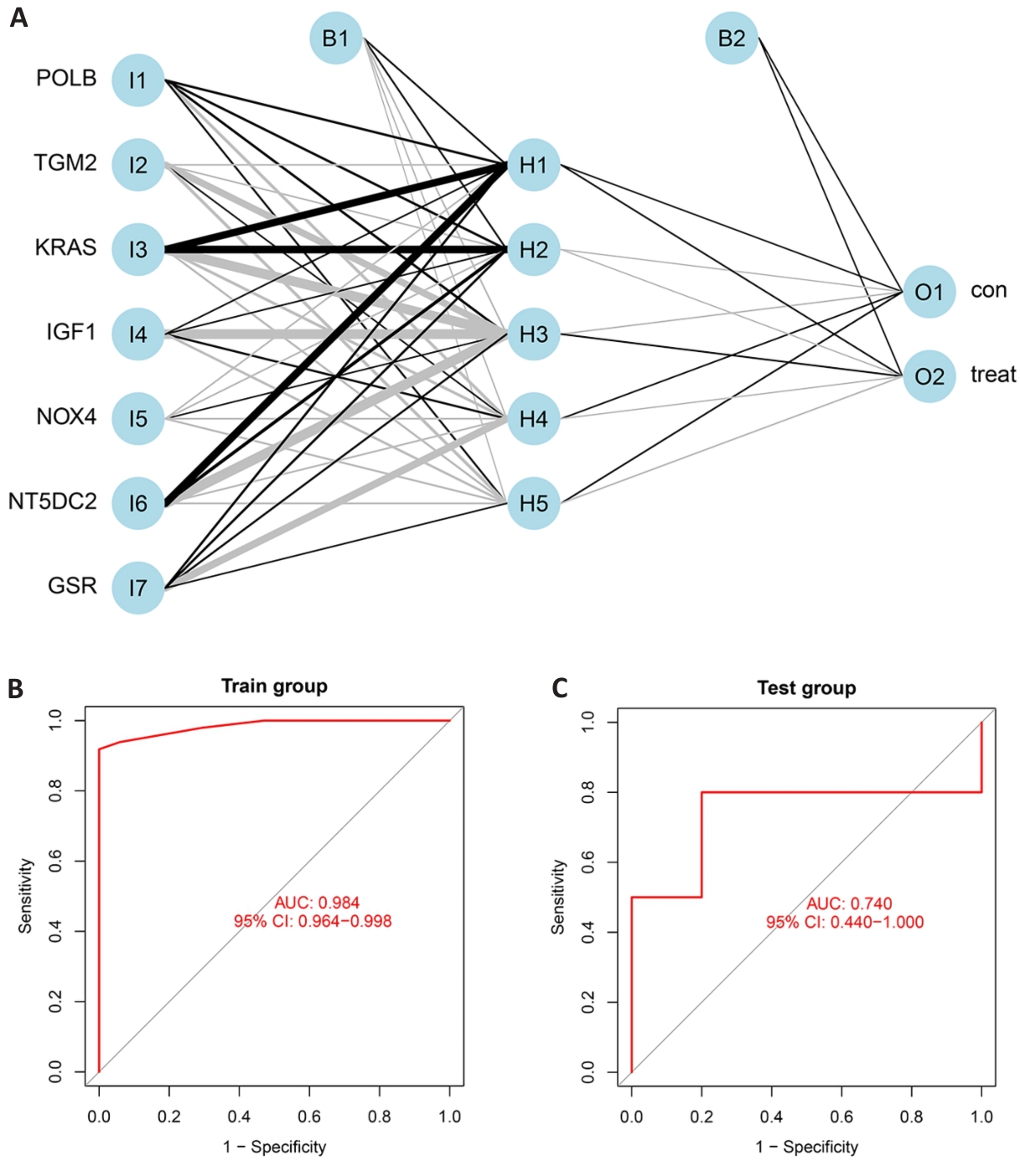
图7 人工神经网络模型的构建与验证
Fig.7 Construction and verification of artificial neural network model. A: ANN result visualization. B: ROC curves of mitochondria-associated genes in the training dataset. C: ROC curves for mitochondria-related genes in the verification dataset.
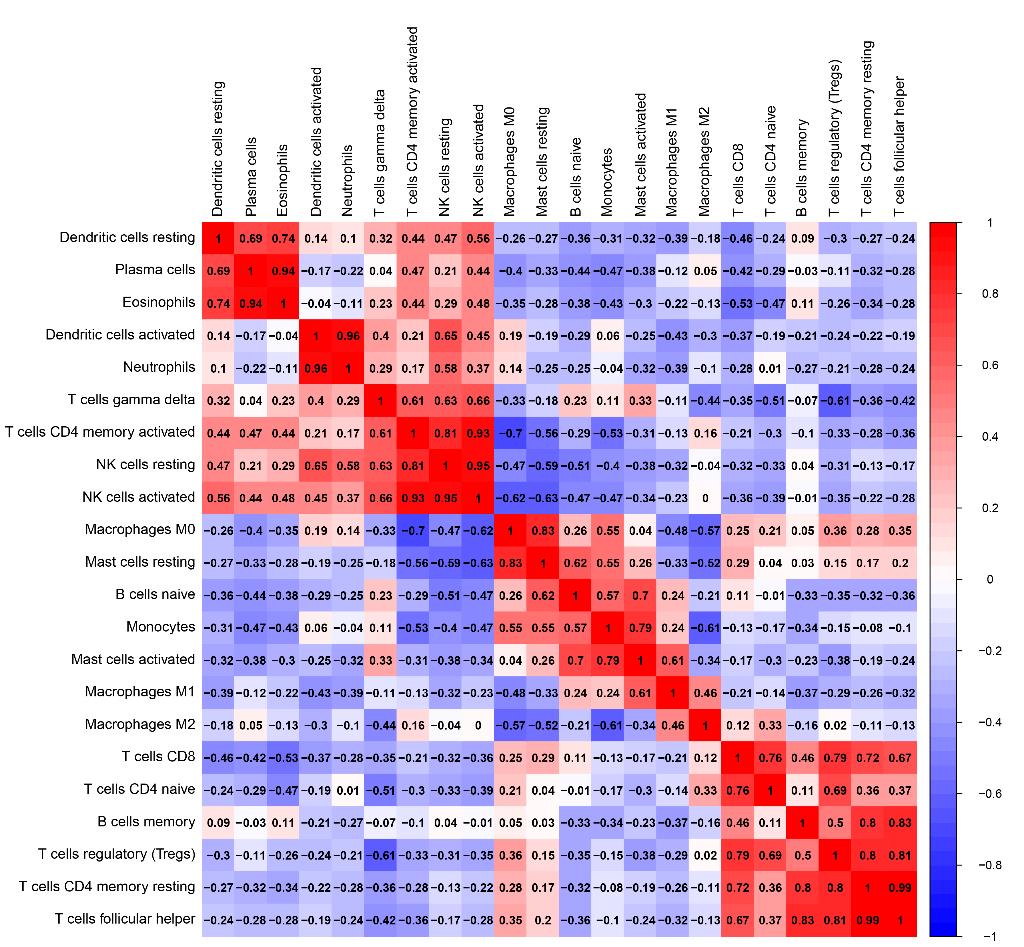
图9 免疫细胞表达相对含量相关性分析
Fig.9 Correlation analysis of immune cell expression. Red indicates a positive correlation and blue indicates a negative correlation.
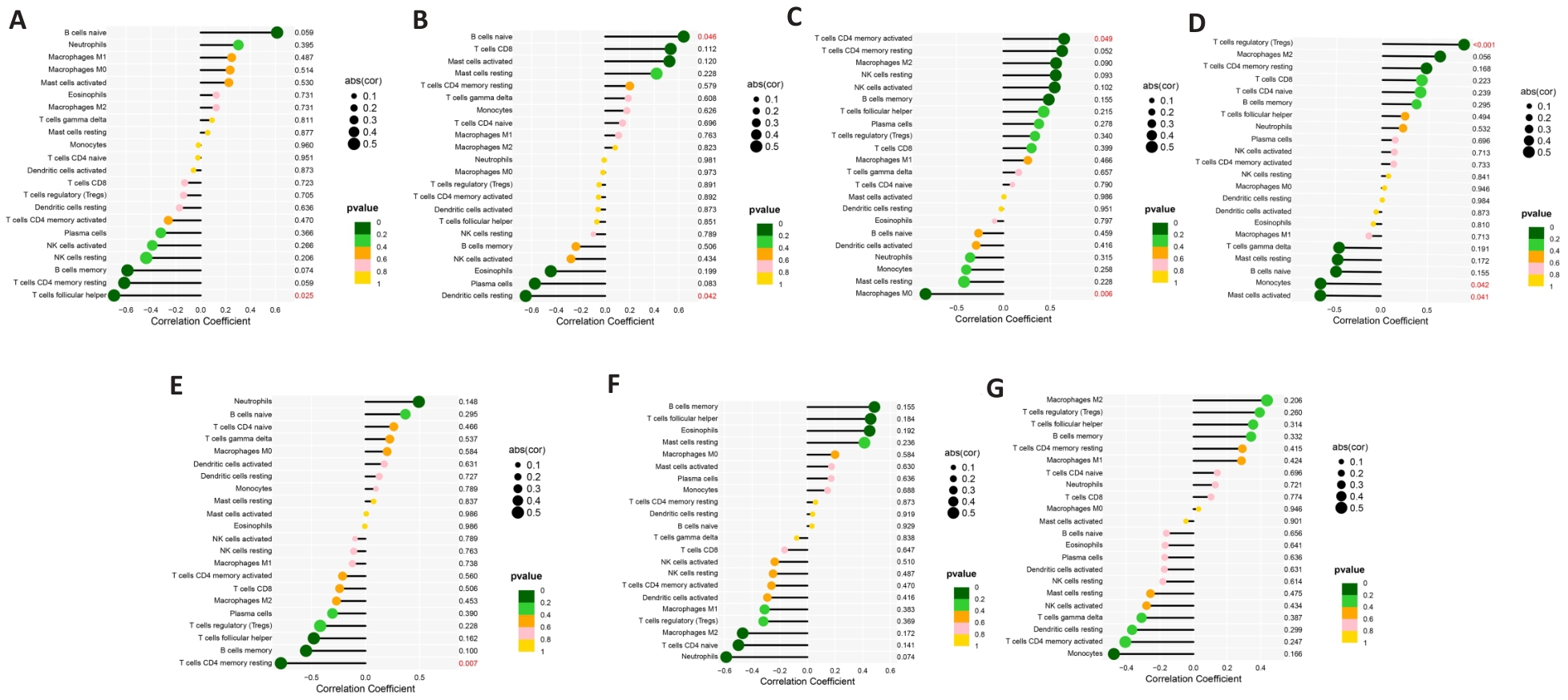
图10 基因潜在生物标志物与浸润性免疫细胞的相关性
Fig.10 Correlation of the genetic biomarkers with the infiltrating immune cells. A: GSR. B: IGF1. C: KRAS. D: NOX4. E: NT5DC2. F: POLB. G: TGM2. P<0.05 indicates a significant correlation between immune cells and genes.
| 1 | Orteu CH, Ong VH, Denton CP. Scleroderma mimics - Clinical features and management[J]. Best Pract Res Clin Rheumatol, 2020, 34(1): 101489. DOI: 10.1016/j.berh.2020.101489 |
| 2 | Li SC. Scleroderma in children and adolescents: localized Scleroderma and systemic sclerosis[J]. Pediatr Clin North Am, 2018, 65(4): 757-81. DOI: 10.1016/j.pcl.2018.04.002 |
| 3 | Tsou PS, Sawalha AH. Unfolding the pathogenesis of scleroderma through genomics and epigenomics[J]. J Autoimmun, 2017, 83: 73-94. DOI: 10.1016/j.jaut.2017.05.004 |
| 4 | Ge YZ, Zhou L, Chen ZX, et al. Identification of differentially expressed genes, signaling pathways and immune infiltration in rheumatoid arthritis by integrated bioinformatics analysis[J]. Hereditas, 2021, 158(1): 5. DOI: 10.1186/s41065-020-00169-3 |
| 5 | Swanson MB, Weidemann DK, Harshman LA. The impact of rural status on pediatric chronic kidney disease[J]. Pediatr Nephrol, 2024, 39(2): 435-46. DOI: 10.1007/s00467-023-06001-0 |
| 6 | Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism[J]. Nat Cell Biol, 2018, 20(7): 745-54. DOI: 10.1038/s41556-018-0124-1 |
| 7 | Uzhachenko R, Shanker A, Yarbrough WG, et al. Mitochondria, calcium, and tumor suppressor Fus1: At the crossroad of cancer, inflammation, and autoimmunity[J]. Oncotarget, 2015, 6(25): 20754-72. DOI: 10.18632/oncotarget.4537 |
| 8 | Henderson J, Duffy L, Stratton R, et al. Metabolic reprogramming of glycolysis and glutamine metabolism are key events in myofibroblast transition in systemic sclerosis pathogenesis[J]. J Cell Mol Med, 2020, 24(23): 14026-38. DOI: 10.1111/jcmm.16013 |
| 9 | Zhou X, Trinh-Minh T, Tran-Manh C, et al. Impaired mitochondrial transcription factor A expression promotes mitochondrial damage to drive fibroblast activation and fibrosis in systemic sclerosis[J]. Arthritis Rheumatol, 2022, 74(5): 871-81. DOI: 10.1002/art.42033 |
| 10 | Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments[J]. Bioinformatics, 2012, 28(6): 882-3. DOI: 10.1093/bioinformatics/bts034 |
| 11 | Chen BY, Li YX, Yan YP, et al. Construction and analysis of heart failure diagnosis model based on random forest and artificial neural network[J]. Medicine, 2022, 101(41): e31097. DOI: 10.1097/md.0000000000031097 |
| 12 | Rongvaux A. Innate immunity and tolerance toward mitochondria[J]. Mitochondrion, 2018, 41: 14-20. DOI: 10.1016/j.mito.2017.10.007 |
| 13 | Kaufman BA, Van Houten B. POLB: a new role of DNA polymerase beta in mitochondrial base excision repair[J]. DNA Repair, 2017, 60: A1-A5. DOI: 10.1016/j.dnarep.2017.11.002 |
| 14 | Sykora P, Kanno S, Akbari M, et al. DNA polymerase beta participates in mitochondrial DNA repair[J]. Mol Cell Biol, 2017, 37(16): e00237-17. DOI: 10.1128/mcb.00237-17 |
| 15 | Couto N, Wood J, Barber J. The role of glutathione reductase and related enzymes on cellular redox homoeostasis network[J]. Free Radic Biol Med, 2016, 95: 27-42. DOI: 10.1016/j.freeradbiomed.2016.02.028 |
| 16 | Xia YC, Wang GH, Jiang ML, et al. A novel biological activity of the STAT3 inhibitor stattic in inhibiting glutathione reductase and suppressing the tumorigenicity of human cervical cancer cells via a ROS-dependent pathway[J]. Onco Targets Ther, 2021, 14: 4047-60. DOI: 10.2147/ott.s313507 |
| 17 | Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome[J]. Nat Genet, 2006, 38(3): 294-6. DOI: 10.1038/ng1749 |
| 18 | Schubbert S, Zenker M, Rowe SL, et al. Germline KRAS mutations cause Noonan syndrome[J]. Nat Genet, 2006, 38(3): 331-6. DOI: 10.1038/ng1748 |
| 19 | Groesser L, Herschberger E, Ruetten A, et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome[J]. Nat Genet, 2012, 44(7): 783-7. DOI: 10.1038/ng.2316 |
| 20 | Aslam A, Salam A, Griffiths CE, et al. Naevus sebaceus: a mosaic RASopathy[J]. Clin Exp Dermatol, 2014, 39(1): 1-6. DOI: 10.1111/ced.12209 |
| 21 | Li RQ, Liu RQ, Zheng SY, et al. Comprehensive analysis of prognostic value and immune infiltration of the NT5DC family in hepatocellular carcinoma[J]. J Oncol, 2022, 2022: 2607878. DOI: 10.1155/2022/2607878 |
| 22 | Li KS, Zhu XD, Liu HD, et al. NT5DC2 promotes tumor cell proliferation by stabilizing EGFR in hepatocellular carcinoma[J]. Cell Death Dis, 2020, 11(5): 335. DOI: 10.1038/s41419-020-2549-2 |
| 23 | Qiu LX, Gong GC, Wu WJ, et al. A novel prognostic signature for idiopathic pulmonary fibrosis based on five-immune-related genes[J]. Ann Transl Med, 2021, 9(20): 1570. DOI: 10.21037/atm-21-4545 |
| 24 | Jiménez SA, Castro SV, Piera-Velázquez S. Role of growth factors in the pathogenesis of tissue fibrosis in systemic sclerosis[J]. Curr Rheumatol Rev, 2010, 6(4): 283-94. DOI: 10.2174/157339710793205611 |
| 25 | Piera-Velazquez S, Makul A, Jiménez SA. Increased expression of NAPDH oxidase 4 in systemic sclerosis dermal fibroblasts: regulation by transforming growth factor Β[J]. Arthritis Rheumatol, 2015, 67(10): 2749-58. DOI: 10.1002/art.39242 |
| 26 | Cho SY, Oh Y, Jeong EM, et al. Amplification of transglutaminase 2 enhances tumor-promoting inflammation in gastric cancers[J]. Exp Mol Med, 2020, 52(5): 854-64. DOI: 10.1038/s12276-020-0444-7 |
| 27 | Wang K, Zu CH, Zhang Y, et al. Blocking TG2 attenuates bleomycin-induced pulmonary fibrosis in mice through inhibiting EMT[J]. Respir Physiol Neurobiol, 2020, 276: 103402. DOI: 10.1016/j.resp.2020.103402 |
| 28 | Tabata K, Mikita N, Yasutake M, et al. Up-regulation of IGF-1, RANTES and VEGF in patients with anti-centromere antibody-positive early/mild systemic sclerosis[J]. Mod Rheumatol, 2021, 31(1): 171-6. DOI: 10.1080/14397595.2020.1726599 |
| 29 | Hamaguchi Y, Fujimoto M, Matsushita T, et al. Elevated serum insulin-like growth factor (IGF-1) and IGF binding protein-3 levels in patients with systemic sclerosis: possible role in development of fibrosis[J]. J Rheumatol, 2008, 35(12): 2363-71. DOI: 10.3899/jrheum.080340 |
| 30 | Jin W, Zheng Y, Zhu P. T cell abnormalities in systemic sclerosis[J]. Autoimmun Rev, 2022, 21(11): 103185. DOI: 10.1016/j.autrev.2022.103185 |
| 31 | DJrMesquita, Cruvinel WM, Resende LS, et al. Follicular helper T cell in immunity and autoimmunity [J]. Braz J Med Biol Res, 2016, 49(5): e5209. DOI: 10.1590/1414-431x20165209 |
| 32 | Jaguin M, Fardel O, Lecureur V. AhR-dependent secretion of PDGF-BB by human classically activated macrophages exposed to DEP extracts stimulates lung fibroblast proliferation[J]. Toxicol Appl Pharmacol, 2015, 285(3): 170-8. DOI: 10.1016/j.taap.2015.04.007 |
| 33 | Cardamone C, Parente R, Feo GD, et al. Mast cells as effector cells of innate immunity and regulators of adaptive immunity[J]. Immunol Lett, 2016, 178: 10-4. DOI: 10.1016/j.imlet.2016.07.003 |
| 34 | Lescoat A, Ballerie A, Jouneau S, et al. M1/M2 polarisation state of M-CSF blood-derived macrophages in systemic sclerosis[J]. Ann Rheum Dis, 2019, 78(11): e127. DOI: 10.1136/annrheumdis-2018-214333 |
| 35 | Maehara T, Kaneko N, Perugino CA, et al. Cytotoxic CD4+ T lymphocytes may induce endothelial cell apoptosis in systemic sclerosis[J]. J Clin Invest, 2020, 130(5): 2451-64. DOI: 10.1172/jci131700 |
| [1] | 赵华轩, 张桂潮, 刘家荣, 莫富添, 李韬恩, 类成勇, 吕世栋. 肾透明细胞癌患者的免疫细胞浸润特征与临床病理参数具有相关性[J]. 南方医科大学学报, 2025, 45(6): 1280-1288. |
| [2] | 张璐, 丁焕章, 许浩燃, 陈珂, 许博文, 杨勤军, 吴迪, 童佳兵, 李泽庚. 参芪补中方通过激活AMPK/SIRT1/PGC-1α改善COPD肺脾气虚证大鼠线粒体功能障碍[J]. 南方医科大学学报, 2025, 45(5): 969-976. |
| [3] | 陈梅妹, 王洋, 雷黄伟, 张斐, 黄睿娜, 杨朝阳. 基于多种机器学习算法和语音情绪特征的阈下抑郁辨识模型构建[J]. 南方医科大学学报, 2025, 45(4): 711-717. |
| [4] | 刘露玉, 公茂伟, 廖国松, 赵维星, 傅强. 高血压通过UCP2下调介导的线粒体功能障碍加重大鼠术后学习记忆损伤[J]. 南方医科大学学报, 2025, 45(4): 725-735. |
| [5] | 廖茗, 钟文华, 张冉, 梁娟, 徐文陶睿, 万文珺, 吴超, 李曙. 源自蛇毒的蛋白C激活剂通过调控HIF-1α抑制BNIP3活性氧生成保护人脐静脉内皮细胞免受缺氧-复氧损伤[J]. 南方医科大学学报, 2025, 45(3): 614-621. |
| [6] | 郭克磊, 李颖利, 宣晨光, 侯紫君, 叶松山, 李林运, 陈丽平, 韩立, 卞华. 益气养阴化浊通络方通过调控miR-21a-5p/FoxO1/PINK1介导的线粒体自噬减轻糖尿病肾病小鼠的足细胞损伤[J]. 南方医科大学学报, 2025, 45(1): 27-34. |
| [7] | 展俊平, 黄硕, 孟庆良, 范围, 谷慧敏, 崔家康, 王慧莲. 缺氧微环境下补阳还五汤通过抑制BNIP3-PI3K/Akt通路抑制类风湿关节炎滑膜成纤维细胞的线粒体自噬[J]. 南方医科大学学报, 2025, 45(1): 35-42. |
| [8] | 陈莉莉, 吴天宇, 张铭, 丁子夏, 张妍, 杨依清, 郑佳倩, 张小楠. 类风湿关节炎的潜在生物标志物及其免疫调控机制:基于GEO数据库[J]. 南方医科大学学报, 2024, 44(6): 1098-1108. |
| [9] | 申采玉, 王帅, 周锐盈, 汪雨贺, 高琴, 陈兴智, 杨枢. 慢性心力衰竭合并肺部感染患者院内死亡风险预测:基于可解释性机器学习方法[J]. 南方医科大学学报, 2024, 44(6): 1141-1148. |
| [10] | 王梓凝, 杨 明, 李双磊, 迟海涛, 王军惠, 肖苍松. 心肌梗死后心肌纤维化小鼠心肌线粒体功能和能量代谢重塑相关性的转录组学分析[J]. 南方医科大学学报, 2024, 44(4): 666-674. |
| [11] | 姜诚诚, 李洋洋, 段可欣, 詹婷婷, 陈子龙, 王永雪, 赵蕊, 马彩云, 郭俣, 刘长青. Parkin通过介导PINK 1/Parkin线粒体自噬信号通路加速小鼠帕金森病发展及加剧神经炎症发生[J]. 南方医科大学学报, 2024, 44(12): 2359-2366. |
| [12] | 王磊, 卞芬兰, 马飞扬, 方舒, 凌梓涵, 刘梦然, 孙红燕, 付程文, 倪诗垚, 赵晓阳, 冯心茹, 孙正宇, 卢国庆, 康品方, 吴士礼. 激活ALDH2通过上调SIRT1/PGC-1α信号通路减轻小鼠缺氧性肺动脉高压[J]. 南方医科大学学报, 2024, 44(10): 1955-1964. |
| [13] | 黄晓茵, 陈凤莲, 张煜, 梁淑君. 多参数多区域MRI影像组学特征与临床信息联合模型可有效预测脑胶质瘤患者生存期[J]. 南方医科大学学报, 2024, 44(10): 2004-2014. |
| [14] | 何慧珊, 郭二嘉, 蒙文仪, 王 彧, 王 雯, 何文乐, 吴元魁, 阳 维. 基于磁共振图像机器学习放射组学模型预测脑胶质瘤的强化[J]. 南方医科大学学报, 2024, 44(1): 194-200. |
| [15] | 叶红伟, 张钰明, 云 琦, 杜若丽, 李 璐, 李玉萍, 高 琴. 白藜芦醇可减轻高糖诱导的心肌细胞肥大:基于促进SIRT1表达维持线粒体稳态[J]. 南方医科大学学报, 2024, 44(1): 45-51. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||